Abstract
The Nef protein of HIV-1 is important for AIDS pathogenesis, but it is not targeted by current antiviral strategies. Here, we describe a single-domain antibody (sdAb) that binds to HIV-1 Nef with a high affinity (Kd = 2 × 10−9M) and inhibited critical biologic activities of Nef both in vitro and in vivo. First, it interfered with the CD4 down-regulation activity of a broad panel of nef alleles through inhibition of the Nef effects on CD4 internalization from the cell surface. Second, it was able to interfere with the association of Nef with the cellular p21-activated kinase 2 as well as with the resulting inhibitory effect of Nef on actin remodeling. Third, it counteracted the Nef-dependent enhancement of virion infectivity and inhibited the positive effect of Nef on virus replication in peripheral blood mononuclear cells. Fourth, anti-Nef sdAb rescued Nef-mediated thymic CD4+ T-cell maturation defects and peripheral CD4+ T-cell activation in the CD4C/HIV-1Nef transgenic mouse model. Because all these Nef functions have been implicated in Nef effects on pathogenesis, this anti-Nef sdAb may represent an efficient tool to elucidate the molecular functions of Nef in the virus life cycle and could now help to develop new strategies for the control of AIDS.
Introduction
The advent of monoclonal antibodies was a major breakthrough for the development of antibody-based therapies against various diseases, including virus infection.1 However, expression in the cytoplasm or nucleus of eukaryotic cells of full-length immunoglobulin G or single-chain antibody variable domain fragments aiming at blocking an intracellular target is poorly efficient because of the reducing nature of these compartments. The engineering of single-chain antibody variable domain fragments (scFvs) that consist in a tandem fusion of the VH and VL domains of a specific antibody partially overcame this limitation. Numerous scFvs have shown their potency for inhibition of the function of their target proteins in cells. In particular, a HIV-1 Tat-specific scFv was shown to neutralize the trans-activating function of this viral protein and to prevent full expression of the viral genome in infected cells.2 ScFvs specific for HIV-1 matrix, integrase, or regulatory proteins such as Tat, Rev, and Vif are also able to interfere with HIV-1 replication.3-9 However, disulfide bonds between VH and VL chains of scFvs are unlikely to form when scFvs are targeted to reducing compartments such as the cytoplasm or the nucleus, resulting in scFvs with lower or no affinity for their antigen.10
Recently, a major improvement was brought by the use of single-domain antibodies (sdAbs) derived from camelids. In addition to conventional antibodies, camelids express antibodies composed of heavy chains only, with a single variable domain (VHH) capable of recognizing their cognate antigens.11 These variable domains, called sdAbs, are endowed with many attractive features.12 The absence of requirement for disulfide bond formation in sdAbs is particularly interesting when targeting of proteins found in reducing cell compartments is considered.13 In contrast to conventional antibodies, these 13-kDa sdAb fragments can penetrate in cavities located on the surface of antigens (for review, see Nguyen et al14 ). These cryptic sites are often more conserved than exposed epitopes on viral proteins and are a target of choice for blocking antibodies. Interestingly, sdAb fragments targeting the HIV-1 Rev or Vif proteins have been already characterized and have displayed antiviral activity in cell-culture assays.15,16
In the present study, we characterized an sdAb from a llama immunized with a recombinant form of Nef, an HIV-1 nonstructural protein found both in the cytoplasm of infected cells and in association with cellular membranes. Considering Nef as a potential target for antiviral therapy arose from the findings that this HIV-1 protein is important for AIDS pathogenesis in vivo (for review, see Foster and Garcia17 ). Nef is abundantly expressed early after virus infection and perturbs the trafficking of several membrane proteins through action on the endocytic pathway. This leads to the modulation of cell surface expression of some receptors, including CD4 and major histocompatibility complex class I (MHC-I) molecules (for review, see Foster and Garcia17 ). Nef-induced cell surface down-regulation of CD4 depends on the integrity of a di-Leu motif found in the C-terminal flexible loop of HIV-1 Nef, which allows interactions with clathrin-associated adaptor protein (AP) complexes that participate in the vesicular transport within the endocytic pathway.18 In contrast, the downmodulation of MHC-I is determined by distinct motifs located in the N-terminal part of Nef,19 indicating that the Nef-mediated down-regulation of either CD4 or MHC-I are related to different mechanisms.
Nef also alters the activation state of infected cells via modulation of signal transduction processes such as T-cell receptor (TCR) signaling.20 In response to TCR engagement, multiple effects of Nef have been observed.21 For example, Nef, via its ability to associate with the cellular p21-activated kinase 2 (PAK2), deregulates the actin-severing factor cofilin,20 affects TCR-induced cytoskeleton organization leading to a reduction of actin polymerization in infected T lymphocytes.22,23
In vivo, HIV-1 or simian immunodeficiency virus (SIV) Nef is required for high virus replication and rapid disease progression in humans24 and macaques,25 respectively, although AIDS developed in macaques infected with SIV Nef-deletion mutants.26 In addition, Nef expression in T cells of transgenic (Tg) mice was found to induce T-cell depletion.27,28 Moreover, Nef expression in mature and immature T cells as well as in cells of the dendritic/macrophage lineage of CD4C/HIVNef Tg mice led to the development of a severe AIDS-like disease.29
So far, only a few Nef inhibitors have been described. Chemical compounds capable of interfering with the ability of Nef to interact with SH3 domains30-32 or activate the Hck tyrosine kinase33,34 have been identified. Although some inhibitors were too cytotoxic for cellular assays,30 others were only confirmed in cellular and biochemistry-based assays, but critical functions of Nef such as CD4 down-regulation and infectivity increase were not investigated. Here, we report the characterization of a specific sdAb displaying a high affinity for HIV-1 Nef. When expressed as an intracellular antibody, this anti-Nef sdAb inhibited important biologic functions of Nef both in vitro and in vivo in CD4C/HIV-1Nef Tg mice.
Methods
Llama immunization, construction, and screening of the sdAb library
A llama (Lama glama) was immunized by 4 subcutaneous injections of 250 μg of the purified recombinant Nef protein (fragment 57-205), performed every 3 weeks. Blood samples were taken 15 days after the last injection, and peripheral blood mononuclear cells were isolated as described.35 Llama management, inoculation, and sample collection were conducted by trained personnel under the supervision of a veterinarian, in accordance with protocols approved by the Centre National de la Recherche Scientifique's ethical committee of animal welfare.
Plasmids
The phage clone showing the most robust interaction with Nef by enzyme-linked immunoabsorbent assay (ELISA; phage-sdAb19) was selected, and the sdAb sequence was amplified with specific primers and subcloned into the XhoI sites of pcDNA3.1+ vector (Invitrogen). Plasmids encoding wild-type (WT) or mutated Nef-hemagglutinin (HA) and Nef–green fluorescent protein (GFP) have been described previously.37,38 Plasmids for expression of HIV-1 nef alleles (SF33, 2693BA, CK1.62, RBF168, 13127K2, HJ256, pCMO2.5, RP4-11, and NP34) were a kind gift from Frank Kirchhoff and Nicoletta Casartelli and have been described previously39,40 ; plasmids for expression of NA7 and YBF30 nef alleles have been also described.41 WT and nef-deleted proviral infectious clones of the NL4.3 virus isolate (HIV-1 NL4.3 WT and HIV-1 NL4.3 ΔNef, respectively) have been described.42
Cell culture and transfection
293T and HeLa cells were grown in Dulbecco minimal essential medium supplemented with 10% fetal calf serum (FCS), 100 IU penicillin/mL, and 100 μg streptomycin/mL (Invitrogen). HeLa-CD4 cells, clone P4.2, were grown in complete medium supplemented with G418, 0.2 mg/mL (PAA Laboratories). CEM, HPB-ALL, and Jurkat CD4+ T cells were grown in RPMI supplemented with 10% FCS, 100 IU penicillin/mL, and 100 μg streptomycin/mL. All cells were grown at 37°C with 5% CO2. Culture media for fetal liver cells consisted of Iscove, 15% FCS (StemCell Technologies), interleukin-6 (IL-6; Fischer), stem cell factor, and Flt3 ligand.
Viral production and infectivity assays
Single-round HIV-1 carrying the GFP gene and replication-competent viruses were produced in 293T cells as described.44 Constructs encoding Nef-HA and sdAb19 were added when indicated in figure legends. Infectivity of HIV-1 HXBc2 and vesicular stomatitis virus envelope glycoprotein (VSV-G)–pseudotyped viruses was analyzed on HeLa-CD4 and HPB-ALL cells as described.43 Human peripheral blood mononuclear cells (PBMCs) were isolated from blood of healthy donors by Ficoll density gradient centrifugation. Cells were cultured in RPMI, 10% FCS, 1% antibiotics supplemented with phytohemagglutinin (5 μg/mL) for 48 hours. Cells were then washed and suspended in complete medium supplemented with 10 ng/mL IL-2. Cells (107) were infected with 0.5 μg of CAp24 of either WT or ΔNef virus particles produced in the presence of sdAb19. Sampling of cell culture supernatants was done immediately after washing (day 0) and at subsequent times. Amounts of CAp24 produced were determined by ELISA (Innogenetics).
For preparation of murine retroviruses expressing sdAb19, a pMSCV-based plasmid expressing sdAb19 upstream of internal ribosome entry site (IRES)–GFP was constructed and transfected into the 293 GPG packaging cell line to produce amphotropic retrovirus. Ecotropic virus was produced with the use of the GP+E86 packaging cell line. The pMSCV vector as well as the 293 GPG and the GP+E86 packaging cell lines were a gift of Guy Sauvageau.
Mice, fetal liver-cell infection, and transplantation
Mouse management, inoculation, and sample collection were conducted by trained personnel under the supervision of a veterinarian, in accordance with protocols approved by the Clinical Research Institute of Montreal (Montreal, Canada) ethical committee of animal welfare.
The CD4C/HIVMutG (designated here CD4C/HIVNef) Tg mice have been described.29 Single-cell suspensions from fetal liver from non-Tg (CD45.1+ CD45.2+) and Tg (CD45.1+ or CD45.2+) 14.5-day-old embryos were preactivated in vitro overnight with IL-6 (10 ng/mL), stem cell factor (100 ng/mL), and Flt3 ligand (50 ng/mL). They were subsequently plated onto irradiated infected GP+E86 cells. Cells were infected for 24 hours and injected into lethally irradiated hosts. Donor cell inoculum consisted of 3.8 × 106 fetal liver cells together with 0.2 × 106 syngenic WT bone marrow cells. Eight- to 12-week-old C3H/HeN (H-2k) (CD45.2+) non-Tg were lethally irradiated (9.5 Gy [950 rads]) and were injected with 4 × 106 cells. Chimeras were analyzed 1-5 months after transplantation. Statistical analysis was performed with the Student t test.
Immunoprecipitation and immunoblot analysis
Immunoprecipitations were carried out on lysates of transfected 293T cells as described43 with the use of anti-GFP (7.1/13.1; Roche) or anti–c-Myc (9E10 clone; Roche) antibodies. Immunoprecipitated proteins were then analyzed by Western blot with anti–c-Myc (9E10; Roche) and anti-GFP (sc-8334; Santa Cruz Biotechnology Inc).
Flow cytometry
Surface staining and internalization assays of CD4 have been described previously.37 RPA-T4 clone phycoerythrin-cyanin 5 (PE-Cy5)–conjugated anti-CD4 antibody was purchased from BD Biosciences. Flow cytometry on mouse cells was performed as previously described.45 PE-, PE-Cy7-, Biotin-, or allophycocyanin (APC)–conjugated antibodies against mouse CD4, CD8α, TCRβ, CD25, CD44, and CD45.1 were purchased from Cedarlane or BD Biosciences. GFP negativity/positivity was determined on the basis of fluorescence of cells from uninfected Tg chimeras. Gating of GFP−, GFPhi, and GFPlow cells was based on distribution of cell populations with distinct GFP intensities as well as levels of CD4 in Tg sdAb19+ chimeras. Acquisition was performed on FACSCalibur (BD Biosciences) or BD-LSR (BD Biosciences). Cell sorting was performed on a Mo-Flo cell sorter (Cytomation).
Immunofluorescence
HeLa cells were seeded in 24-well plates on coverslips and then transfected by Lipofectamin 2000 (Invitrogen). After 24 hours, cells were washed twice in phosphate-buffered saline (PBS), fixed 20 minutes at 4°C in PBS supplemented with paraformaldehyde, 4% (Sigma-Aldrich), and washed twice in PBS supplemented with 0.1% bovine serum albumin (PBS-BSA). Coverslips were then incubated with PBS-BSA supplemented with Triton X-100, 0.1% (Sigma-Aldrich), and the anti-c-Myc 9E10 antibody. Cells were incubated 60 minutes on ice, washed twice in PBS-BSA, and incubated 45 minutes on ice in PBS-BSA supplemented with Alexa 594–coupled goat antibodies directed against mouse immunoglobulin G (Invitrogen). Samples were examined under an epifluorescence microscope (Leica DMB) with a cooled charge-coupled device camera (Micromax 1300Y/HS; Roper Princeton Instruments), using a Plan APO 100×. The acquisition of images was done with MetaMorph 7.6 (Molecular Devices).
In vitro kinase assay, actin remodeling, and phospho-cofilin immunofluorescence analysis
In vitro kinase assay was performed as described38 on Jurkat cells transfected with expression plasmids for WT or mutant Nef-GFP fusion proteins and variable amounts of the sdAb19 plasmid. After 24 hours, cell lysates were immunoprecipitated with a rabbit anti-GFP antibody. After washing, the immunoprecipitates were resuspended in KAB (50mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 8, 150mM NaCl, 5mM EDTA [ethylenediaminetetraacetic acid], 0.02% Triton X-100, and 10mM MgCl2) containing 10 μCi (0.37 MBq) of [γ-32P]adenosine triphosphate. After incubation for 10 minutes, bound proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and subjected to autoradiography.
Analysis of TCR-mediated cell spreading and actin ring formation was carried out as described23 on transfected Jurkat cells with phalloidin (0.5 μg/mL phalloidin–tetramethyl rhodamine isothiocyanate; Sigma-Aldrich) to show F-actin. Staining of p-cofilin was performed with the use of a rabbit anti-Ser3 phosphorylated cofilin (77G2; Cell Signaling/NEB).20
Results
Characterization of a sdAb targeting Nef
After immunization of a llama with a recombinant fragment of Nef (aa 57-205), a phage library of sdAbs was built from peripheral mononuclear cells of the immunized animal and screened with the phage-display technique. A phage-sdAb (sdAb19) that yielded the highest ELISA signal on immobilized Nef was thus isolated and selected for further investigation (supplemental Figure 1). The affinity of sdAb19 for Nef was determined by surface plasmon resonance, yielding a ka (on rate) of 0.89 ± 0.12 × 105 M−1S−1, a kd (off rate) of 1.81 ± 0.04 × 10−4 s−1 and a calculated Kd (dissociation constant) of 2nM (supplemental Figure 1E).
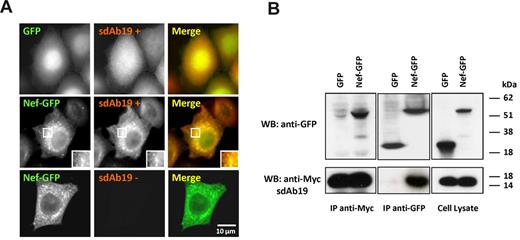
To explore the ability of sdAb19 to interfere with the functions of Nef, the open reading frame encoding sdAb19 was subcloned in a vector with a c-Myc tag to facilitate detection in mammalian cells. The ability of sdAb19 to target native Nef (HIV-1Lai strain) was first investigated by immunofluorescence in cells coexpressing sdAb19 and Nef-GFP (Figure 1A). When expressed with GFP, sdAb19 randomly distributed in the cytoplasm and the nucleus (Figure 1A top). On the contrary, in Nef-expressing cells (Figure 1A middle), Nef-GFP and sdAb19 colocalized in cytoplasmic dotted structures that were mainly concentrated in the perinuclear region and resembled the endocytic compartments we described previously.46
Association of sdAb19 with Nef. HeLa (A) or 293T (B) cells were transfected with the vector for expression of the c-Myc–tagged sdAb19 in combination with vector for expression of either Nef-GFP or GFP. (A) Intracellular distribution of Nef-GFP and sdAb19. sdAb19 was detected by indirect immunofluorescence with anti–c-Myc in Nef-GFP– (middle and bottom) or GFP-expressing cells (top). (B) Coimmunoprecipitation of Nef-GFP and sdAb19. Cell lysates (right) were submitted to immunoprecipitation with anti–c-Myc (left) or anti-GFP (center). Immunoprecipitates were analyzed by Western blotting with the use of anti-c-Myc (bottom) or anti-GFP (top). IP indicates immunoprecipitate.
Association of sdAb19 with Nef. HeLa (A) or 293T (B) cells were transfected with the vector for expression of the c-Myc–tagged sdAb19 in combination with vector for expression of either Nef-GFP or GFP. (A) Intracellular distribution of Nef-GFP and sdAb19. sdAb19 was detected by indirect immunofluorescence with anti–c-Myc in Nef-GFP– (middle and bottom) or GFP-expressing cells (top). (B) Coimmunoprecipitation of Nef-GFP and sdAb19. Cell lysates (right) were submitted to immunoprecipitation with anti–c-Myc (left) or anti-GFP (center). Immunoprecipitates were analyzed by Western blotting with the use of anti-c-Myc (bottom) or anti-GFP (top). IP indicates immunoprecipitate.
To confirm that this colocalization was related to the association of sdAb19 with Nef, coimmunoprecipitation experiments were performed from transfected cells. As shown in Figure 1B, the anti–c-Myc antibody was able to coprecipitate Nef-GFP, but not GFP, with sdAb19 (Figure 1B left). Conversely, anti-GFP efficiently coprecipitated sdAb19 from cells expressing Nef-GFP but not from cell expressing GFP (Figure 1B center). Interestingly, sdAb19 also coprecipitated with another HIV-1 Nef allele (SF2 strain) but not with the Nef protein from the SIVmac239 strain (supplemental Figure 2A). However, sdAb19 failed to bind to deletion mutants covering the N-terminal (aa 1-61), the core (aa 58-189), and the C-terminal (aa 160-206) regions of HIV-1 Nef (supplemental Figure 2B), suggesting that it rather recognizes a conformational structure preserved in the core domain of HIV-1 Nef proteins.
sdAb19 inhibits Nef-induced down-regulation and endocytosis of CD4
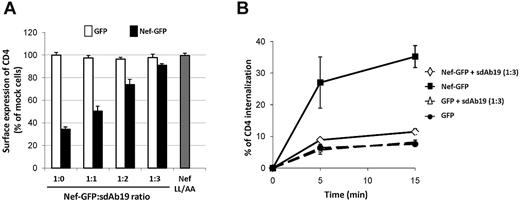
To investigate the effect of sdAb19 on the ability of Nef to down-regulate cell surface CD4, HPB-ALL CD4+ T cells were cotransfected with constructs encoding sdAb19 and Nef-GFP and analyzed for cell surface CD4 expression by flow cytometry (Figure 2A). In the absence of sdAb19, efficient CD4 down-regulation was measured in Nef-GFP–expressing cells (1:0 ratio). When cells were cotransfected with increasing amounts of sdAb19-encoding plasmid (1:1 to 1:3 ratios), CD4 down-regulation by Nef-GFP was largely inhibited in a dose-dependent manner.
sdAb19 inhibits Nef-induced CD4 down-regulation. HPB-ALL cells were transfected with plasmids for the expression of either Nef-GFP or GFP in combination with increasing amounts of the plasmid for expression of sdAb19. (A) sdAb19 activity on Nef-induced CD4 cell surface down-regulation. Transfected cells were stained with PE-Cy5–conjugated anti-CD4 at 4°C, and surface expression of CD4 in Nef-GFP– or GFP-expressing cells was measured by flow cytometry. Results are expressed as the percentage of the MFI determined in GFP-positive cells relative to that determined in GFP-negative cells. Values are the means of 3 independent experiments. Error bars represent 1 SD from the mean. The NefLL/AA mutant (▩) mutated in the AP-binding di-Leu motif (LL164/165) was used as negative control. (B) sdAb19 activity on Nef-induced acceleration of CD4 internalization. Transfected cells were stained at 4°C with PE-Cy5–conjugated anti-CD4 and then incubated at 37°C for 5 or 15 minutes to allow internalization. Cell surface-bound anti-CD4 was then stripped by acidic wash, and cell-associated CD4 staining was measured by flow cytometry on GFP-positive cells. The percentage of internalized CD4 at each time point was calculated as described.43
sdAb19 inhibits Nef-induced CD4 down-regulation. HPB-ALL cells were transfected with plasmids for the expression of either Nef-GFP or GFP in combination with increasing amounts of the plasmid for expression of sdAb19. (A) sdAb19 activity on Nef-induced CD4 cell surface down-regulation. Transfected cells were stained with PE-Cy5–conjugated anti-CD4 at 4°C, and surface expression of CD4 in Nef-GFP– or GFP-expressing cells was measured by flow cytometry. Results are expressed as the percentage of the MFI determined in GFP-positive cells relative to that determined in GFP-negative cells. Values are the means of 3 independent experiments. Error bars represent 1 SD from the mean. The NefLL/AA mutant (▩) mutated in the AP-binding di-Leu motif (LL164/165) was used as negative control. (B) sdAb19 activity on Nef-induced acceleration of CD4 internalization. Transfected cells were stained at 4°C with PE-Cy5–conjugated anti-CD4 and then incubated at 37°C for 5 or 15 minutes to allow internalization. Cell surface-bound anti-CD4 was then stripped by acidic wash, and cell-associated CD4 staining was measured by flow cytometry on GFP-positive cells. The percentage of internalized CD4 at each time point was calculated as described.43
Because the Nef-induced down-regulation of cell surface CD4 is largely related to an acceleration of the internalization rate of CD4 from the cell surface in CD4+ T cells,43 CD4 internalization was measured in cells coexpressing Nef-GFP and sdAb19 (Figure 2B). As expected, Nef-GFP induced a 5-fold increase in the rate of CD4 internalization; however, when Nef-GFP and sdAb19 were coexpressed, the rate of CD4 internalization was not significantly different from that measured in the absence of Nef-GFP. These data indicate that the inhibitory effect of sdAb19 on Nef-induced CD4 down-regulation measured at steady state can be explained, at least in part, by the inhibition of CD4 internalization induced by Nef. Similarly, the kinetic of internalization from the plasma membrane of a CD8-Nef chimera, in which Nef was fused to the extracellular and transmembrane domains of CD8, was greatly decreased in cells coexpressing sdAb19 compared with that measured in the absence of sdAb19 (supplemental Figure 3A). This inhibition of the internalization of CD8-Nef correlated with a redistribution of the chimera at the plasma membrane as evidenced by immunofluorescence (supplemental Figure 3B) and flow cytometric analyses (supplemental Figure 3C).
Because both in trans CD4 down-regulation by Nef and in cis down-regulation of CD8-Nef require the AP-binding Leu-based motif of Nef,37 these results indicate that the sdAb19 activity is probably related to an action on the interaction of Nef with the AP complex machinery. However, expression of increasing levels of sdAb19 failed to inhibit the Nef-induced cell surface down-regulation of MHC-I molecules (supplemental Figure 4). This latter result confirms that the Nef effect on MHC-I trafficking is induced by a mechanism different from that involved in CD4 downmodulation.
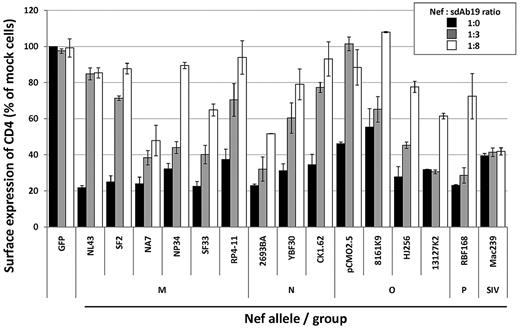
Finally, we evaluated whether sdAb19 was able to cross-react with the Nef proteins of different HIV-1 groups through analysis of its ability to inhibit the CD4 down-regulation activity of a panel of nef alleles from both primary and laboratory-adapted HIV-1 strains. CEM T cells were cotransfected with the vector for expression of the different nef alleles in combination with increasing amounts of sdAb19-encoding plasmid (1:0, 1:3, and 1:8 ratios) and then analyzed for cell surface CD4 expression by flow cytometry. As recapitulated in Figure 3, sdAb19 was able to counteract most of the HIV-1 nef alleles analyzed in a dose-dependent manner, including Nef proteins from groups M, N, O, and P; as a control, sdAb19 was not able to inhibit the CD4 down-regulation activity of the Nef protein from the SIVmac239 strain. Of note, some of the HIV-1 nef alleles were inhibited only when high levels of sdAb19 (1:8 ratio) were coexpressed with Nef, suggesting that sdAb19 recognized these Nef proteins with distinct affinities. However, these data indicate that sdAb19 was able to cross-react with a broad panel of Nef proteins from different HIV-1 groups.
Activity of sdAb19 on CD4 down-regulation induced by HIV-1 Nef alleles. CEM T cells were cotransfected with plasmids for expression of nef alleles in combination with increasing amounts of the plasmid for expression of sdAb19 (1:3 and 1:8 ratios) or an irrelevant sdAb (1:0 ratio). Twenty-four hours later, cells were stained with PE-Cy5–conjugated anti-CD4 at 4°C, and cell surface expression was measured by flow cytometry. Results are expressed as the percentage of the MFI determined in GFP-positive cells relative to that determined in GFP-negative cells. Values are the means of 3 independent experiments. Error bars represent 1 SD from the mean.
Activity of sdAb19 on CD4 down-regulation induced by HIV-1 Nef alleles. CEM T cells were cotransfected with plasmids for expression of nef alleles in combination with increasing amounts of the plasmid for expression of sdAb19 (1:3 and 1:8 ratios) or an irrelevant sdAb (1:0 ratio). Twenty-four hours later, cells were stained with PE-Cy5–conjugated anti-CD4 at 4°C, and cell surface expression was measured by flow cytometry. Results are expressed as the percentage of the MFI determined in GFP-positive cells relative to that determined in GFP-negative cells. Values are the means of 3 independent experiments. Error bars represent 1 SD from the mean.
sdAb19 inhibits association of Nef with the cellular kinase PAK2 and reverts effects of Nef on actin remodeling and cofilin phosphorylation
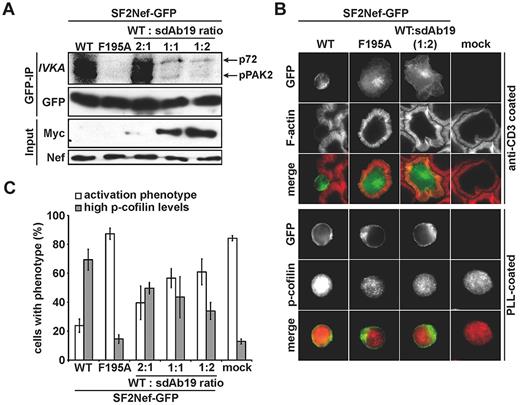
Nef also interferes with multiple signal transduction processes in infected T lymphocytes, including TCR signaling that leads to profound actin cytoskeleton rearrangements.22,23 Inhibition of TCR-induced actin remodeling requires the association of Nef with PAK2 activity.23,47 Because the PAK2 association varies among HIV-1 nef alleles, we used the Nef protein from the SF2 strain, which associated with sdAb19 as efficiently as LAI Nef (supplemental Figure 2A), to investigate whether sdAb19 was able to interfere with Nef/PAK2 association and the related effects on actin cytoskeleton. As shown in Figure 4A, immunoprecipitation of Nef from Jurkat cells and subsequent in vitro kinase assay showed the coprecipitation of associated PAK2 activity that resulted in the phosphorylation of PAK2 itself (pPAK2; 62 kDa) and an unidentified substrate (p72; 72 kDa). Expectedly, no kinase activity was coprecipitated from cells expressing the NefF195A mutant that lacks the ability to associate with PAK2.38 Similarly, Nef-associated kinase activity was almost completely abolished when cells coexpressed WT Nef with sdAb19 at the 1:1 and 1:2 plasmid ratios.
sdAb19 blocks Nef-PAK2 association and restores actin remodeling after TCR engagement and cofilin deregulation. (A) Jurkat cells expressing WT or F195A Nef-GFP and increasing amounts of sdAb19 were subjected to anti-GFP immunoprecipitation and subsequent in vitro kinase assay. Nef-associated PAK2 activity is shown by the phosphorylated 62-kDa band (p-PAK2, IVKA). (B) Representative micrographs of the cells used in panel A. Cells were plated onto anti-CD3 or poly-L-lysine (PLL)–coated cover glasses (top and bottom panels, respectively), fixed and stained either with phalloidin to reveal F-actin (top) or for p-cofilin (bottom). (C) Frequency of the cells shown in panel B that are able to form F-actin–rich circumferential rings and with high p-cofilin levels. Values are the means of 3 independent experiments, and error bars represent 1 SD from the mean; ≥ 100 cells were analyzed per transfection.
sdAb19 blocks Nef-PAK2 association and restores actin remodeling after TCR engagement and cofilin deregulation. (A) Jurkat cells expressing WT or F195A Nef-GFP and increasing amounts of sdAb19 were subjected to anti-GFP immunoprecipitation and subsequent in vitro kinase assay. Nef-associated PAK2 activity is shown by the phosphorylated 62-kDa band (p-PAK2, IVKA). (B) Representative micrographs of the cells used in panel A. Cells were plated onto anti-CD3 or poly-L-lysine (PLL)–coated cover glasses (top and bottom panels, respectively), fixed and stained either with phalloidin to reveal F-actin (top) or for p-cofilin (bottom). (C) Frequency of the cells shown in panel B that are able to form F-actin–rich circumferential rings and with high p-cofilin levels. Values are the means of 3 independent experiments, and error bars represent 1 SD from the mean; ≥ 100 cells were analyzed per transfection.
We next investigated the effect of sdAb19 on the ability of Nef to inhibit actin remodeling after TCR engagement. Jurkat cells expressing WT Nef-GFP or the NefF195A-GFP mutant, either alone or with sdAb19, were seeded on coverslips coated with CD3-specific antibodies, and F-actin was then stained with fluorescent phalloidin. In the absence of Nef, circumferential F-actin rings were formed on adhesion of cells to the substratum (Figure 4B top). In contrast, neither formation of circumferential F-actin–rich rings nor cell spreading was observed when cells expressed WT Nef-GFP. As expected,20 the Nef F195A mutant failed to inhibit F-actin polymerization on TCR engagement. When cells coexpressed Nef and sdAb19, a significant dose-response inhibition of actin remodeling interference by Nef was observed, resulting in the formation of circumferential F-actin–rich rings in > 60% of Nef-expressing cells at a 1:2 ratio of Nef and sdAb19 expression vectors (Figure 4C white bars). Comparable inhibition of Nef activity on actin remodeling by sdAb19 was observed when analyzing the inhibition of SDF-1α–induced membrane ruffling20 (data not shown). In line with these results and the role of cofilin hyperphosphorylation in the inhibition of actin remodeling by Nef,20 sdAb19 was able to reduce in a dose-dependent manner the frequency of Nef-expressing cells with elevated levels of phosphorylated, inactive cofilin (Figure 4B bottom and 4C gray bars). These results show that sdAb19 is able to reverse the inhibitory effect of Nef on actin remodeling through inhibition of the Nef/PAK2 interaction and thereby preventing deregulation of cofilin.
sdAb19 inhibits virus infectivity in a Nef-dependent manner and counteracts the positive effect of Nef on virus replication
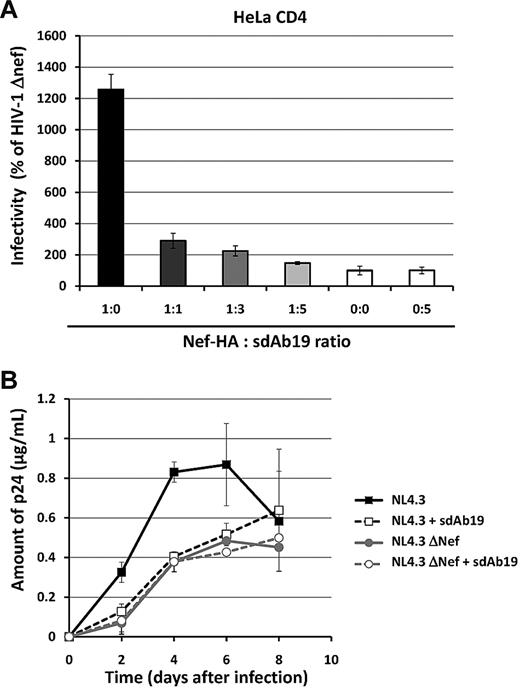
Because nef-deleted viruses are consistently less infectious than their WT counterparts,44,46 the ability of sdAb19 to affect virus infectivity of new progeny virions produced in the presence of Nef was first investigated in a single-round infection assay. Reporter viruses carrying the gene encoding GFP were produced in 293T cells in the absence or presence of increasing concentrations of sdAb19, and their infectivity was assayed on either HeLa-CD4 or HPB-ALL T cells (Figure 5A and supplemental Figure 5B, respectively). As expected, there was a net increase of virus infectivity when HA-tagged Nef was expressed during production of HIV-1 Env-pseudotyped viruses. As shown in Figure 5A, an ∼ 80% decrease of this specific effect of Nef was observed when equivalent amounts of plasmids (1:1 ratio) were used for expression of Nef and sdAb19 during virus production. The inhibition was almost complete when highest concentrations of sdAb19 were expressed in combination with Nef in virus-producing cells (1:3 and 1:5 ratios). We also confirmed the specificity of the effect of Nef and sdAb19 in this assay by analyzing the infectivity of VSV-G–pseudotyped viruses produced in similar conditions. Neither Nef nor sdAb19, alone or in combination, affected the infectivity of VSV-G–pseudotyped virions (supplemental Figure 5A-B). These data show that the inhibition of HIV-1 infectivity by sdAb19 results from the inhibition of a specific function of Nef.
sdAb19 inhibits Nef-mediated enhancement of virus infectivity and the Nef-positive effect on virus replication. (A) Single-round GFP reporter viruses were produced in 293T cells in the absence or presence of increasing amounts of the plasmid expressing sdAb19. Forty-eight hours later, viruses were pelleted from cell culture supernatants and were used to infect HeLa-CD4 cells. The percentages of GFP-positive infected cells were then measured by flow cytometry 60 hours later. Viral infectivity was normalized to that of viruses produced in the absence of Nef. (B) WT (black curves) or ΔNef (gray curves) replication-competent viruses were produced in 293T cells in the absence (plain lines) or the presence of sdAb19 (dashed lines) in a 1:1 ratio and were used to infect PBMCs. Aliquots of cell culture supernatant were collected 2, 4, 6, and 8 days after infection for CAp24 quantification.
sdAb19 inhibits Nef-mediated enhancement of virus infectivity and the Nef-positive effect on virus replication. (A) Single-round GFP reporter viruses were produced in 293T cells in the absence or presence of increasing amounts of the plasmid expressing sdAb19. Forty-eight hours later, viruses were pelleted from cell culture supernatants and were used to infect HeLa-CD4 cells. The percentages of GFP-positive infected cells were then measured by flow cytometry 60 hours later. Viral infectivity was normalized to that of viruses produced in the absence of Nef. (B) WT (black curves) or ΔNef (gray curves) replication-competent viruses were produced in 293T cells in the absence (plain lines) or the presence of sdAb19 (dashed lines) in a 1:1 ratio and were used to infect PBMCs. Aliquots of cell culture supernatant were collected 2, 4, 6, and 8 days after infection for CAp24 quantification.
Interestingly, sdAb19 was incorporated into purified viral particles only when cells coexpressed Nef (supplemental Figure 5C), indicating that a Nef/sdAb19 interaction in virion-producing cells mediates the incorporation of sdAb19 into viral particles. Of note, expression of sdAb19 in virus-producing cells did not affect the pattern of Gag either in virion-producing cells or in viral particles (supplemental Figure 5C), confirming that the inhibitory effect of sdAb19 on virus infectivity was not because of a major inhibition of virion maturation but reflects the inhibition of a specific function of Nef.
We then evaluated whether sdAb19 was also active for inhibition of the Nef effect on replication-competent viruses by comparing the kinetics of replication of viruses produced in the presence or absence of sdAb19. WT and nef-deleted viruses (NL4.3 WT and NL4.3 ΔNef, respectively) were thus produced in 293T cells expressing sdAb19 and used to infect human PBMCs. Virus production was then monitored by measuring the CAp24 antigen every 2 days (Figure 5B). As expected, NL4.3 WT-expressing Nef replicated efficiently in PBMCs with a rapid increase in CAp24 production as soon as 2 days after infection, whereas NL4.3 ΔNef had a replication delay, with significant lower levels of CAp24 detected in cell culture medium 2, 4, and 6 days after infection. Importantly, NL4.3 WT viruses produced in the presence of sdAb19 displayed the same delayed kinetic of virus replication than the NL4.3 ΔNef viruses. Together, these data show that sdAb19 is able to counteract both the Nef-dependent enhancement of virion infectivity and the positive effect of Nef on virus replication.
sdAb19 inhibits Nef-mediated T-cell phenotypes of CD4C/HIVNef Tg mice in vivo
Next, we tested whether sdAb19 was able to neutralize Nef effects in vivo. For this purpose, we used the CD4C/HIVNef Tg mice showing CD4 cell surface down-regulation, altered thymic CD4+ T-cell development, and profound peripheral CD4+ T-cell depletion.29 Because these phenotypes are transplantable, fetal liver cells from Tg or non-Tg mice were infected with retroviruses encoding sdAb19-IRES-GFP and were injected into lethally irradiated host animals. GFP levels were used as an indication of the level of sdAb19 expression.
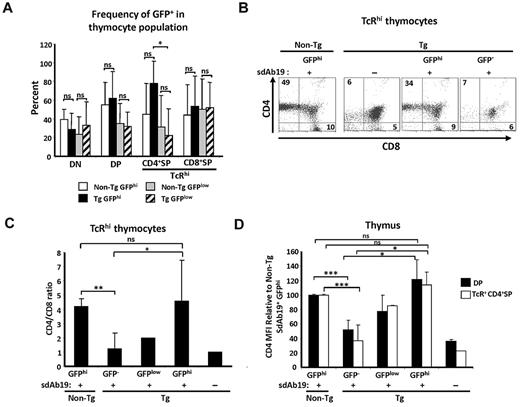
Evaluation of chimeric mice was done 1-5 months after transplantation, and all analyses were performed on donor (CD45.1+) cells. These analyses showed expression of GFP at different intensities in a significant portion of thymocyte subsets: from the least mature CD4−CD8− (double negative), through the intermediate CD4+CD8+ (double positive) to the most mature TcRhi CD4+SP (single positive) and TcRhi CD8+SP (Figure 6A; supplemental Figure 6A). The average percentage of GFPlow and GFPhi populations was similar between non-Tg sdAb19+ and Tg sdAb19+ chimeras compared within all thymocyte populations, except for TcRhi CD4+SP T cells, which were enriched in GFPhi cells in Tg sdAb19+ chimeras (Figure 6A). This result suggests a generation/survival advantage for TcR+ CD4+SP Tg thymocytes expressing high levels of sdAb19. Consistent with this interpretation, we found that, among TcRhi thymocytes, GFPhi but not GFP− cells had an increase of CD4+SP percentage and of the CD4/CD8 ratio (Figure 6B-C), suggesting that sdAb19 had reverted this thymic maturation defect. Moreover, GFPhi cells (both double positive and TcRhi CD4+SP) had a CD4 mean fluorescence intensity (MFI) comparable to levels observed on thymocytes from non-Tg chimeras (Figure 6D; supplemental Figure 6A), in contrast to GFP−/low thymocytes that exhibited significant CD4 down-regulation. This reversal of CD4 down-regulation by sdAb19 was not caused by the absence of Nef in these transduced GFPhi thymocytes, because they harbored high levels of Nef as detected by Western blot (supplemental Figure 6B).
sdAb19 rescues Nef-mediated thymic CD4 maturation block in vivo. Fetal liver cells from CD4C/HIVNef Tg and non-Tg 14.5-day-old embryos were infected in vitro with retrovirus coding sdAb19-IRES-GFP or were uninfected and were injected into syngenic lethally irradiated hosts. Thymuses from host animals were analyzed 1-5 months after reconstitution. (A) Percentage of GFPhi and GFPlow cells measured by flow cytometry within designated thymocyte subsets. Gating of GFP−, GFPhi, and GFPlow cells was based on distribution of cell populations with distinct GFP intensities as well as levels of CD4 in Tg sdAb19+ chimeras (supplemental Figure 6A). (B-C) Thymocytes were labeled with antibodies against TCR, CD4, and CD8, and percentages of SP thymocytes as well as average CD4+SP to CD8+SP ratios among TcRhi thymocytes were determined by flow cytometry. (D) Average CD4 MFI on thymocytes was compared with average CD4 MFI found on thymocytes in non-Tg chimeras. CD4 MFI in Tg GFP− cells was determined on CD4low cells. Data represent ≥ 2 distinct experiments with a total of 3-8 mice per group. Statistical analysis was performed with the Student t test (*P < .5, **P < .01, and ***P < .001; ns indicates not significant). Error bars represent 1 SD from the mean.
sdAb19 rescues Nef-mediated thymic CD4 maturation block in vivo. Fetal liver cells from CD4C/HIVNef Tg and non-Tg 14.5-day-old embryos were infected in vitro with retrovirus coding sdAb19-IRES-GFP or were uninfected and were injected into syngenic lethally irradiated hosts. Thymuses from host animals were analyzed 1-5 months after reconstitution. (A) Percentage of GFPhi and GFPlow cells measured by flow cytometry within designated thymocyte subsets. Gating of GFP−, GFPhi, and GFPlow cells was based on distribution of cell populations with distinct GFP intensities as well as levels of CD4 in Tg sdAb19+ chimeras (supplemental Figure 6A). (B-C) Thymocytes were labeled with antibodies against TCR, CD4, and CD8, and percentages of SP thymocytes as well as average CD4+SP to CD8+SP ratios among TcRhi thymocytes were determined by flow cytometry. (D) Average CD4 MFI on thymocytes was compared with average CD4 MFI found on thymocytes in non-Tg chimeras. CD4 MFI in Tg GFP− cells was determined on CD4low cells. Data represent ≥ 2 distinct experiments with a total of 3-8 mice per group. Statistical analysis was performed with the Student t test (*P < .5, **P < .01, and ***P < .001; ns indicates not significant). Error bars represent 1 SD from the mean.
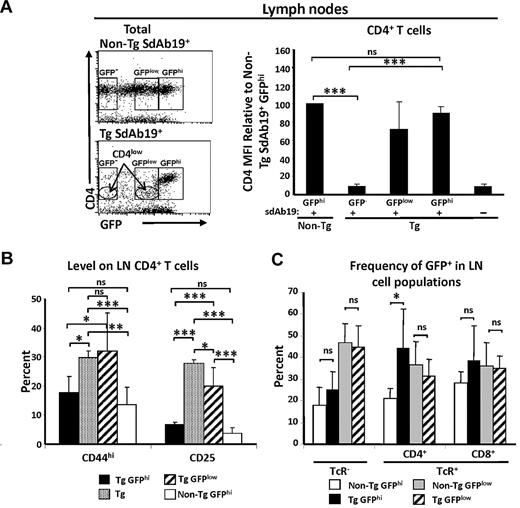
As in the thymus, cell populations of different GFP intensities were detected in peripheral lymphoid organs (not shown). CD4 staining in chimeras also showed the presence of CD4hi GFPhi and CD4low GFPlow Tg populations (Figure 7A), again indicating that high levels of sdAb19 can neutralize Nef-mediated CD4 down-regulation. Previously, we reported that Nef-expressing Tg CD4+ T cells are CD25+ and exhibit an activated “effector/memory” phenotype (CD44hi).45 Analysis of these markers on lymph node (LN) cells of Tg chimeras expressing sdAb19 showed that GFPhi CD4hi cells had CD44 and CD25 levels similar to those of non-Tg cells, whereas a higher percentage of GFPlow CD4low cells were CD44hi and CD25+ and thus resembled phenotypically untransduced Tg CD4+ cells (Figure 7B). These data indicate that sdAb19 is able to reverse Nef-mediated effector/memory induction in peripheral CD4+ T cells. The percentage of GFPlow and GFPhi populations was similar between non-Tg sdAb19+ and Tg sdAb19+ chimeras compared within the TcR− and TcR+ CD8+ T-cell subsets (Figure 7C). However, TcR+ CD4+ T cells were enriched in the GFPhi population of Tg SdAb19+ chimeras, and this difference was statistically significant (Figure 7C). Intriguingly, despite the capacity of sdAb19 to fully reverse CD4 down-regulation and “activation” in peripheral Tg CD4+ T cells, only a negligible reversal of their depletion and of the low CD4/CD8 cell ratio was observed (supplemental Figure 6C-D).
sdAb19 reverses Nef effects on peripheral CD4+ T cells in vivo. Peripheral lymph nodes were analyzed 4-5 months after reconstitution, gating on donor (CD45.1+) cells. Gating of GFP−, GFPhi, and GFPlow cells was based on distribution of cell populations with distinct GFP intensities as well as levels of CD4 in Tg sdAb19+ chimeras. (A) Average CD4 MFI on pLN CD4+ T cells was compared with the average CD4 MFI found on CD4+ T cells in non-Tg chimeras. CD4 MFI in Tg GFP− cells was determined on CD4low cells. (B) Average percentage of cells expressing CD44 or CD25 among pLN CD4+ T cells showing CD4 and GFP high or low levels or among CD4low T cells in chimeras reconstituted with uninfected Tg fetal liver. (C) Percentage of GFPhi and GFPlow cells within designated subsets. Statistical analysis was performed with the Student t test (*P < .5, **P < .01, and ***P < .001; ns indicates not significant). Error bars represent 1 SD from the mean.
sdAb19 reverses Nef effects on peripheral CD4+ T cells in vivo. Peripheral lymph nodes were analyzed 4-5 months after reconstitution, gating on donor (CD45.1+) cells. Gating of GFP−, GFPhi, and GFPlow cells was based on distribution of cell populations with distinct GFP intensities as well as levels of CD4 in Tg sdAb19+ chimeras. (A) Average CD4 MFI on pLN CD4+ T cells was compared with the average CD4 MFI found on CD4+ T cells in non-Tg chimeras. CD4 MFI in Tg GFP− cells was determined on CD4low cells. (B) Average percentage of cells expressing CD44 or CD25 among pLN CD4+ T cells showing CD4 and GFP high or low levels or among CD4low T cells in chimeras reconstituted with uninfected Tg fetal liver. (C) Percentage of GFPhi and GFPlow cells within designated subsets. Statistical analysis was performed with the Student t test (*P < .5, **P < .01, and ***P < .001; ns indicates not significant). Error bars represent 1 SD from the mean.
Together, these results show that sdAb19 is able to rescue Nef-mediated thymic CD4+ T-cell maturation defect and peripheral CD4+ T-cell activation phenotypes of the CD4C/HIV-1Nef Tg mouse model.
Discussion
In the present study, we describe a Nef-specific sdAb that recognizes an epitope conserved in most of the HIV-1 Nef proteins. Intracellular expression of sdAb19 resulted in the inhibition of Nef functions such as down-regulation of cell surface CD4, PAK2-dependent actin remodeling on TCR stimulation, the Nef-dependent enhancement of HIV-1 infectivity and replication, as well as in vivo CD4+ T-cell activation and inhibition of CD4+ thymocyte maturation. On the contrary, sdAb19 had no effect on Nef-induced down-regulation of cell surface MHC-I.
Using deletion mutants covering the N-terminal, the core, and the C-terminal regions of Nef, we failed to determine which region of Nef was recognized by sdAb19 (supplemental Figure 2B). Interestingly, sdAb19 was not able to coprecipitate with the (58-189) Nef fragment that corresponds to the core region of the protein. The core and C-terminal regions contain most of the motifs needed for interaction with cellular proteins and involved in the functions of Nef, including the nonstructured loop of Nef containing the di-Leu and di-Asp signals required for association with AP complexes of the endocytic pathway. Because sdAb19 still reacted with the Nef-LL164/165AA and DD174/175AA mutants (supplemental Figure 2C; data not shown), this indicates that these residues, required for both Nef-mediated CD4 down-regulation and enhancement of virus infectivity, are not directly in contact with sdAb19. Together, these results indicate that sdAb19 binding needs the maintenance of the full structure of the protein. These observations suggest that it rather recognizes a conformational epitope conserved in a broad panel of HIV-1 nef alleles from groups M, N, O, and P, in relation with the relative conservation of the core region of HIV-1 Nef proteins.
One of the earliest characterized functions of Nef is its ability to down-regulate cell surface levels of CD4 in infected cells (for review, see Foster and Garcia17 ). This property strongly correlates with the effect of Nef on virus replication and the rate of disease progression, both in infected patients and in animal models.48 The ability of Nef to relocate CD4 from the cell surface to endosomal compartments requires the interaction of Nef with AP complexes that are involved in vesicle formation and cargo sorting within the endocytic pathway.49 In T cells, Nef increases the rate of CD4 endocytosis and targets internalized CD4 molecules to the lysosome-dependent degradation pathway.43 In this work, sdAb19 had no obvious effect on the subcellular localization of Nef, yet it efficiently inhibited Nef-induced mouse and human CD4 down-regulation in primary Tg T lymphocytes and in T-cell lines, respectively. Such activity could result from the inhibition of the Nef/CD4 interaction, and sdAb19 should efficiently compete for CD4 binding on Nef and prevent CD4 down-regulation. Alternatively, sdAb19 could rather prevent the targeting of Nef to clathrin-coated pits (CCP) where CD4 endocytosis takes place.37 Although our data show that sdAb19 does not directly target the AP-binding di-Leu motif of Nef (supplemental Figure 2C) required for both Nef localization in CCP and CD4 down-regulation,37 it could indirectly impose steric hindrance to the Nef/AP-2 interaction required for CD4 down-regulation. Similarly, sdAb19 was also able to prevent the internalization of a CD8-Nef chimera, resulting in the enrichment of the chimera at the plasma membrane (supplemental Figure 3). This was expected because in trans down-regulation of CD4 by Nef and in cis internalization of CD8-Nef both require the AP-binding di-Leu motif of Nef.46 However, the inhibitory effect of sdAb19 on CD8-Nef internalization could not be explained by the disruption of the interaction between Nef and its cargo. Rather, sdAb19 probably interferes with in cis down-regulation of CD8-Nef by preventing the targeting of cell surface CD8-Nef to CCP and the recruitment of AP-2 to the di-Leu motif of Nef, as evidenced by the accumulation of CD8-Nef at the plasma membrane in cells coexpressing sdAb19 (supplemental Figure 3).
The ability of Nef to down-regulate CD4 and MHC-I relies on genetically distinct determinants of Nef.50 The Nef-induced MHC-I down-regulation is independent of the di-Leu motif and rather depends of motifs found in the N-terminal part of the protein. The fact that sdAb19 did not inhibit Nef-induced MHC-I down-regulation (supplemental Figure 4) confirms that independent mechanisms are involved in CD4 and MHC-I down-regulation. It is possible that the interaction between Nef and sdAb19 masks Nef domains involved in CD4 or AP2 binding, leaving the N-terminal α-helix (aa 17-26), as well as the 62EEEE65 and 72PxxP75 motifs required for Nef-induced MHC-I down-regulation,19,51 fully accessible.
Neutralizing activity of sdAb19 was also investigated for the ability of Nef to precipitate PAK2 kinase activity. Nef/PAK2 association leads to elevated cellular levels of the inactive, phosphorylated form of the actin-severing factor cofilin, a deregulation that is instrumental for the inhibition of TCR- or chemokine-induced F-actin remodeling.20,22,23 Our experiments show that coexpression of sdAb19 with WT Nef was phenotypically comparable to the expression of the NefF195A alone. Because this Nef mutant specifically lacks association with PAK2 activity without impairment of any other Nef function,52 these results suggest a direct interference of sdAb19 upstream or at the level of PAK2 recruitment by Nef. Conceivably, this inhibition may result from blocking of protein interactions of the F195 residue that is in close proximity with the di-Leu motif required for CD4 internalization. Interestingly, despite an ∼ 90% decrease of Nef-associated kinase activity when a 1:2 ratio of Nef/sdAb19 was used, only ∼ 50% of Nef-expressing cells were defective for actin remodeling on TCR engagement or cofilin hyperphosphorylation. These results are reminiscent of those obtained after silencing of endogenous PAK220 and suggest that low amounts of residual Nef-associated kinase activity can be sufficient for the induction of downstream functions such as inhibition of F-actin remodeling and deregulation of cofilin.
The inhibitory effect of sdAb19 was analyzed on the ability of Nef to increase HIV-1 infectivity and to affect virus replication in PBMCs. The mechanism responsible for the positive effect of Nef on virus infectivity is presently unknown, but it is related, at least in part, to the functional perturbations of the intracellular trafficking induced by Nef at the level of the endocytic pathway.46,48 The fact that similar amounts of Nef are incorporated into viral particles regardless of the presence of sdAb19 confirms that some features of the trafficking of Nef are not altered by sdAb19. Of note, the binding of sdAb19 to Nef did not prevent Nef cleavage by HIV-1 protease.44 Given that the major protease cleavage site in Nef and the putative CD4 binding site partially overlap, this domain of Nef might be fully accessible in the Nef/sdAb19 complex. In any case, the fact that sdAb19 reverted the Nef-related HIV-1 infectivity suggests that the sdAb19/Nef interaction is detrimental to this unknown mechanism. We and others have suggested that Nef increases HIV-1 infectivity by “modifying” virions in the course of their biogenesis.44,53 These modifications could include posttranslational modifications of viral proteins or incorporation/exclusion of cellular factors into/from viral particles, and sdAb19 might prevent such effects of Nef.
Finally, with the use of the CD4C/HIV-1Nef Tg mouse model,29 we showed that sdAb19 was able to revert some in vivo Nef-mediated phenotypes, notably the CD4 down-regulation, the developmental block of CD4+ SP thymocytes, and the activation of peripheral CD4+ T cells. Because we have recently found that the Tg thymic SP CD4+ T-cell developmental block is largely the impairment of CD4/Lck function,54 the reversion of the Nef-mediated CD4 down-regulation by sdAb19 probably explains the reversal of thymic developmental block of CD4+ T cells. In peripheral LN cells, sdAb19 was found to efficiently reverse not only CD4 down-regulation but also the up-regulation of CD44 and CD25. In contrast, peripheral CD4+ T-cell depletion was only marginally affected by sdAb19, indicating that distinct domains of Nef may be responsible for peripheral CD4+ T-cell depletion on one hand and activation as well as CD4 down-regulation on the other hand.
Together, our data show that the interaction between sdAb19 and Nef inhibits highly conserved functions of Nef both in vitro and in vivo. Because distinct domains of Nef are involved in the various functions of this viral protein, we hypothesize that the functions inhibited by sdAb19 are those in which the sdAb19/Nef complex can prevent Nef from interacting with cellular partners, while leaving other Nef interfaces readily accessible for binding to other cellular effectors and for inducing phenotypes, such as MHC-I down-regulation even in the presence of sdAb19. The delineation of the sdAb19 binding domain on Nef will help to understand the selective inhibitory effect of sdAb19 and will shed light on the many mechanisms used by Nef to usurp cellular pathways. X-ray analysis of the complex formed between recombinant forms of sdAb19 and Nef is in progress to delineate the interface recognized by sdAb19.
In summary, we used the specific advantages of the sdAbs from camelids for the development of a specific reagent that targets Nef, a viral factor essential during HIV-1 infection for virus replication and disease progression. This sdAb could constitute an efficient tool to analyze the molecular and cellular functions of Nef in vitro in infected cells, as well as in vivo in primary target cells of different organs of the CD4C/HIV-1Nef Tg animals. For example, sdAb19 will be used as a tool to try to define, at the molecular level, how it is able to revert most of the Nef functions and to interfere with the interactions of Nef with host cell factors. Moreover, our data show that it is possible to inhibit most important functions of Nef with a single component; therefore, the characterization of this anti-Nef sdAb, active on a large panel of nef alleles from different HIV-1 groups, could now help to develop new antiviral strategies for the control of AIDS. The first strategy could be to use sdAb19 as a recombinant protein fused to a short cell-penetrating sequence and able to pass through the biologic membranes to access the cytoplasmic compartment where Nef is localized. Alternatively, characterization of small chemical compounds that mimic the action of sdAb19 and counteract the biologic properties of Nef could represent a first step in the development of specific inhibitors of Nef. High-throughput screening of molecules able to target the Nef/sdAb19 interaction is in progress for isolation of chemical compounds that bind to the molecular interface recognized by sdAb19; such molecules may thus reproduce the inhibitory effects of sdAb19 on the Nef functions during virus replication. In conclusion, the characterization of the anti-Nef sdAb19 described in the present study showed that it is possible to target most of the functions of Nef with a single ligand.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Frank Kirchhoff (University of Ulm) and Nicoletta Casartelli (Pasteur Institute) for the generous gift of reagents. We thank Stéphanie Charles, Martine Chartier, Isabelle Corbin, Ginette Masse, and Benoit Laganiere for technical skills and Martine Pugnière for SPR experiments. We also thank Alexandre Benmerah for discussion and constant support and Zaher Hanna for help with preparing figures. We thank Eric Massicotte and Martine Dupuis from the Cell Biology Core (IRCM).
This work was supported by Inserm, CNRS, Université Paris-Descartes, and the French National Agency for AIDS Research (ANRS; S.B.), as well as the Deutsche Forschungsgemeinschaft (grant GRK1188; B.S.) and (SFB638 and EXO81; O.T.F.).
Authorship
Contribution: J.B., S.E.B., P. Chrobak, B.S., O.T.F., P. Chames, P.J., S.B., and D.B. designed research; J.B., S.E.B., P. Chrobak, B.S., and N.B. performed research; J.B., S.E.B., P. Chrobak, B.S., O.T.F., P. Chames, P.J., S.B., and D.B. analyzed data; and J.B., S.E.B., P. Chrobak, O.T.F., P. Chames, P.J., S.B., and D.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Serge Benichou, Institut Cochin, 27 Rue du Faubourg Saint-Jacques, 75014 Paris, France; e-mail: serge.benichou@inserm.fr; or Daniel Baty, Inserm U624, 163 Avenue de Luminy, 13288 Marseille Cedex 09 France; e-mail: daniel.baty@inserm.fr.
References
Author notes
J.B., S.E.B., and P. Chrobak contributed equally to this study.
S.B. and D.B. are senior coauthors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal