Abstract
Because lymphoid progenitors can give rise to natural killer (NK) cells, NK ontogeny has been considered to be exclusively lymphoid. Here, we show that rare human CD34+ hematopoietic progenitors develop into NK cells in vitro in the presence of cytokines (interleukin-7, interleukin-15, stem cell factor, and fms-like tyrosine kinase-3 ligand). Adding hydrocortisone and stromal cells greatly increases the frequency of progenitor cells that give rise to NK cells through the recruitment of myeloid precursors, including common myeloid progenitors and granulocytic-monocytic precursors to the NK-cell lineage. WNT signaling was involved in this effect. Cells at more advanced stages of myeloid differentiation (with increasing expression of CD13 and macrophage colony-stimulating factor receptor [M-CSFR]) could also differentiate into NK cells in the presence of cytokines, stroma, and hydrocortisone. NK cells derived from myeloid precursors (CD56−CD117+M-CSFR+) showed more expression of killer immunoglobulin-like receptors, a fraction of killer immunoglobulin–like receptor-positive–expressing cells that lacked NKG2A, a higher cytotoxicity compared with CD56−CD117+M-CSFR− precursor-derived NK cells and thus resemble the CD56dim subset of NK cells. Collectively, these studies show that NK cells can be derived from the myeloid lineage.
Introduction
Hematopoietic stem cells (HSCs) give rise to all blood lineages.1 As HSCs differentiate along one lineage, they gradually lose the ability to develop into other lineages.2 Hematopoietic differentiation involves lineage commitment, defined here as the initiation of developmental program(s) that lead to a particular cell fate. The accompanying inability to differentiate into other lineages has been referred to as lineage maintenance.3 Lineage commitment and lineage maintenance are complementary processes that guide cell fate decisions. Thus, cells committed to a particular lineage have alternative developmental choices until lineage maintenance is complete.4
Hematopoietic differentiation has been schematically depicted as a “tree of hematopoiesis,”1 outlining the possible developmental choices. According to this prevailing schema, the decision between lymphoid and myeloid lineages occurs very early. However, alternative views have been proposed, including the existence of a common myelo-lymphoid progenitor.5,6 Elucidation of hematopoietic developmental pathways and extrinsic stimuli that influence them is instrumental to understanding both normal and malignant hematopoiesis. In particular, factors that favor natural killer (NK)–cell development could be used to exploit their activity against malignancies.
NK cells are innate immune effector cells. Their derivation from either lymphoid or myeloid lineages was debated early in their discovery.7 Further research showed that NK cells can be derived from common lymphoid progenitors (CLPs) and hence have been considered separate from myeloid lineage.8,9 However, some studies question this and have shown that progenitors expressing myeloid antigens can develop into NK cells.10,11 NK-cell differentiation from hematopoietic progenitor cells (HPCs) can be studied in vitro.12,13 This process depends on cytokines, notably interleukin-2 (IL-2) or IL-15, whereas other factors (stem cell factor [SCF], fms-like tyrosine kinase-3 ligand [FLT-3L]) induce early HPC expansion and responsiveness to IL-2 and IL-15 signaling.14
CD34+ HPCs are heterogeneous and include cells at various levels of differentiation. Multipotent HPCs with long-term repopulation potential are contained within the CD34+CD38− subset.15 More advanced lineage precursors, included in the CD34+CD38+ fraction,15 have been categorized as common myeloid progenitor (CMP; CD34+CD38+CD123+CD45RA−), granulocytic-monocytic precursor (GMP; (CD34+CD38+CD123+CD45RA+), and megakaryocyte-erythroid precursor (CD34+CD38+CD123−CD45RA−).16 Subsets of CD34+ precursors have also been distinguished by their ability to readily differentiate into NK cells. Surface receptors that define NK precursors include CD7,13 CD122,17 CD161,18 integrin β7, and CD45RAhigh.19
Stromal cell layers have been used to differentiate HPCs into NK cells. Sources of stroma include bone marrow,13,20 murine fetal liver cell lines,21 and human splenic fibroblasts.22 Stroma increase the efficiency of NK-cell generation and advance the maturational status of HPC-derived NK cells.21 Physiologic concentrations of hydrocortisone (HDC) also advances NK-cell development from CD34+ HPCs.10
We previously described discrete stages of human NK-cell differentiation by studying CD34+ cells cultured on the murine fetal liver stromal cell line, EL08.1D2.23 Notably, the stages we defined in vitro closely resemble those in human lymph nodes.24 This culture system results in a strikingly high efficiency of NK-cell differentiation from CD34+ HPCs.23 Here, we investigate the mechanism of stroma-induced NK-cell differentiation to better understand the origins of NK cells. We demonstrate that fetal liver stroma and HDC enhance NK differentiation by recruiting CMP and GMP to the NK lineage. Likewise, precursors at more advanced stages of myeloid differentiation retain NK cell–generating capacity. The concept that NK cells can be derived from the myeloid lineage advances our understanding of NK ontogeny and the relationship to other blood lineages, creating the potential for therapeutic applications.
Methods
CD34+ cells isolation
CD34+ cells were positively selected from umbilical cord blood (UCB) mononuclear cells (Miltenyi Biotec). Selected cells were > 97% pure and lacked CD56+ cells (not shown). When specified, CD34+ cells underwent fluorescence-activated cell sorting (FACS) into CD34+NKlin+, CD34+CD38−NKlin−, and CD34+CD38+NKlin− (NK lineage markers were CD161, CD122, CD7, integrin β7; and CD45RAhigh; cells negative for all were considered NKlin−). Granulo-monocytic precursors (CD34+CD38+CD64+),25 CMPs (CD34+CD38+CD123+CD45RA−), and GMPs (CD34+CD38+CD123+CD45RA+) were FACS purified from UCB CD34+ cells.16 After the initial sort, the CMP and GMP fractions were resorted and deposited directly into culture wells at 1 or 10 cells per well.
NK-cell differentiation cultures
The embryonic liver cell line EL08.1D2 was cultured to confluence and was irradiated (30 Gy [3000 rad]).23 CD34+ cells or specified subsets were cultured with/without a monolayer of EL08.1D2 cells in Ham F12 plus Dulbecco modified Eagle medium (1:2 ratio) with 20% human AB sera, ethanolamine (50μM), ascorbic acid (20 mg/L), 5 μg/L sodium selenite (Na2SeO3), β-mercaptoethanol (24μM), and penicillin (100 U/mL)/streptomycin (100 U/mL). At the start of cultures, IL-3 (5 ng/mL), IL-7 (20 ng/mL), IL-15 (10 ng/mL), SCF (20 ng/mL), and FLT-3L (10 ng/mL) were added. Where specified, HDC (10−6M; Stem Cell Technologies), macrophage colony-stimulating factor (M-CSF), or Dickkopf-1 (R&D Systems) were added. Cultures were refed weekly by 50% volume change of media, supplemented with the above-listed cytokines (except IL-3), with or without HDC. After 14, 21, and 28 days of culture, 3 replicate wells were harvested, counted, and analyzed.
NK-cell precursor frequency assay
CD34+ HPC or specified subsets were plated at 200, 100, 50, and 25 cells/well, 24 replicates/dilution. Conditions included media with or without stroma (EL08.1D2) and with or without HDC (10−6M). After 4 weeks cellular outgrowth was scored, and split-well analysis was performed in which one-half of each well was tested for K562 cytotoxicity. If cytotoxicity exceeded a predetermined cutoff value (mean spontaneous release + 3× SD), wells were considered positive. The fraction of negative wells at each dilution was used to calculate the NK-cell precursor (NKp) frequency as determined by Poisson kinetics.26
CMP and GMP cultures
CMP and GMP (n = 5) were double-sorted and deposited at 1 and 10 cells per well (n = 60 replicates per condition). Conditions included (1) cytokines, (2) cytokines and stroma, (3) cytokines and stroma and HDC, and (4) methylcellulose enriched with growth factors (MethoCult; StemCell Technologies). The latter condition was included to control for their colony-generating potential (CMP or GMP). Cultures were maintained for 17-21 days then tested for expression of NK-cell markers (CD56, CD94) in combination with myeloid markers (CD15, CD14, CD33). The colony-forming potential of sorted fractions was assessed after 14-21 days of methylocellulose culture by 2 researchers, including an independent (blinded) investigator not otherwise involved in the study.
Methylocellulose assay and myeloid cell culture
CD34+ HPCs were plated at 1 cell/well in methylocellulose enriched with growth factors (MethoCult GF+H4435; StemCell Technologies). After 20-21 days individual granulocyte-macrophage colony-forming units (CFU-GMs) were isolated and plated into culture in medium as for NK differentiation (IL-15, IL-7, SCF, FLT-3L) with stromal cells and HDC (10−6M).
Isolation of CD13+ cells and cells positive for M-CSF receptor
NK-differentiation cultures were harvested after 14 days and depleted of CD56+ cells with the use of using magnetically activated cell-sorting beads. This CD56− population underwent FASC into CD56−CD33+/−CD13low/−, CD56−CD33+CD13intermediate, and CD56−CD33+CD13high fractions. Sorted populations were cultured (in triplicates) at 10 000 cells/well in 96-well plates in NK-differentiation medium with/without stroma and with/without HDC. After 18-21 days cells were analyzed. Similarly, cultures started with CD34+ HPCs were harvested at day 18, CD56+ cells were depleted, and CD117+ cells were isolated from CD56− fraction with the use of magnetically activated cell-sorting beads. The resulting populations were then sorted into CD56−CD94−CD117+M-CSFR+ (myeloid precursors) and CD56−CD94−CD117+M-CSFR− (nonmyeloid, presumed to include lymphoid precursors). Sorted fractions were cultured as described earlier. In selected experiments M-CSF was added.
FACS analysis
The following antibodies were used: CD7-FITC (fluorescein isothiocyanate), CD13-PE (phycoerythrin), CD1aFITC, CD14-FITC or PE, CD16 (FITC, PE, or peridinin chlorophyll protein and cyanine 5.5 [PerCp-Cy5.5]), CD33 PerCp-Cy5.5, CD34 (PerCp, PE, and allophycocyanin [APC]), CD38-APC, CD56-APC, CD94-FITC, CD117 (PE or PerCp-Cy5.5), CD122-PE, CD161 (FITC or PE), CD158a-PE, CD158b-PE, CD158e-PE (DX-9), CD64-PE, Granzyme-B–FITC, Perforin-FITC, CD68 (macrosialin)–PE, and myeloperoxidase-FITC. For CMP, GMP, and megakaryocyte-erythroid precursor subsets included CD123-PE, CD34–PerCp-Cy5.5, CD45RA-Horizon450 (all from BD Biosciences), CD38-APC, and Lineage cocktail, including FITC-conjugated CD2, CD3, CD4, CD7, CD8, CD10, CD14, CD16, CD19, CD11b, CD20, CD56, GPA (e-Bioscience). Additional antibodies included CD115-PE, CD45RA-FITC (Biolegend), CD13-FITC (Serotec), lysozyme-FITC (Caltag), NKp30-PE, NKp44-PE, and NKp46-PE (Beckman Coulter). Intracellular staining was performed with cytofix/cytoperm (BD Biosciences). FACS was performed on a FACSCalibur. Data were analyzed with WinMDI (FACS Core Facility, The Scripps Research Institute) or FlowJo (TreeStar Inc). Sorting was performed on FACSAria, and postsort reanalysis usually showed > 99% purity (not shown).
51Cr release assay
K562 or B721.221 lymphoblastoid cell line targets were used in killing assays.23
Statistical analysis
For grouped analysis, experimental values were normalized by log transformation before 1-way (or 2-way) analysis of variance, as indicated. Individual groups were compared with Bonferroni posttest. The χ2 analysis was used to compare fractions of methylcellulose (CFU-GM) colonies that gave rise to NK cells.
Results
Stromal cells and HDC additively enhance NK-cell differentiation
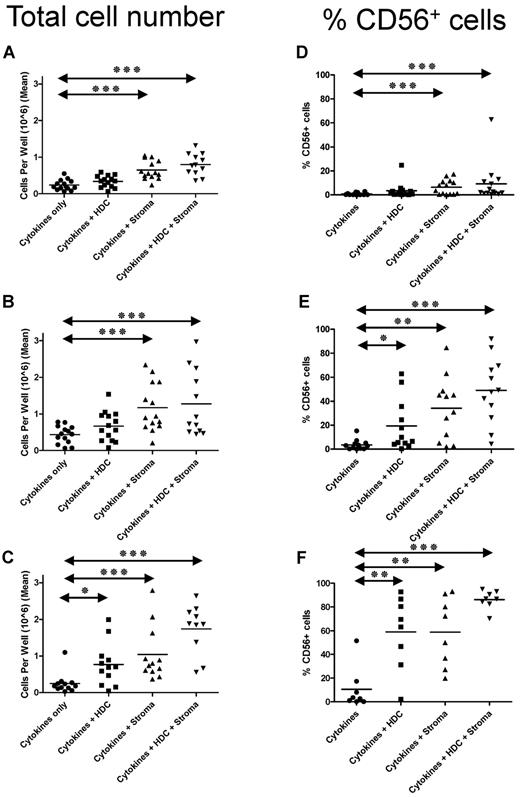
We investigated how stroma, HDC, and the combination of the two affect NK differentiation. CD34+ cells were cultured in media containing cytokines (defined here as IL-3 for the first week, SCF, FLT-3L, IL-7, and IL-15), and, for comparison, cells were cultured with (1) cytokines and stroma, (2) cytokines and HDC, or (3) the combination of all three. The addition of stroma or HDC or both significantly increased cell numbers and the percentage of NK cells (CD56+ cells) at 2 (Figure 1A,D), 3 (Figure 1B,E), and 4 (Figure 1C,F) weeks (P < .05). The 2 factors (stroma and HDC) were additive because the combination was significantly better than either one alone (P < .05 at 4 weeks).
HDC and stroma increase the number of NK cells generated from CD34+ HPCs. Five hundred CD34+ cells were plated per well in 24-well cell culture plates. Where indicated, plates were previously coated with stromal cells (EL08.1D2) and irradiated (30 Gy). Where indicated, HDC (10−6M) was added to the starting, as well as, refreshing medium (added weekly). Average number of viable cells/well (of 3 replicates) after (A) 14, (B) 21, and (C) 28 days of culture. Percentage of CD56+ cells (by FACS) after (D) 14, (E) 21, and (F) 28 days. Horizontal bars represent the means of 12 separate donors in various conditions. Groups showing significant differences in comparison with the cytokines-alone group are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), as determined by repeated-measures analysis of variance on log-transformed values with Bonferroni posttest.
HDC and stroma increase the number of NK cells generated from CD34+ HPCs. Five hundred CD34+ cells were plated per well in 24-well cell culture plates. Where indicated, plates were previously coated with stromal cells (EL08.1D2) and irradiated (30 Gy). Where indicated, HDC (10−6M) was added to the starting, as well as, refreshing medium (added weekly). Average number of viable cells/well (of 3 replicates) after (A) 14, (B) 21, and (C) 28 days of culture. Percentage of CD56+ cells (by FACS) after (D) 14, (E) 21, and (F) 28 days. Horizontal bars represent the means of 12 separate donors in various conditions. Groups showing significant differences in comparison with the cytokines-alone group are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), as determined by repeated-measures analysis of variance on log-transformed values with Bonferroni posttest.
With the addition of either HDC or stroma, all wells contained NK cells (not shown). However, when CD34+ HPCs were cultured without stroma or HDC, we observed considerable variability between replicates. For instance, there were individual wells in which NK cells developed, whereas in others they did not. This suggested that the CD34+ HPC fraction contained precursors capable of NK differentiation with cytokines alone (ie, without HDC or stroma). Given the well-to-well variability with initial input of 500 CD34+ cells/well, such NK precursors appear to be rare. At least 2 nonexclusive mechanisms can explain these findings. First, HDC and stroma might increase the efficiency of NK-cell generation from progenitors already “committed” to the NK lineage. Second, they might act on progenitors that might not otherwise become NK cells. One method to verify if the latter possibility was operational is to measure the NKp frequency with the use of a limiting dilution assay. If stroma or HDC or both increase the frequency of CD34+ HPCs giving rise to NK cells in limiting dilution assays, then the logical explanation would be that these factors act on CD34+ HPCs to recruit them to the NK lineage.
NKp frequency in UCB-derived CD34+ cells
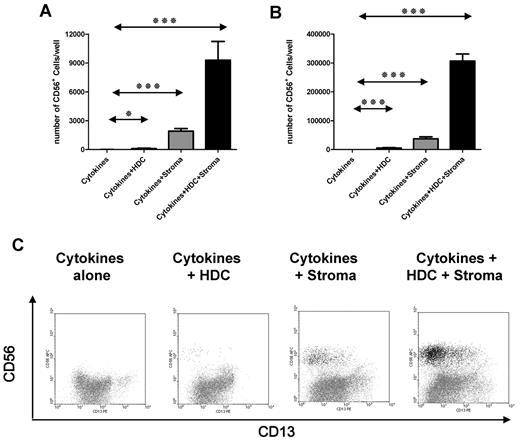
To measure the NKp frequency serial, 2-fold dilutions of CD34+ cells (25-200 cells/well, 24 replicates/dilution) were plated as in Figure 1 (ie, cytokines alone, cytokines and stroma, cytokines and HDC, and cytokines and HDC and stroma). Because the hallmark NK characteristic is unprimed effector function (eg, K562 cytotoxicity), we defined an NKp as a CD34+ HPC that could give rise to effectors with K562 cytotoxicity at 4 weeks of culture. Wells showing no cytotoxicity were assumed to contain no NKps (ie, precursors capable of NK-cell development in the given condition). The fractions of wells showing no killing (Figure 2A) were used to calculate the NKp frequency (Figure 2B).26 Compared with CD34+ HPCs cultured in cytokines alone, the same cell suspension cultured with stroma, HDC, or the combination of both showed a higher NKp frequency (P < .001; n = 8; Figure 2B).
EL08.1D2 stroma and HDC increase the NKp frequency in the CD34+ UCB fraction. (A) Semi-log plot presenting the results of a limiting dilution experiment with CD34+ HPCs cultured in cytokines alone (◇, long-dashed line), cytokines and HDC (■, gray line), cytokines and stroma (▴, black line), and cytokines and HDC and stroma (○, short-dashed line). Data for a single donor are shown and are representative of 8 donors tested. The steeper slope of regression line indicates increasing NKp frequency in different conditions. (B) NKp frequency/106 CD34+ HPCs cultured in the above-described conditions (n = 8 donors). Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), by repeated-measures analysis of variance on log-transformed values with Bonferroni posttest. (C) Results of microscopic scoring of cell density in limiting dilution experiment at day 28. Each graph represents the results of a limiting dilution experiment with the use of the same CD34+ HPC cell suspension in different conditions (ie, cytokines alone, cytokines and HDC, cytokines and stroma, cytokines and HDC and stroma). The total height of each bar represents 24 replicate wells at a given serial dilution (25 cells/well to 200 cells/well). Each well was inspected with microscopy, and cellular density was scored with a scale from 0 (no or very few cells) to 5 (densely populating the entire well). Results of a single representative donor are shown (n = 8).
EL08.1D2 stroma and HDC increase the NKp frequency in the CD34+ UCB fraction. (A) Semi-log plot presenting the results of a limiting dilution experiment with CD34+ HPCs cultured in cytokines alone (◇, long-dashed line), cytokines and HDC (■, gray line), cytokines and stroma (▴, black line), and cytokines and HDC and stroma (○, short-dashed line). Data for a single donor are shown and are representative of 8 donors tested. The steeper slope of regression line indicates increasing NKp frequency in different conditions. (B) NKp frequency/106 CD34+ HPCs cultured in the above-described conditions (n = 8 donors). Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), by repeated-measures analysis of variance on log-transformed values with Bonferroni posttest. (C) Results of microscopic scoring of cell density in limiting dilution experiment at day 28. Each graph represents the results of a limiting dilution experiment with the use of the same CD34+ HPC cell suspension in different conditions (ie, cytokines alone, cytokines and HDC, cytokines and stroma, cytokines and HDC and stroma). The total height of each bar represents 24 replicate wells at a given serial dilution (25 cells/well to 200 cells/well). Each well was inspected with microscopy, and cellular density was scored with a scale from 0 (no or very few cells) to 5 (densely populating the entire well). Results of a single representative donor are shown (n = 8).
Before cytotoxicity testing, wells were microscopically scored for outgrowth. Similar to the NKp results, cell density was least when CD34+ HPCs were cultured with only cytokines (Figure 2C). Combining the cytotoxicity with the outgrowth data, we observed wells with the following patterns: (1) no (or few) cells and no cytotoxicity, (2) cells present but no cytotoxicity, and (3) cells present and cytotoxicity present. Representative wells were analyzed by FACS. Cytotoxicity was uniformly associated with the presence of NK cells at a functional stage of development (ie, CD56+CD94+CD117low/−).23,27 Frequently, these cells were intermixed with cells at earlier stages of development (CD56+CD94−CD117high; not shown). Conversely, wells with no cytotoxicity lacked NK cells and instead contained cells of myeloid origin by forward scatter (FSC) and side scatter (SSC) characteristics (not shown).
Collectively, these studies show that a small but measurable fraction of CD34+ HPCs give rise to NK cells under the influence of only cytokines. Conversely, the majority of HPCs cultured with the above cytokine combination do not generate functional NK cells. Thus, the increased NKp frequency in the presence of stroma, HDC, or both, strongly suggest that these agents induce NK differentiation by acting on HPCs (and their progeny), which do not generate NK cells when cultured with cytokines only.
Identification of CD34+ HPCs that require HDC or stroma for in vitro NK-cell differentiation
Our aim was to isolate the subset of CD34+ HPCs that required stroma or HDC or both for NK differentiation. Several surface receptors are associated with NK lineage commitment, including CD7, CD122, CD161, integrin β7, and CD45RAhigh.13,17-19 These were expressed on nonfully overlapping CD34+ subsets (not shown). We hypothesized that CD34+ HPCs lacking all of these NK lineage markers (CD34+NKlin−) might be “uncommitted” to the NK lineage (ie, unable to give rise to NK cells with cytokines alone). Two NKlin− subsets (CD34+CD38−NKlin−, CD34+CD38+NKlin−) were tested in the NKp frequency assay with and without stroma and HDC. CD34+ cells expressing any of the above NK lineage markers were used for comparison (CD34+NKlin+). In the absence of stroma or HDC or a combination, both CD34+NKlin− populations (CD38− and CD38+) yielded few wells with K562 cytotoxicity, resulting in a flat slope of the semi-log–limiting dilution plot (Figure 3A). In contrast, the CD34+NKlin+ population cultured in only cytokines produced a noticeable fraction of wells with K562 cytotoxicity, resulting in a steeper slope (Figure 3A).
Stroma and HDC induce NK differentiation in the CD34+ HPC subset that could not differentiate into NK cells with cytokines alone. (A) Semi-log plot showing the fraction of negative wells (wells containing no NK precursors) as a function of input cells per well. Three subsets of CD34+ cells were tested in limiting dilution culture with only cytokines (IL-15, IL-7, SCF, FLT-3L, and IL-3): CD34+CD38−NKlin− (□, dashed line), CD34+CD38+NKlin− (◇, dotted line), and CD34+NKlin+ (▴, solid line). No positive wells were observed for NKlin− subsets cultured with cytokines alone, and the plot therefore shows a flat slope for these conditions. In contrast, CD34+NKlin+ subset showed a measurable fraction of positive wells in culture with cytokines alone, thus creating a steeper slope of the regression line. A representative donor (n = 4) is shown. (B) Calculated NKp frequencies for the 3 CD34+ HPC subsets cultured with the above-mentioned cytokines (n = 4 donors), expressed as number of NKp/106 cells. (C) Semi-log plot of the fraction of negative wells as a function of input cells per well for the CD34+CD38+NKlin− fraction cultured with cytokines alone (◇, long-dashed line), cytokines and HDC (■, gray line), cytokines and stroma (▴, black line), and cytokines and HDC and stroma (○, short-dashed line). A representative donor (n = 4) is shown. (D) Calculated NKp/106 cells for CD34+CD38+NKlin− fraction cultured in different conditions (n = 4 donors tested). (E) Semi-log plot of the fraction of negative wells as a function of input cells per well for the CD34+CD38−NKlin− fraction cultured with cytokines alone (◇, long-dashed line), with cytokines and HDC (■, gray line), with cytokines and stroma (▴, black line), and with cytokines and HDC and stroma (○, short-dashed line). A representative donor (n = 4) is shown. (F) NKp frequency per 106 cells in the CD34+CD38−NKlin− population cultured in different conditions; n = 4 donors tested. Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 1-way analysis of variance on log-transformed values with Bonferroni posttest. (G) Fold change in mRNA levels coding for T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors and their downstream targets (WISP and CyclinD1) induced in NKlin−CD34+ HPCs after 14 days of culture of with (1) cytokines, stromal cells, and HDC versus (2) cytokines only. (H) Effect of Dickkopf-1 (DKK-1) on the generation of NK cells from CD34+ HPCs in the culture with cytokines, stroma, and HDC. Shown are the average number of CD56+ cells/well after 21 days of culture ± SEM. The difference between no addition of DKK-1 and addition at 10 ng/mL is statistically significant (P = .012). Results are the mean of 6 wells for each condition and are representative of independent 2 donors.
Stroma and HDC induce NK differentiation in the CD34+ HPC subset that could not differentiate into NK cells with cytokines alone. (A) Semi-log plot showing the fraction of negative wells (wells containing no NK precursors) as a function of input cells per well. Three subsets of CD34+ cells were tested in limiting dilution culture with only cytokines (IL-15, IL-7, SCF, FLT-3L, and IL-3): CD34+CD38−NKlin− (□, dashed line), CD34+CD38+NKlin− (◇, dotted line), and CD34+NKlin+ (▴, solid line). No positive wells were observed for NKlin− subsets cultured with cytokines alone, and the plot therefore shows a flat slope for these conditions. In contrast, CD34+NKlin+ subset showed a measurable fraction of positive wells in culture with cytokines alone, thus creating a steeper slope of the regression line. A representative donor (n = 4) is shown. (B) Calculated NKp frequencies for the 3 CD34+ HPC subsets cultured with the above-mentioned cytokines (n = 4 donors), expressed as number of NKp/106 cells. (C) Semi-log plot of the fraction of negative wells as a function of input cells per well for the CD34+CD38+NKlin− fraction cultured with cytokines alone (◇, long-dashed line), cytokines and HDC (■, gray line), cytokines and stroma (▴, black line), and cytokines and HDC and stroma (○, short-dashed line). A representative donor (n = 4) is shown. (D) Calculated NKp/106 cells for CD34+CD38+NKlin− fraction cultured in different conditions (n = 4 donors tested). (E) Semi-log plot of the fraction of negative wells as a function of input cells per well for the CD34+CD38−NKlin− fraction cultured with cytokines alone (◇, long-dashed line), with cytokines and HDC (■, gray line), with cytokines and stroma (▴, black line), and with cytokines and HDC and stroma (○, short-dashed line). A representative donor (n = 4) is shown. (F) NKp frequency per 106 cells in the CD34+CD38−NKlin− population cultured in different conditions; n = 4 donors tested. Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 1-way analysis of variance on log-transformed values with Bonferroni posttest. (G) Fold change in mRNA levels coding for T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors and their downstream targets (WISP and CyclinD1) induced in NKlin−CD34+ HPCs after 14 days of culture of with (1) cytokines, stromal cells, and HDC versus (2) cytokines only. (H) Effect of Dickkopf-1 (DKK-1) on the generation of NK cells from CD34+ HPCs in the culture with cytokines, stroma, and HDC. Shown are the average number of CD56+ cells/well after 21 days of culture ± SEM. The difference between no addition of DKK-1 and addition at 10 ng/mL is statistically significant (P = .012). Results are the mean of 6 wells for each condition and are representative of independent 2 donors.
For the NKlin− populations (CD38− and CD38+) the NKp frequency was not significantly different from 0 (CD34+CD38−NKlin−, 1102/106 cells; 95% confidence interval [CI], −598 to 2728; CD34+CD38+NKlin−, 474/106 cells, 95% CI, −637 to 2153). In contrast, CD34+NKlin+ cells showed a higher NKp frequency (median, 7370/106 cells; 95% CI, 3891-13 178; n = 4; P < .01) (Figure 3B). The addition of HDC or stroma or both induced the capacity of CD34+NKlin− populations (both CD38− and CD38+) to give rise to functional NK cells (Figure 3D,F), changing the slopes of the semi-log plots (Figure 3C,E). Thus, we defined a subset of HPCs critically dependent on HDC or stroma or both to differentiate into NK cells (ie, CD34+NKlin−).
To understand the mechanisms of stroma and HDC here, CD34+NKlin− cells were cultured in cytokines or in cytokines, stroma, and HDC. Transcriptional profiling studies showed up-regulation of the WNT signaling pathway in response to stroma and HDC (Figure 3G). To verify whether WNT signaling plays a role in the effect of stroma and HDC, we added Dickkopf-1, a canonical WNT pathway inhibitor, resulting in a dose-dependent decrease in the number of NK cells generated (Figure 3H; P = .012).
Although the above NKp frequency assay has a functional endpoint (ie, cytotoxicity), it offers no insight into the developmental events during the 4-week culture. Prior studies show that HDC increases NK expansion,28 perhaps through proliferation or antiapoptotic activity. Thus, HDC or stroma or both might simply induce proliferation or maintain survival of immature NK cells. To address this, cultures were evaluated at days 14 and 21. CD34+NKlin+ cells cultured in only cytokines (ie, no stroma or HDC) generated first CD56+ NK cells within 14 days, whereas CD34+NKlin− cells did not (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In an alternative approach, small numbers of CD34+NKlin− cells (100 cells/well, 12 replicates) were tested at days 14 and 21. In the presence of cytokines alone, CD56+ cells were not observed. In contrast, HDC or stroma or both induced CD56+ development after 14 and 21 days (Figure 4A and B, respectively). Collectively these results show that CD34+NKlin− cells did not develop into CD56+ cells throughout the culture with cytokines alone, excluding the possibility that NK cells merely failed to expand or survive or both in the absence of HDC or stroma or both.
HDC and stroma induce CD34+CD38−NKlin− progenitors to become NK cells. (A-B) CD34+CD38−NKlin− cells were cultured at 100 cells/well in 12 replicates. After (A) 14 and (B) 21 days one-half the contents of each well were harvested and analyzed for the presence of CD56+ cells by FACS (enumeration was performed with polystyrene beads). The average number of CD56+ cells/well is presented. A representative donor is shown (n = 4). Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 1-way analysis of variance on log-transformed values with Bonferroni posttest. (C) Phenotype of cells derived from CD34+CD38−NKlin− cells less than various conditions. Representative examples of individual wells after 14 days of culture with cytokines alone (IL-15, IL-7, SCF, FLT-3L, and IL-3; left), cytokines and HDC (second column), cytokines and stroma (third column), and cytokines and HDC and stroma (right).
HDC and stroma induce CD34+CD38−NKlin− progenitors to become NK cells. (A-B) CD34+CD38−NKlin− cells were cultured at 100 cells/well in 12 replicates. After (A) 14 and (B) 21 days one-half the contents of each well were harvested and analyzed for the presence of CD56+ cells by FACS (enumeration was performed with polystyrene beads). The average number of CD56+ cells/well is presented. A representative donor is shown (n = 4). Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 1-way analysis of variance on log-transformed values with Bonferroni posttest. (C) Phenotype of cells derived from CD34+CD38−NKlin− cells less than various conditions. Representative examples of individual wells after 14 days of culture with cytokines alone (IL-15, IL-7, SCF, FLT-3L, and IL-3; left), cytokines and HDC (second column), cytokines and stroma (third column), and cytokines and HDC and stroma (right).
Transient myeloid antigen expression on developing NK cells
CD34+NKlin− cells cultured with only cytokines showed growth after 14 and 21 days, but they did not differentiate into NK cells (Figure 4C; supplemental Figure 1). The majority of these cells expressed markers associated with the myeloid lineage (CD13 and CD33) (Figure 4C). When the same CD34+NKlin− cells were cultured with HDC or stroma or both, some CD56+ NK cells coexpressed CD13 and CD33 (Figure 4C; supplemental Figure 1). Thus, CD56+ cells might arise from CD13+CD33+ precursors, prompting us to speculate that stroma and HDC could recruit myeloid progenitors to differentiate into NK cells.
CD13+CD33+ cells in these cultures were tested for characteristics unique to the myeloid lineage. CD13+ cells expressed intracellular enzymes (myeloperoxidase, lysozyme, macrosialin [CD68]) and surface receptors (CD14 and CD1a), consistent with myeloid maturation (supplemental Figure 2). We next tested whether precursors with various amounts of CD13 could give rise to NK cells. Not only the CD13low/− but also the CD13int and CD13high subsets differentiated into NK cells after culture with cytokines and stroma (Figure 5A). The efficiency of NK-cell generation decreased with increasing CD13 expression (P < .05) (Figure 5B). Thus, cells following myeloid differentiation as indicated by increasing CD13 staining intensity29 gradually lose the ability to give rise to NK cells.
CD13+CD56−cells can give rise to NK cells. (A) Cells with distinct levels of CD13 expression: CD13low/neg (bottom), CD13intermediate (middle), and CD13high (top) were FACS purified from NK differentiation cultures (at day 14) and cultured further with cytokines and stroma. All 3 subsets gave rise to CD56+ NK cells, some of which coexpressed CD13. (B) Quantitative yield of CD56+ NK cells/104 CD13+ cells undergoing FACS. There was decreasing ability for NK-cell generation with increasing CD13 expression. A representative donor is shown (n = 4). Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 1-way analysis of variance on log-transformed values with Bonferroni posttest. (C) Freshly isolated GMPs (CD34+CD38+CD123+CD45RA−) were double-sorted from UCB and deposited into 96-well plates with stroma, HDC, and cytokines (FLT-3L, SCF, IL-7, and IL-15). After ∼ 21 days progeny were analyzed by FACS. Shown are the cells falling within the lymphoid gate by FSC versus SSC.
CD13+CD56−cells can give rise to NK cells. (A) Cells with distinct levels of CD13 expression: CD13low/neg (bottom), CD13intermediate (middle), and CD13high (top) were FACS purified from NK differentiation cultures (at day 14) and cultured further with cytokines and stroma. All 3 subsets gave rise to CD56+ NK cells, some of which coexpressed CD13. (B) Quantitative yield of CD56+ NK cells/104 CD13+ cells undergoing FACS. There was decreasing ability for NK-cell generation with increasing CD13 expression. A representative donor is shown (n = 4). Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 1-way analysis of variance on log-transformed values with Bonferroni posttest. (C) Freshly isolated GMPs (CD34+CD38+CD123+CD45RA−) were double-sorted from UCB and deposited into 96-well plates with stroma, HDC, and cytokines (FLT-3L, SCF, IL-7, and IL-15). After ∼ 21 days progeny were analyzed by FACS. Shown are the cells falling within the lymphoid gate by FSC versus SSC.
Individual CFU-GM colonies give rise to NK cells under the influence of stroma, HDC, and cytokines but not cytokines alone
To further test whether myeloid cells could give rise to NK cells, CD34+ cells were plated in methylcellulose. Individual CFU-GM colonies were isolated and cultured in NK-supporting conditions. A fraction of CFU-GM colonies gave rise to NK cells (CD56+ lymphocytes by FACS) after culture with cytokines, stroma, and HDC but not with cytokines alone (9 of 54 vs 0 of 36; P = .01; supplemental Figure 3).
In the presence of HDC or stroma or both, freshly isolated CMPs and GMPs can develop into NK cells
Prior studies have identified human hematopoietic progenitors with the capacity for myeloid and erythroid differentiation (CMPs, CD34+CD38+IL-3RlowCD45RA−), as well as descendants that are restricted to the myeloid lineage (GMP, CD34+CD38+IL-3RlowCD45RA+).16 We examined whether freshly isolated CMPs and GMPs could develop into NK cells. Because purity was critical, cells were double-sorted directly into 96-well plates. As a control, CMPs and GMPs were also deposited into plates containing methylcellulose. As previously described,16 single CMPs gave rise to myeloid, erythroid, or mixed (granulocyte, erythroid, megakaryocyte, and macrophage) colonies (cloning efficiency, 61.7%-91.7%; average, 76.7%), whereas GMPs yielded exclusively myeloid colonies (cloning efficiency, 43.3%-82.3%; average, 62.7%) (Table 1), and none of the wells showed 2 separate colonies. GMPs plated at 10 cells/well gave virtually no erythroid or mixed colonies (1 mixed colony in 300 wells; n = 5 donors). When cultured in NK-supporting cytokines (ie, IL-3, SCF, FLT-3L, IL-7, and IL-15), CMPs and GMPs very rarely yielded CD56+ cells (0.7% of wells; Table 1). These CD56+ cells were at an immature stage of NK differentiation (CD56+CD117highCD94−)19,23 (not shown). When the same populations were cultured with cytokines and stroma with/without HDC, a significant fraction generated CD56+ cells (Table 1). These CD56+ cells fell in the lymphocytic gate (by FSC/SSC) and coexpressed CD94, corresponding to mature, functional NK cells.19,23 Myeloid cells (CD15+) coexisted with NK cells in some cultures (Figure 5C), showing that a single CMP or GMP could produce both myeloid cells and NK cells. As in the above-described experiments, single CFU-GM colonies (derived from the control-sorted CMPs and GMPs cultured in methylocellulose) could give rise to NK cells if isolated and further cultured in cytokines and stroma and HDC (2 of 30 colonies; not shown). These results show that on a single-cell level, myeloid progenitors (CMPs and GMPs) can produce NK cells.
CMPs and GMPs can give rise to NK cells under the influence of stroma and HDC
| . | 1 cell/well . | 10 cells/well . | 1 cell/well in methylcellulose . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytokines only . | Stroma + cytokines . | HDC + stroma + cytokines . | Cytokines only . | Stroma + cytokines . | HDC + stroma + cytokines . | Erythroid colonies . | Myeloid colonies . | Mixed colonies . | Cloning efficiency . | |
| CMP | ||||||||||
| BG118 | 0 | 1.67 | 3.33 | 0 | 30 | 51.67 | 15 | 40 | 8.3 | 63.3 |
| BG119 | 3.33 | 35 | 27.5 | 8.33 | 100 | 100 | 35 | 46.7 | 10 | 91.7 |
| BG120 | 0 | NE | 17.5 | NE | 86.67 | 100 | 30 | 43.3 | 11.7 | 85 |
| BG121 | 0 | 3.33 | 8.33 | 1.67 | 68.33 | 55 | 26.7 | 23.3 | 11.7 | 61.7 |
| BG122 | 0 | 23.33 | 34.17 | 5 | 98.15 | 100 | 18.3 | 56.7 | 6.7 | 81.7 |
| Median | 0 | 13.3 | 17.5 | 3.3 | 86.7 | 100 | 26.7 | 43.3 | 10 | 81.7 |
| Average | 0.7 | 15.8 | 18.2 | 3.7 | 76.6 | 81.3 | 25 | 42 | 9.7 | 76.7 |
| GMP | ||||||||||
| BG118 | 0 | 8.33 | 15 | 0 | 50 | 36.7 | 0 | 55 | 0 | 55 |
| BG119 | 1.7 | 21.7 | 25 | 5 | 100 | 100 | 0 | 83.3 | 0 | 83.3 |
| BG120 | 0 | 31.7 | 29.2 | ND | 75 | 88 (5/well) | 0 | 61.7 | 0 | 61.7 |
| BG121 | 0 | 25 | 5 | 5 | 73.3 | 30 | 0 | 43.3 | 0 | 43.3 |
| BG122 | 1.7 | 15 | 22.3 | 16.7 | 88.1 | 95 | 0 | 70 | 0 | 70 |
| Median | 0 | 21.7 | 22.3 | 5 | 75 | 88 | 0 | 61.7 | 0 | 61.7 |
| Average | 0.7 | 20.3 | 19.3 | 6.7 | 77.3 | 69.9 | 0 | 62.7 | 0 | 62.7 |
| . | 1 cell/well . | 10 cells/well . | 1 cell/well in methylcellulose . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytokines only . | Stroma + cytokines . | HDC + stroma + cytokines . | Cytokines only . | Stroma + cytokines . | HDC + stroma + cytokines . | Erythroid colonies . | Myeloid colonies . | Mixed colonies . | Cloning efficiency . | |
| CMP | ||||||||||
| BG118 | 0 | 1.67 | 3.33 | 0 | 30 | 51.67 | 15 | 40 | 8.3 | 63.3 |
| BG119 | 3.33 | 35 | 27.5 | 8.33 | 100 | 100 | 35 | 46.7 | 10 | 91.7 |
| BG120 | 0 | NE | 17.5 | NE | 86.67 | 100 | 30 | 43.3 | 11.7 | 85 |
| BG121 | 0 | 3.33 | 8.33 | 1.67 | 68.33 | 55 | 26.7 | 23.3 | 11.7 | 61.7 |
| BG122 | 0 | 23.33 | 34.17 | 5 | 98.15 | 100 | 18.3 | 56.7 | 6.7 | 81.7 |
| Median | 0 | 13.3 | 17.5 | 3.3 | 86.7 | 100 | 26.7 | 43.3 | 10 | 81.7 |
| Average | 0.7 | 15.8 | 18.2 | 3.7 | 76.6 | 81.3 | 25 | 42 | 9.7 | 76.7 |
| GMP | ||||||||||
| BG118 | 0 | 8.33 | 15 | 0 | 50 | 36.7 | 0 | 55 | 0 | 55 |
| BG119 | 1.7 | 21.7 | 25 | 5 | 100 | 100 | 0 | 83.3 | 0 | 83.3 |
| BG120 | 0 | 31.7 | 29.2 | ND | 75 | 88 (5/well) | 0 | 61.7 | 0 | 61.7 |
| BG121 | 0 | 25 | 5 | 5 | 73.3 | 30 | 0 | 43.3 | 0 | 43.3 |
| BG122 | 1.7 | 15 | 22.3 | 16.7 | 88.1 | 95 | 0 | 70 | 0 | 70 |
| Median | 0 | 21.7 | 22.3 | 5 | 75 | 88 | 0 | 61.7 | 0 | 61.7 |
| Average | 0.7 | 20.3 | 19.3 | 6.7 | 77.3 | 69.9 | 0 | 62.7 | 0 | 62.7 |
CMP and GMP were double-sorted from UCB and deposited in 96-well plates at 1 cell/well or 10 cells/well (see “CD34+ cells isolation” for details). Culture conditions included cytokines alone (IL-3, IL-7, IL-15, SCF, and FLT-3L), cytokines and stroma, cytokines and stroma and HDC, or methylcellulose enriched with growth factors. Sixty replicates were performed for each donor (n = 5). At the end of the culture period each well was assayed for NK cells with the use of FACS. Shown is the percentage of wells that contained NK cells. Cells cultured in methylcellulose were examined for outgrowth of erythroid, myeloid, and mixed colonies. The median and average for all the experiments (n = 300 wells with the use of 5 separate donors for each condition) are shown.
NE indicates not evaluable; ND, not done.
To reaffirm this conclusion, granulo-monocytic precursors were purified by FACS according to an alternative definition.25 CD34+CD38+CD64+ myeloid precursors also differentiated into NK cells less than the influence of cytokines and stroma with a frequency similar to the remaining CD34+CD38+NKlin− precursors (supplemental Figure 4).
NK cells derived from myeloid precursors
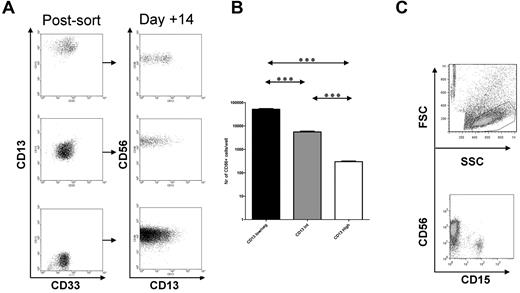
We next isolated myeloid progenitors on the basis of M-CSF receptor expression. The M-CSF receptor (M-CSFR, CSF-1R, CD115) is expressed by granulo-monocytic precursors30 and renders hematopoietic cells responsive to M-CSF, a growth factor promoting monocytic differentiation.31 Therefore, M-CSFR is not merely a phenotypic marker, but it is also functionally linked to the monocytic lineage. We previously determined that CD34+ HPCs cultured on stroma continue to generate NK cells when depleted of CD56+ cells after 2-3 weeks of culture. The cells responsible for this are contained in the CD56−CD117+ subset23 (not shown). These cells variably express M-CSFR (Figure 6A). Therefore, CD56−CD117+M-CSFR+ and CD56−CD117+M-CSFR− cells were isolated from NK differentiation cultures to test their potential to generate NK cells. Both populations gave rise to CD56+ cells on further culture (Figure 6A). To examine their maturation status, cells were tested for CD94 expression.23,24 CD56−CD117+M-CSFR− precursors cultured in only cytokines readily differentiated into functional NK cells (CD56+CD94+). In contrast, CD56−CD117+M-CSFR+ precursors cultured in cytokines alone gave rise to CD56+ cells lacking CD94 (Figure 6A). The addition of HDC or stroma or both to CD56−CD117+M-CSFR+ myeloid precursors increased the CD94 and killer immunoglobulin–like receptor (KIR)–expressing cells (Figure 6B). Thus, CD56−CD117+M-CSFR+ myeloid precursors require HDC or stroma or both to advance to a functional stage of NK differentiation, whereas the M-CSFR− counterparts do not.
M-CSFR+ precursors can become NK cells but have differential developmental requirements. (A) The CD56−CD117+ cell fraction was isolated from day 18 NK-cell differentiation culture of CD34+ cells and labeled for M-CSFR (CD115) expression. FACS gates for the M-CSFR+ and M-CSFR− subsets are shown. On further culture for 2 weeks with cytokines alone (IL-15, IL-7, SCF, FLT-3L) the M-CSFR+ subset gives rise to CD56+CD94− NK cells (top plot), whereas M-CSFR− subset generates predominantly CD56+CD94+ NK cells (bottom plot). (B) The addition of HDC, stroma, or both to CD56−CD117+M-CSFR+ myeloid precursors induces increasing acquisition of CD94 (top) and KIR (bottom). (C) The CD56−CD117+M-CSFR+ subset generates increasing numbers of CD14+ cells on the addition of M-CSF (□, no M-CSF; ▩, 10 ng/mL; ■, 50 ng/mL). The number of CD14+ cells is shown after 1 week of culture with NK-supporting cytokines (IL-15, IL-7, SCF, FLT-3L) and stroma (left) or cytokines and stroma and HDC (right). (D) M-CSF at high doses abolishes NK-cell development of CD56−CD117+M-CSFR+ precursors. Shown are the number of CD56+ cells when CD56−CD117+M-CSFR+ precursors are cultured in NK-supporting cytokines (IL-15, IL-7, SCF, FLT-3L) and in stroma (left) or cytokines, stroma, and HDC (right) and cultured with/without M-CSF (□ indicates no M-CSF; ▩, 10 ng/mL; ■, 50 ng/mL). The number of NK cells at 2 weeks is shown. (E) The CD56−CD117+M-CSFR+ subset cultured with low concentrations (0, 1, 10, and 20 ng/mL) of M-CSF induces CD56+ NK cells with coexpression of CD14 (percentages of CD14+ in CD56+ fraction indicated). Cells were present in the lymphocyte gate by SSC versus FSC characteristics (not shown).Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 2-way analysis of variance on log-transformed values with Bonferroni posttest.
M-CSFR+ precursors can become NK cells but have differential developmental requirements. (A) The CD56−CD117+ cell fraction was isolated from day 18 NK-cell differentiation culture of CD34+ cells and labeled for M-CSFR (CD115) expression. FACS gates for the M-CSFR+ and M-CSFR− subsets are shown. On further culture for 2 weeks with cytokines alone (IL-15, IL-7, SCF, FLT-3L) the M-CSFR+ subset gives rise to CD56+CD94− NK cells (top plot), whereas M-CSFR− subset generates predominantly CD56+CD94+ NK cells (bottom plot). (B) The addition of HDC, stroma, or both to CD56−CD117+M-CSFR+ myeloid precursors induces increasing acquisition of CD94 (top) and KIR (bottom). (C) The CD56−CD117+M-CSFR+ subset generates increasing numbers of CD14+ cells on the addition of M-CSF (□, no M-CSF; ▩, 10 ng/mL; ■, 50 ng/mL). The number of CD14+ cells is shown after 1 week of culture with NK-supporting cytokines (IL-15, IL-7, SCF, FLT-3L) and stroma (left) or cytokines and stroma and HDC (right). (D) M-CSF at high doses abolishes NK-cell development of CD56−CD117+M-CSFR+ precursors. Shown are the number of CD56+ cells when CD56−CD117+M-CSFR+ precursors are cultured in NK-supporting cytokines (IL-15, IL-7, SCF, FLT-3L) and in stroma (left) or cytokines, stroma, and HDC (right) and cultured with/without M-CSF (□ indicates no M-CSF; ▩, 10 ng/mL; ■, 50 ng/mL). The number of NK cells at 2 weeks is shown. (E) The CD56−CD117+M-CSFR+ subset cultured with low concentrations (0, 1, 10, and 20 ng/mL) of M-CSF induces CD56+ NK cells with coexpression of CD14 (percentages of CD14+ in CD56+ fraction indicated). Cells were present in the lymphocyte gate by SSC versus FSC characteristics (not shown).Groups showing significant differences are indicated by asterisks (***P < .001, **P = .001-.01, and *P = .01-.05), 2-way analysis of variance on log-transformed values with Bonferroni posttest.
To investigate whether myeloid and NK differentiation represent 2 alternative developmental pathways for CD56−CD117+M-CSFR+ precursors, M-CSF was added to NK developmental cultures. M-CSF increased the generation of CD14+ (CD56−) monocytes and decreased NK-cell development in a dose-dependent manner (P < .001) (Figure 6C and D, respectively). High concentrations of M-CSF (≥ 50 ng/mL) abolished NK-cell development from M-CSFR+ precursors, despite conditions optimal for NK-cell development (cytokines, stromal cells, and HDC) (Figure 6D). Thus, development of NK cells (CD56+) or monocytes (CD14+CD56−) from M-CSFR–expressing myeloid precursors are alternative outcomes, influenced by M-CSF. Interestingly at lower concentrations (1-20 ng/mL), M-CSF induced low-level CD14 coexpression on a fraction of developing CD56+ lymphocytes (Figure 6E). This provides complementary evidence that the cells that give rise to NK cells also respond to M-CSF. Collectively, these studies show that myeloid progenitors can generate NK cells, strongly supporting a myeloid NK-cell differentiation pathway.
Distinct properties of myeloid precursor–derived NK cells
We tested whether myeloid precursor–derived NK cells differ from the primarily lymphoid (M-CSFR−) fraction. Both CD56−CD117+M-CSFR+ and CD56−CD117+M-CSFR− subsets were cultured in conditions optimal for NK-cell generation (cytokines, stroma and HDC). A higher percentage of NK cells derived from CD56−CD117+M-CSFR+ progenitors expressed KIR compared with NK cells derived from CD56−CD117+M-CSFR− precursors (Figure 7A-B; P < .0001). Interestingly, NK cells derived from CD56−CD117+M-CSFR+ progenitors also contained a fraction of NK cells that were NKG2A−KIR+, whereas this fraction was absent in the CD56−CD117+M-CSFR− precursor–derived NK cells (Figure 7C). Both populations showed K562 killing; however, M-CSFR+ precursors consistently showed marginally higher cytotoxicity (Figure 7D). Human leukocyte antigen–deficient Epstein-Barr virus–transformed B-cell targets (721.221) showed a more pronounced difference. NK cells derived from M-CSFR− precursors showed minimal cytotoxicity, whereas M-CSFR+–derived cells readily killed these targets (Figure 7E). There were no consistent differences in perforin and granzyme B protein or in the expression of activating receptors (2B4, NKp30, NKp46, and NKG2D) that would sufficiently explain these results (not shown). Thus, NK cells developing from distinct precursors have diverse phenotypic and functional properties.
NK cells derived from myeloid (M-CSFR+) precursors show abundant KIR expression and higher cytotoxicity compared with lymphoid (M-CSFR−) precursors. (A) Expression of KIR (combination of CD158a, CD158b, and CD158e1) is higher on NK cells derived from myeloid (M-CSFR+) precursors (right) compared with M-CSFR−–derived NK cells (left). A representative donor is shown (n = 4). (B) Percentages of KIR+ NK cells derived from CD56−CD117+M-CSFR+ (▵) and CD56−CD117+M-CSFR− (●) progenitors for 4 individual donors are shown (mean ± SEM; > 3 replicates for each donor). P < .001, by 2-way analysis of variance. (C) Expression of NKG2A and KIR on CD56-gated NK cells derived from CD56−CD117+M-CSFR− (left) and CD56−CD117+M-CSFR+ (right) progenitors. Shown is the presence of a potentially alloreactive population of NKG2A-KIR+ population in the population derived from the CD56−CD117+M-CSFR+ progenitors. (D) Cytotoxicity of M-CSFR+–derived (▵) and M-CSFR−–derived (■) NK cells against K562 targets and (E) against 721.221 human leukocyte antigen–negative B-lymphoblastoid cell line. Data points represent average cytotoxicity of triplicates ± SD at the specified effector-to-target (E:T) ratios. A representative donor is shown (n = 3).
NK cells derived from myeloid (M-CSFR+) precursors show abundant KIR expression and higher cytotoxicity compared with lymphoid (M-CSFR−) precursors. (A) Expression of KIR (combination of CD158a, CD158b, and CD158e1) is higher on NK cells derived from myeloid (M-CSFR+) precursors (right) compared with M-CSFR−–derived NK cells (left). A representative donor is shown (n = 4). (B) Percentages of KIR+ NK cells derived from CD56−CD117+M-CSFR+ (▵) and CD56−CD117+M-CSFR− (●) progenitors for 4 individual donors are shown (mean ± SEM; > 3 replicates for each donor). P < .001, by 2-way analysis of variance. (C) Expression of NKG2A and KIR on CD56-gated NK cells derived from CD56−CD117+M-CSFR− (left) and CD56−CD117+M-CSFR+ (right) progenitors. Shown is the presence of a potentially alloreactive population of NKG2A-KIR+ population in the population derived from the CD56−CD117+M-CSFR+ progenitors. (D) Cytotoxicity of M-CSFR+–derived (▵) and M-CSFR−–derived (■) NK cells against K562 targets and (E) against 721.221 human leukocyte antigen–negative B-lymphoblastoid cell line. Data points represent average cytotoxicity of triplicates ± SD at the specified effector-to-target (E:T) ratios. A representative donor is shown (n = 3).
Discussion
Here, we show that a small fraction of CD34+ HPCs expressing any of the lineage markers previously associated with NK-cell development (ie, CD122, CD161, integrin β7, and CD45RAhigh and CD7)13,17-19 can develop into NK cells under the influence of cytokines (IL-15, IL-7, SCF, FLT-3L, and IL-3) and do not require HDC or stroma. However, most HPCs lack these markers (ie, NKlin−) and are unable to differentiate into NK cells with these cytokines. Rather, NKlin− cells follow myeloid differentiation, as evidenced by surface markers (CD13, CD33, CD14, and CD1a) and intracellular enzymes (lysozyme, myeloperoxidase, and macrosialyn [CD68]). These myeloid precursors could be recruited to the NK lineage by HDC and stroma. Perhaps not surprisingly, they gradually lost the ability to develop into NK cells as they progressed along myeloid differentiation (indicated by increasing CD13 staining). We also show that freshly isolated single CMPs or GMPs or both gave rise to NK cells in the presence of stroma and cytokines but not cytokines alone. The observations that (1) single CMPs and GMPs could generate NK cells, (2) myeloid cells coexisted with NK cells in cultures derived from a single CMP or GMP, and (3) some granulocyte-macrophage colonies from these precursors (grown in methylocellulose) could subsequently differentiate into NK cells provide solid evidence that myeloid precursors can give rise to NK cells. We also show that stroma and HDC mediate these properties, in part, through WNT activation. This is in agreement with the known role of T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors in lymphocyte development and the ability of fetal liver stromal cells to trigger this pathway.32
We also isolated myeloid precursors (CD56−CD117+M-CSFR+) arising in vitro and demonstrated their potential to generate NK cells. The observation that low concentrations of M-CSF induced CD14 expression by NK cells developing from myeloid precursors further supports the assertion that cells responding to M-CSF give rise to NK cells. These CD56+CD14+ cells appear to be NK cells (and not monocytes) because they fell in the lymphoid gate (by SSC vs FSC) and expressed other NK antigens, including CD94 and KIR (not shown). Higher doses of M-CSF, however, enforced monocytic differentiation, prohibiting NK-cell development, suggesting a lineage choice between myeloid and NK-cell fates. These choices are influenced by the environment (stroma) and growth factors (cytokines).
Our results also indicate that myeloid precursor–derived NK cells have distinct properties. For instance, M-CSFR+ precursors, but not M-CSFR− counterparts, require stroma or HDC for CD94 acquisition, a marker of maturation. Likewise, when these 2 starting populations are cultured with cytokines, stroma, and HDC. the resulting NK cells differ. Myeloid-derived NK cells show a significantly higher KIR expression, a KIR+NKG2A− cell fraction and higher cytotoxicity. KIR acquisition has been used as a measure of maturity,27 but the factors that lead to KIR expression are largely unknown. We recently found that myc binds to KIR promoters, driving expression.33 Interestingly, myc expression is linked to M-CSFR signaling and myeloid development.34 Recently, a thymic pathway of NK-cell development was shown in rodents. Compared with the majority of NK cells, thymic-derived NK cells were uniquely dependent on IL-7 and GATA-3 for development. The thymic-derived NK cells are characterized by low cytotoxicity but high cytokine secretion potential.35 These findings, along with the data presented here, support the concept that various developmental trajectories, governed by specific transcription factors, influence the phenotypic and functional characteristics of the resulting population.
According to the widely accepted model of hematopoiesis, a multipotent HSC gives rise to CMPs or CLPs, committing the first choice in hematopoiesis between myeloid/erythroid and lymphoid fates. NK cells are believed to be exclusively derived from CLPs, which are, by definition, devoid of myeloid potential.8 In disagreement with this, we find that NK cells can be derived from freshly isolated CMPs, GMPs, and CD34+CD64+ granulo-monocytic precursors25 as well as CD33+CD13+, and M-CSFR+ progeny of cultured CD34+ HPCs. Whether this is unique to NK cells or extends to other lymphocyte subsets requires further investigation. However, neither T- nor B-cell differentiation occurred in our cultures (not shown). We hypothesize that CMP and GMP progenitors could give rise to other lymphocyte populations less than appropriate, permissive conditions (DLL-1–expressing stroma and optimal cytokines for T-cell development). In line with these observations, alternative models for lymphoid ontogeny have been proposed. Through the identification of common lympho-myeloid progenitors5,6 and thymic T-cell progenitors that retain myeloid potential, it has been concluded that alternative schemes of hematopoietic lineage commitment should be considered.36,37 An important caveat is that the model of hematopoiesis is largely based on in vivo transplantation studies into irradiated mice.38 The in vivo behavior of distinct progenitors is determined, in part, by the environment after transplantation, which is influenced by their trafficking. Although there are clear strengths of the transplantation assay, it does not always show the intrinsic abilities observed in vitro.39
We show that myeloid precursors can differentiate into NK cells and that less than certain circumstances NK cells could be derived from seemingly restricted myeloid progenitors. Whether this occurs in vivo and to what extent it contributes to human physiology needs to be addressed in future studies. Given the absence of NK cells in common γ-chain deficiency,40 myeloid precursors that develop into NK cells would conceivably acquire (and depend on) this cytokine receptor for NK-cell development.
In humans, 2 NK subsets can be distinguished, CD56bright and CD56dim.41 The CD56bright subset has low cytotoxicity and readily produces cytokines in response to IL-12 and IL-18.42 Only a small percentage of CD56bright cells show expression of KIR and CD16, but they do express c-kit receptor (CD117). In comparison, the CD56dim subset has higher cytotoxicity, abundant CD16 and KIR expression, and lack CD117. The developmental relationship of the 2 subsets has not been unambiguously resolved. The majority of NK cells derived in vitro from CD34+ HPCs in our21,23 as well as in other12,20,22,43 systems resemble CD56bright NK cells. It is thus noteworthy that NK cells derived from myeloid precursors are distinguished by higher cytotoxicity and KIR expression, as well a fraction of KIR-expressing cells that lack NKG2A and lower levels of CD117 (not shown), features of the CD56dim subset.
A myeloid origin of NK cells was formerly considered and debated.7,44 However, the unequivocal demonstration of a common T/NK precursor resulted in the abandonment of this concept.45 More recently, it has been shown that dendritic cells (DCs) and NK cells share a common developmental progenitor,11 whereas others have described a rare CD14+ cell in UCB (but not adult blood) that gives rise to NK cells less than the influence of HDC, FLT-3L, and IL-15.10 These results are in line with the findings present in this study. Here we posit that, similar to DCs,46 NK cells can have either a lymphoid or a myeloid origin. The developmental relationship between NK cells and DCs may be closer than previously recognized in that both can be derived from common precursors. Whether a stable cell type exists which constitutively shares the function of NK cells and DCs47,48 is a matter of debate.49,50 Nevertheless, in certain conditions DCs can acquire cytotoxicity, characteristic for NK cells.51 Conversely NK cells can acquire antigen-presenting ability.52 The notion that myeloid precursors previously known to give rise to monocyte/macrophage and DCs53 are also capable of NK-cell differentiation puts the recent findings in a new perspective.54 In summary we present evidence that NK cells can be derived from myeloid precursors.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Linda Kluge and BioE for providing some of the UCB units used in these studies.
This work was supported by the American Cancer Society (M.R.V.), Children's Cancer Research Fund (B.G. and M.R.V.), Leukemia Research Foundation (M.R.V.), and the National Institutes of Health (grants P01 CA65493 [J.S.M.], P01 111412 [M.R.V. and J.S.M.], R01 HL55417 [J.S.M.], R01 CA72669 [B.R.B.], and PO1067493 [J.S.M. and B.R.B.]).
National Institutes of Health
Authorship
Contribution: B.G. designed, performed, and analyzed experiments and wrote the paper; Nandini Kataria and Niketa Kataria performed and analyzed experiments; B.R.B. and J.S.M. analyzed data and contributed to writing the paper; and M.R.V. designed and analyzed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael R. Verneris, Department of Pediatrics, Division of Blood and Marrow Transplantation, University of Minnesota, 425 E River Rd, Ste 660, Minneapolis, MN 55455; e-mail: verneris@umn.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal