Abstract
The transcription factor STAT5 is an essential mediator of the pathogenesis of chronic myelogenous leukemia (CML). In CML, the BCR/ABL fusion kinase causes the constitutive activation of STAT5, thereby driving the expression of genes promoting survival. BCR/ABL kinase inhibitors have become the mainstay of therapy for CML, although CML cells can develop resistance through mutations in BCR/ABL. To overcome this problem, we used a cell-based screen to identify drugs that inhibit STAT-dependent gene expression. Using this approach, we identified the psychotropic drug pimozide as a STAT5 inhibitor. Pimozide decreases STAT5 tyrosine phosphorylation, although it does not inhibit BCR/ABL or other tyrosine kinases. Furthermore, pimozide decreases the expression of STAT5 target genes and induces cell cycle arrest and apoptosis in CML cell lines. Pimozide also selectively inhibits colony formation of CD34+ bone marrow cells from CML patients. Importantly, pimozide induces similar effects in the presence of the T315I BCR/ABL mutation that renders the kinase resistant to presently available inhibitors. Simultaneously inhibiting STAT5 with pimozide and the kinase inhibitors imatinib or nilotinib shows enhanced effects in inhibiting STAT5 phosphorylation and in inducing apoptosis. Thus, targeting STAT5 may be an effective strategy for the treatment of CML and other myeloproliferative diseases.
Introduction
Activating mutations of tyrosine kinases are common events in cancer pathogenesis.1 These mutant kinases trigger a series of signaling events that culminate in the activation of genes that drive the malignant behavior of a cell. The identification of transcription factors that mediate the effect of activated kinases would provide an attractive target for cancer therapy. One family of transcription factors activated by tyrosine kinases is the signal transducer and activator of transcription (STAT) family. These transcription factors are latent proteins residing in the cytoplasm that are activated by phosphorylation on a critical tyrosine residue. After traveling to the nucleus, STATs regulate transcription of their target genes, which include genes involved in survival, proliferation, and differentiation.2
One STAT family member, STAT5, is constitutively active in many forms of hematologic cancers, including chronic myelogenous leukemia (CML).3,4 CML cells are characterized by the BCR/ABL fusion gene, which generates a constitutively activated tyrosine kinase. BCR/ABL causes the activation of STAT5, which leads to increased expression of genes driving cell cycle progression and promoting survival.4,5 The development of the BCR/ABL inhibitor imatinib mesylate represented a paradigm shift in the treatment of CML patients.6 However, patients can develop resistance to this drug through point mutations in BCR/ABL that decrease the binding of imatinib to the active site of the kinase.7,8 One such mutation, T315I, renders CML cells completely resistant not only to imatinib but also to the second-generation BCR/ABL inhibitors nilotinib and dasatinib.9 Therefore, targeting STAT5 directly is an attractive approach to overcome resistance to BCR/ABL kinase inhibitors.
Given that constitutive STAT activation is a common pathogenic event in tumorigenesis, we undertook a screen to isolate STAT inhibitors that may be useful for cancer therapy. We used a transcriptionally based assay, which provides a nonbiased approach for the identification of inhibitors targeting any part of the STAT-signaling pathway.10 To accelerate the identification of drugs that could be used in proof-of-concept clinical trials, we used a chemical library that contained compounds known to be safe in humans. Here we describe the identification and characterization of the STAT5 inhibitory activity of the neuroleptic drug pimozide, which potently induces apoptosis in CML cells. These effects are enhanced when pimozide is combined with the kinase inhibitors imatinib or nilotinib. Importantly, pimozide is equally effective against imatinib-sensitive and -resistant cells. These data provide the framework to consider clinical trials of STAT5 inhibition for CML patients with and without resistance to kinase inhibitors.
Methods
Cells
KU812 cells were obtained from ATCC; K562 cells were obtained from Daniel G. Tenen (Beth Israel Deaconess Medical Center, Boston, MA); the generation of Ba/F3.p210, Ba/F3.p210-T315I, 32d.p210, and 32d.p210-T315I was described previously.11 Ba/F3 cells expressing constitutively active STAT5a1*6 under the control of a doxycycline-inducible promoter12 were grown in 1 ng/mL murine interleukin-3 (PeproTech). To express STAT5a1*6, the cells were washed to remove interleukin-3 and then cultured in the presence of 1 μg/mL doxycycline (Sigma-Aldrich). All cells were cultured in RPMI media supplemented with 10% fetal calf serum. To measure transcription factor-dependent luciferase activity, we used STAT-luc/U3A cells for STAT3 activity, STAT-luc/2FTGH cells for STAT1 activity, NCAM2-luc/T47D for STAT5 activity, and NF-κB-luc/293 for nuclear factor-κB (NF-κB) activity.10 Bone marrow mononuclear cells from patients with untreated CML were obtained through a Dana-Farber Cancer Institute Institutional Review Board-approved protocol for which patients gave written informed consent in accordance with the Declaration of Helsinki. Peripheral blood mononuclear cells from healthy platelet donors were obtained through an Institutional Review Board-approved protocol for which donors gave written informed consent, and were isolated by Ficoll-Hypaque density sedimentation. Primary cells were cultured in RPMI media containing 10% fetal calf serum.
Colony formation assays
CD34+ hematopoietic cells were isolated from bone marrow samples using the CD34 MicroBead Kit according to the manufacturer's instructions (Miltenyi Biotec). Four days after thawing and 2 days after addition of drug, 10 000 cultured CD34+ hematopoietic cells were plated in methylcellulose containing erythropoietin, stem cell factor, granulocyte-macrophage colony-stimulating factor, and interleukin-3 (H4434; StemCell Technologies). Colony formation was assessed after 12 to 14 days of culture. Evaluation of colonies was performed by a person blinded to the experimental conditions.
Compounds
Pimozide was obtained from Sigma-Aldrich. Imatinib and nilotinib were from Novartis Pharma AG. Jak Inhibitor 1 and U0126 were obtained from Calbiochem. All drugs were dissolved in dimethyl sulfoxide and were diluted to a final concentration of 0.1% dimethyl sulfoxide in all experiments.
Antibodies
Antibodies recognizing STAT5 (sc-835), MCL1 (sc-819), and ABL1 (sc-887) were obtained from Santa Cruz Biotechnology. Antibodies to phosphotyrosine (9411 and 9416), phospho-STAT5 (9359), phospho-ABL1 (5300), and the PathScan BCR/ABL Activity Assay antibody cocktail (7130) were obtained from Cell Signaling Technology. Anti-actin (A5316) was obtained from Sigma-Aldrich.
Immunoblotting and immunoprecipitation
Immunoblots and immunoprecipitations were performed as described using the appropriate antibodies.10 Where indicated, band intensity was quantitated using ImageJ software (National Institutes of Health). Each immunoblot shown is representative of a minimum of 2 experiments.
Quantitation of viable cell number
Viable cells were measured by adenosine triphosphate (ATP)–dependent bioluminescence using the CellTiter-Glo assay (Promega).
Quantitative RT-PCR
RNA was harvested using an RNeasy Mini Kit from QIAGEN. cDNA was generated using the SuperScript First Stand Synthesis kit (Invitrogen) or the TaqMan Reverse Transcription kit (Applied Biosystems), and quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed using primers as described.13 Data are expressed as mean fold change ± SE of 3 replicates.
Statistical analysis
The combinatorial index14 to measure the effects of drug combinations on the number of viable cells was calculated using CalcuSyn software (Conservion).
Cell-cycle and apoptosis assays
Cell suspensions were washed twice with phosphate-buffered saline. Staining and counterstaining with annexin-V–fluorescein isothiocyanate and propidium iodide, respectively, was done using the annexin-V–FLUOS Staining Kit (Roche Applied Science) following the manufacturer's instructions. Samples were analyzed on the BD FACSCanto II Flow Cytometer (BD Biosciences) using negative and single-color controls to adjust compensation. Apoptotic cells were quantified and differentiated from viable and necrotic cells using FACSDiva Version 6.1 software (BD Biosciences). For cell-cycle analysis, cells were treated for 48 hours with vehicle or pimozide, after which they were washed twice with cold phosphate-buffered saline. Washed cells were resuspended in 0.5 mL of propidium iodide staining solution (50 μg/mL propidium iodide, 0.1% NP-40, 0.1% sodium citrate). After a 15-minute incubation on ice, the cells were analyzed by flow cytometry. Cell–cycle distribution was determined using the Modfit LT Version 3.2 software.
In vitro kinase assay
Kinase inhibitory activity of pimozide was analyzed using the SelectScreen Kinase Profiling service (Invitrogen).
Results
Identification of pimozide as a STAT5 inhibitor
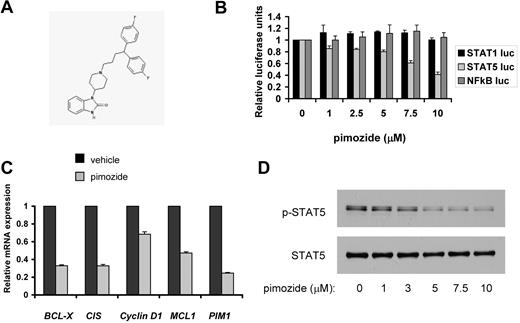
To identify inhibitors of STAT5, we performed a high throughput screen based on STAT transcriptional activity. Cells stably transfected with a STAT-responsive element driving expression of a luciferase reporter gene were used to interrogate the Prestwick collection, which consists of 1120 clinically used drugs and other known bioactives.10 Active agents were confirmed as specific STAT5 inhibitors by analyzing the inhibitory activity of these compounds on STAT5-dependent luciferase activity compared with NF-κB–dependent transcriptional activity. This NF-κB counterscreen was performed to eliminate compounds that showed nonspecific effects or were generally cytotoxic. From this approach, the neuroleptic drug pimozide (Figure 1A) was found to inhibit STAT5-dependent reporter gene expression (Figure 1B). Pimozide had negligible effects on transcriptional activity dependent on NF-κB or the homologous STAT family member STAT1, suggesting that pimozide does not inhibit transcription factors broadly, and is not generally cytotoxic (Figure 1B).
Pimozide inhibits STAT5 activity. (A) Chemical structure of pimozide. (B) Reporter cell lines were treated with the indicated doses of pimozide for 2 hours, after which cytokines were added to activate the appropriate transcription factor. Luciferase activity was quantitated by luminometry 6 hours later. (C) KU812 cells were treated with vehicle or pimozide (5μM) for 18 hours, after which RNA was harvested, and expression of the indicated genes was measured using quantitative RT-PCR and normalized to the expression of β-actin. (D) KU812 cells were treated with the indicated concentrations of pimozide for 3 hours. Immunoblots were performed using antibodies specific for tyrosine phosphorylated STAT5 and total STAT5.
Pimozide inhibits STAT5 activity. (A) Chemical structure of pimozide. (B) Reporter cell lines were treated with the indicated doses of pimozide for 2 hours, after which cytokines were added to activate the appropriate transcription factor. Luciferase activity was quantitated by luminometry 6 hours later. (C) KU812 cells were treated with vehicle or pimozide (5μM) for 18 hours, after which RNA was harvested, and expression of the indicated genes was measured using quantitative RT-PCR and normalized to the expression of β-actin. (D) KU812 cells were treated with the indicated concentrations of pimozide for 3 hours. Immunoblots were performed using antibodies specific for tyrosine phosphorylated STAT5 and total STAT5.
Pimozide reduces the expression of endogenous STAT5 target genes
Having shown that pimozide inhibits the expression of a STAT5-dependent reporter gene (Figure 1B), we next analyzed the expression of endogenous STAT5 target genes. KU812 and K562 CML cells, which display constitutive STAT5 activation, were treated with pimozide or vehicle for 18 hours, after which RNA was harvested. Quantitative RT-PCR revealed a decrease in expression of 5 key STAT5 target genes, including antiapoptotic and progrowth genes (Figure 1C; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Therefore, pimozide treatment leads to the loss of expression of endogenous STAT5 target genes that can promote malignant cellular behavior.
Pimozide decreases STAT5 tyrosine phosphorylation but is not a tyrosine kinase inhibitor
To understand the mechanism by which pimozide inhibits STAT5 activity, KU812 and K562 CML cells were treated with pimozide for 3 hours over a range of doses, after which cells were harvested and STAT5 phosphorylation was determined. Both cell lines showed a dose-dependent loss of the activating STAT5 tyrosine phosphorylation (Figure 1D; supplemental Figure 2). This was reflected in a decrease in phosphorylated (and total) STAT5 within the nucleus (supplemental Figure 3). We next asked whether pimozide affects other STATs in CML cells. We did not detect any constitutive phosphorylation of STAT1 or STAT3 in K562 or KU812 cell lines (data not shown); therefore, we analyzed the effect of pimozide on cytokine-induced STAT1 and STAT3 activation. We found that, in K562 cells, interferon-α can induce the phosphorylation of STAT1 and LIF can induce the phosphorylation of STAT3. Pretreatment of K562 cells with pimozide followed by cytokine stimulation resulted in negligible effects on STAT1 or STAT3 phosphorylation while completely abrogating the constitutive STAT5 phosphorylation (supplemental Figure 4). These results indicate that pimozide has specificity as an inhibitor of STAT5 in CML cells.
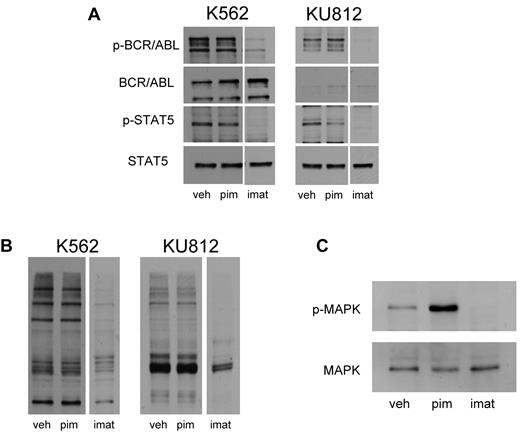
Because STAT5 phosphorylation was reduced, we considered the possibility that pimozide might be an inhibitor of BCR/ABL. We took several approaches to evaluate this possibility. First, activation of BCR/ABL kinase activity is reflected by autophosphorylation of this protein. To determine whether pimozide affects this autophosphorylation, we treated KU812 and K562 cells with either the kinase inhibitor imatinib or pimozide, and we evaluated the phosphorylation status of this protein. Imatinib reduced the phosphorylation of both STAT5 and BCR/ABL. However, whereas pimozide reduced STAT5 phosphorylation by 35% to 55% in these cell lines, BCR/ABL phosphorylation was largely unaffected (Figure 2A). Thus, although both imatinib and pimozide reduce the phosphorylation of STAT5, only imatinib consistently reduces BCR/ABL phosphorylation, suggesting that pimozide does not function as a BCR/ABL kinase inhibitor (Figure 2A).
Pimozide is not a direct inhibitor of BCR/ABL. (A) K562 and KU812 cells were treated with pimozide (10μM) or imatinib (1μM) for 3 hours, after which immunoblot analysis was performed using antibodies to the phosphorylated forms of ABL1 and STAT5, as well as total ABL1 and STAT5. (B) K562 and KU812 cells were treated with pimozide (10μM) or imatinib (1μM) for 3 hours, after which immunoblot analysis was performed using a pan-antiphospho-tyrosine antibody. (C) K562 cells were treated with pimozide and imatinib for 3 hours, and MAPK activation was measured by immunoblot analysis using an antibody to phosphorylated MAPK and a total MAPK antibody.
Pimozide is not a direct inhibitor of BCR/ABL. (A) K562 and KU812 cells were treated with pimozide (10μM) or imatinib (1μM) for 3 hours, after which immunoblot analysis was performed using antibodies to the phosphorylated forms of ABL1 and STAT5, as well as total ABL1 and STAT5. (B) K562 and KU812 cells were treated with pimozide (10μM) or imatinib (1μM) for 3 hours, after which immunoblot analysis was performed using a pan-antiphospho-tyrosine antibody. (C) K562 cells were treated with pimozide and imatinib for 3 hours, and MAPK activation was measured by immunoblot analysis using an antibody to phosphorylated MAPK and a total MAPK antibody.
Second, BCR/ABL phosphorylates multiple substrates on tyrosine residues in CML cells. Treatment of these cells with imatinib led to a loss of phosphorylation not only of STAT5, but of these other substrates as well (Figure 2B). However, at a dose that caused similar inhibition of STAT5 phosphorylation, pimozide did not decrease the phosphorylation of other proteins. One BCR/ABL substrate is Crkl.15 Imatinib treatment for 16 hours led to a 30% to 50% decrease in Crkl phosphorylation in KU812 and K562 cells. However, pimozide treatment had no significant effect on Crkl phosphorylation (supplemental Figure 5). These data further suggest that pimozide is not acting as a generalized inhibitor of BCR/ABL kinase activity.
We next examined another downstream mediator of BCR/ABL, MAP kinase (MAPK). Because KU812 cells have a relatively low level of MAPK activation, we focused our analysis on K562 cells. As expected, imatinib completely inhibited the phosphorylation of MAPK in these cells; however, after treatment with pimozide, the phosphorylation of MAPK increased, possibly because of decreased expression of negative feedback regulators (Figure 2C). Taken together, these data demonstrate that the mechanism of action of pimozide is distinct from that of imatinib. Both compounds decrease STAT5 tyrosine phosphorylation. However, whereas imatinib reduces the function of BCR/ABL by directly inhibiting its kinase activity, pimozide does not inhibit BCR/ABL autophosphorylation, the tyrosine phosphorylation of other cellular substrates, or other downstream effectors of this kinase.
We also considered the possibility that other kinases, in addition to BCR/ABL, may be mediators of STAT5 activation in these cells. We first focused on JAK2 given its importance in cytokine signaling and in other myeloproliferative disorders. However, we did not detect any phosphorylated JAK2 in either KU812 or K562 cells, nor was there any loss of viability of either cell type when JAK2 was inhibited (data not shown). This is consistent with results from other investigators suggesting that JAK2 is not critical for STAT5 signaling in cell lines containing BCR/ABL.16,17 In contrast to JAK2, the SRC family of kinases has been implicated in BCR/ABL signaling in CML cell lines.18 To determine whether pimozide is directly inhibiting SRC family members, we used an in vitro kinase assay to determine its effects on SRC, LYN, and HCK, as well as ABL1. Pimozide, at a dose that significantly inhibited STAT5 phosphorylation, showed minimal inhibition of SRC activity (7% inhibition) but had no effect on the other kinases tested (Table 1). We conclude that pimozide is not a kinase inhibitor; therefore, the mechanism of action of this drug is distinct from that of ABL1 inhibitors, such as imatinib.
Pimozide does not directly inhibit tyrosine kinase activity
| Kinase . | % inhibition of kinase activity . | |
|---|---|---|
| Pimozide . | Imatinib . | |
| ABL1 | < 5 | 40 |
| ABL1 T315I | < 5 | < 5 |
| HCK | < 5 | ND |
| LYN A | < 5 | ND |
| LYN B | < 5 | ND |
| SRC | 7 | ND |
| Kinase . | % inhibition of kinase activity . | |
|---|---|---|
| Pimozide . | Imatinib . | |
| ABL1 | < 5 | 40 |
| ABL1 T315I | < 5 | < 5 |
| HCK | < 5 | ND |
| LYN A | < 5 | ND |
| LYN B | < 5 | ND |
| SRC | 7 | ND |
The effect of pimozide on the activity of selected tyrosine kinases was analyzed using the SelectScreen Kinase Profiling service (Invitrogen).
ND indicates not determined.
Finally, if pimozide was targeting STAT5 specifically, then it might be expected to also inhibit constitutively active mutants of STAT5. Indeed, we found that pimozide decreased gene transcription driven by the STAT5a1*6 mutant,19 and decreased survival of Ba/F3 cells dependent on expression of this form of STAT5 (supplemental Figure 6), further supporting the role of pimozide as a STAT5 inhibitor.
Pimozide induces apoptosis and cell cycle arrest in CML cells
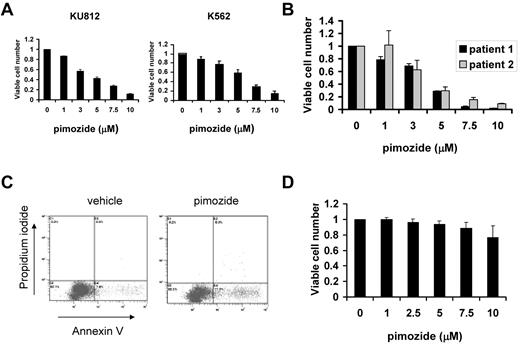
Given that pimozide effectively decreases STAT5 function, we next analyzed the effect of this drug on the viability of cell lines derived from patients with CML, which are dependent on STAT5 for survival. After 48 hours, pimozide caused a dose-dependent decrease in the number of viable cells of both KU812 and K562 cells (Figure 3A), similar to its dose-dependent inhibition of STAT5 phosphorylation. We then determined whether primary CML cells were also sensitive to pimozide. Unfractionated bone marrow cells harvested from patients with untreated CML were exposed to pimozide. As with the cell lines, CML cells from patients showed a dose-dependent decrease in the number of viable cells (Figure 3B). To determine the mechanism for this decrease in viable cell number, we measured the induction of apoptosis as assessed by annexin V staining. Treatment of KU812 cells with pimozide led to a 2- to 3-fold increase in apoptotic cells after 48 hours (Figure 3C). Thus, pimozide decreases viability of CML cells through induction of apoptosis. To exclude the possibility that pimozide was nonspecifically cytotoxic, we harvested peripheral blood mononuclear cells from 7 healthy persons and exposed them to pimozide in vitro. At concentrations up to 10μM, pimozide had little effect on cell survival (Figure 3D).
Pimozide induces apoptosis of CML cells. (A) KU812 and K562 cells were treated with the indicated concentrations of pimozide for 48 hours, after which viable cell number was measured using an ATP-dependent bioluminescence assay. Data are representative of 3 independent experiments. (B) Unfractionated bone marrow mononuclear cells harvested from 2 untreated CML patients were treated with pimozide at the indicated concentrations for 48 hours, and viable cell number was determined. (C) KU812 cells were treated with vehicle or pimozide (5μM) for 48 hours, after which annexin V/propidium iodide staining and flow cytometry were performed. Among vehicle-treated cells, 7.6% showed annexin V staining, whereas among pimozide-treated cells 17.3% were annexin V positive. (D) Peripheral blood mononuclear cells harvested from 7 healthy persons were treated with the indicated concentrations of pimozide for 48 hours, after which viable cell number was measured using an ATP-dependent bioluminescence assay. Data are mean ± SEM.
Pimozide induces apoptosis of CML cells. (A) KU812 and K562 cells were treated with the indicated concentrations of pimozide for 48 hours, after which viable cell number was measured using an ATP-dependent bioluminescence assay. Data are representative of 3 independent experiments. (B) Unfractionated bone marrow mononuclear cells harvested from 2 untreated CML patients were treated with pimozide at the indicated concentrations for 48 hours, and viable cell number was determined. (C) KU812 cells were treated with vehicle or pimozide (5μM) for 48 hours, after which annexin V/propidium iodide staining and flow cytometry were performed. Among vehicle-treated cells, 7.6% showed annexin V staining, whereas among pimozide-treated cells 17.3% were annexin V positive. (D) Peripheral blood mononuclear cells harvested from 7 healthy persons were treated with the indicated concentrations of pimozide for 48 hours, after which viable cell number was measured using an ATP-dependent bioluminescence assay. Data are mean ± SEM.
The loss of viable cell number, as measured by an ATP-dependent bioluminescence assay (Figure 3A), is associated with an increase in apoptosis (Figure 3C). However, because STAT signaling is also involved in proliferation,5 we considered the possibility that pimozide may also cause cell cycle arrest. We examined the cell-cycle distribution of KU812 cells treated with 5μM pimozide for 48 hours. There was a decrease in cells in S phase from 38% to 23% and an increase in cells in G0/G1 from 46% to 72% (supplemental Figure 7). Therefore, the decrease in viable cell number in pimozide-treated cells reflects both cell-cycle arrest and increased apoptosis.
Pimozide inhibits colony formation of primary CD34+ cells from CML patients
Given that CML probably arises from a pluripotent hematopoietic stem cell, we next determined the effect of pimozide on the growth and differentiation of primary human CD34+ cells. CD34+ cells were isolated from the bone marrow of healthy donors and from 2 CML patients. Neither of the patients had been treated with a kinase inhibitor, and 100% of their metaphases showed a 9;22 translocation. The CD34+ cells were treated with 5μM pimozide for 2 days, after which they were plated in methylcellulose containing cytokines, and the resulting colonies were counted. Although pimozide caused a slight reduction in colony formation of normal cells, colony formation was completely abolished in pimozide-treated CML cells (Table 2). Therefore, pimozide selectively inhibits the viability and differentiation of CD34+ CML cells.
The effect of pimozide on myeloid colony formation of CD34+ cells from CML patients and healthy donors
| CD34 source/treatment . | CFU-E . | BFU-E . | CFU-GM . | CFU-GEMM . |
|---|---|---|---|---|
| Healthy donors | ||||
| Vehicle | 59 ± 10 | 139 ± 60 | 36 ± 12 | 12 ± 7 |
| Pimozide | 57 ± 8 | 107 ± 16 | 21 ± 10 | 12 ± 7 |
| CML patients | ||||
| Vehicle | 60 ± 12 | 17 ± 13 | 12 ± 14 | 0 |
| Pimozide | 0 | 0 | 0 | 0 |
| CD34 source/treatment . | CFU-E . | BFU-E . | CFU-GM . | CFU-GEMM . |
|---|---|---|---|---|
| Healthy donors | ||||
| Vehicle | 59 ± 10 | 139 ± 60 | 36 ± 12 | 12 ± 7 |
| Pimozide | 57 ± 8 | 107 ± 16 | 21 ± 10 | 12 ± 7 |
| CML patients | ||||
| Vehicle | 60 ± 12 | 17 ± 13 | 12 ± 14 | 0 |
| Pimozide | 0 | 0 | 0 | 0 |
Cells were treated with vehicle or pimozide (5μM); then 10 000 cells were plated in triplicate and the number of each colony type was counted. Data are number of colonies (mean ± SD).
CFU-E indicates colony-forming unit-erythroid; BFU-E, burst-forming unit-erythroid; CFU-GM, colony-forming unit-granulocyte-macrophage; and CFU-GEMM, colony-forming unit-granulocyte, erythroid, macrophage, megakaryocyte.
Pimozide synergizes with imatinib and nilotinib to decrease STAT5 phosphorylation and induce apoptosis
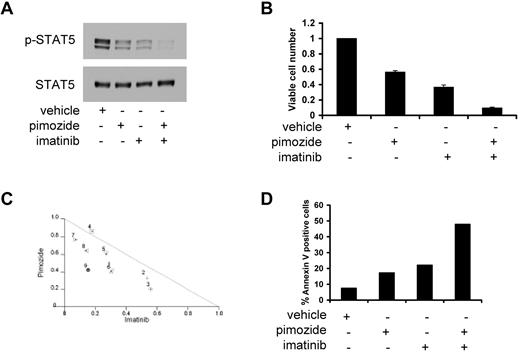
Because kinase inhibitors, such as imatinib and nilotinib, inhibit STAT5 activation by a mechanism distinct from pimozide, we considered the possibility that a combination of these approaches might provide enhanced activity. Treatment of KU812 cells with either pimozide or imatinib resulted in a reduction in STAT5 phosphorylation of approximately 60%. However, when these 2 drugs were combined, STAT5 phosphorylation was inhibited by greater than 90% (Figure 4A). Similar results were seen when pimozide was combined with nilotinib, a newer generation BCR/ABL inhibitor11 that was designed based on the structure of imatinib (supplemental Figure 8A). Therefore, pimozide combined with either imatinib or nilotinib shows enhanced inhibition of STAT5 phosphorylation.
The combination of pimozide and imatinib leads to enhanced effects on STAT5 inhibition and apoptosis. (A) KU812 cells were treated with pimozide (5μM), imatinib (50nM), or both for 3 hours, after which immunoblot analysis was performed for phospho-STAT5 and total STAT5. (B) KU812 cells were treated with pimozide (5μM), imatinib (50nM), or both for 48 hours, after which viable cell number was measured using an ATP-dependent bioluminescence assay. (C) Isobologram analysis was performed based on change in viable cell number for 9 different combinations of pimozide and imatinib. Data points below the line indicate superadditive effects. (D) KU812 cells were treated with pimozide (5μM), imatinib (50nM), or both for 48 hours, after which flow cytometric analysis was performed. The percentage of cells staining with annexin V is shown. Data are representative of 3 independent experiments.
The combination of pimozide and imatinib leads to enhanced effects on STAT5 inhibition and apoptosis. (A) KU812 cells were treated with pimozide (5μM), imatinib (50nM), or both for 3 hours, after which immunoblot analysis was performed for phospho-STAT5 and total STAT5. (B) KU812 cells were treated with pimozide (5μM), imatinib (50nM), or both for 48 hours, after which viable cell number was measured using an ATP-dependent bioluminescence assay. (C) Isobologram analysis was performed based on change in viable cell number for 9 different combinations of pimozide and imatinib. Data points below the line indicate superadditive effects. (D) KU812 cells were treated with pimozide (5μM), imatinib (50nM), or both for 48 hours, after which flow cytometric analysis was performed. The percentage of cells staining with annexin V is shown. Data are representative of 3 independent experiments.
Given that pimozide and kinase inhibitors inhibit STAT5 phosphorylation by distinct mechanisms, we reasoned that they might also show an enhanced effect on inhibiting cell survival when used in combination. When pimozide is combined with imatinib, there is a greater loss in the number of viable cells than when either drug is used alone (Figure 4B). Isobologram analysis (Figure 4C) and analysis of the Chou-Talalay combination index14 (data not shown) demonstrate synergy with these combinations. We also tested whether pimozide and nilotinib had a greater effect on viable cell number when cells were treated with both drugs together. This combination resulted in a greater decrease in viable cells compared with either drug alone, with most combinations resulting in synergy (supplemental Figure 8B-C).
Because pimozide alone can induce apoptosis in CML cells, we analyzed the effect of the combination of pimozide and a kinase inhibitor on the induction of apoptosis. Treatment of KU812 cells for 48 hours with either pimozide or imatinib resulted in an approximately 2.5-fold increase in apoptosis over baseline as measured by annexin V staining (Figure 4D). The combination of these 2 drugs had a much larger effect on apoptosis, with a greater than 6-fold induction over untreated cells. Even more striking synergy was seen with nilotinib. Although nilotinib induced apoptosis approximately 3-fold over vehicle-treated cells, the combination of pimozide and nilotinib induced an approximately 9-fold increase in apoptosis (supplemental Figure 8D). A similar effect was seen with the combination of pimozide and imatinib in K562 cells (supplemental Figure 9). Therefore, the combination of pimozide with either imatinib or nilotinib results in a synergistic decrease in viable cell number, at least in part through enhanced apoptosis.
MAPK inhibition enhances the effect of pimozide on viable cell number
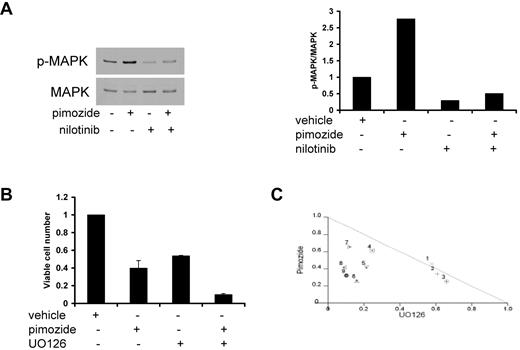
Given that activated MAPK confers prosurvival signals, it is possible that the increase in phosphorylated MAPK on pimozide treatment may counteract the effect of decreased STAT5 activation on cellular viability. Therefore, inhibiting MAPK concurrent with pimozide treatment in CML cells may enhance cell killing compared with pimozide alone. To test this hypothesis, we first analyzed phosphorylation of MAPK when pimozide and nilotinib were combined. Phosphorylated MAPK was reduced after either nilotinib treatment or combination treatment with nilotinib and pimozide (Figure 5A), demonstrating that the enhancement of MAPK activation seen with pimozide can be counteracted by nilotinib. We next analyzed viable cell number in K562 cells treated with pimozide and the MEK inhibitor U0126, which blocks MAPK activation. The combination of these 2 drugs resulted in a greater inhibition of viable cell number compared with either drug alone (Figure 5B), and isobologram analysis demonstrated that this combination is synergistic (Figure 5C). In addition, analysis of the Chou-Talalay combination index,14 showed that most of the drug combinations are synergistic (data not shown). Therefore, the ability of pimozide to decrease viable cell number is enhanced by MAPK inhibition.
Inhibition of MAPK enhances the effects of pimozide on cell viability. (A) K562 cells were treated with pimozide (5μM), nilotinib (3nM), or both for 3 hours, and immunoblots were performed to phospho-MAPK and total MAPK (left); phospho-MAPK/total MAPK band intensity is shown in the right panel. (B) K562 cells were treated with pimozide (7.5μM), the MEK inhibitor U0126 (10μM), or both for 48 hours, after which viable cell number was quantitated by ATP-dependent bioluminescence. (C) Isobologram analysis was performed based on loss of cell viability for 9 different combinations of pimozide and UO126. Data points below the line indicate superadditive effects.
Inhibition of MAPK enhances the effects of pimozide on cell viability. (A) K562 cells were treated with pimozide (5μM), nilotinib (3nM), or both for 3 hours, and immunoblots were performed to phospho-MAPK and total MAPK (left); phospho-MAPK/total MAPK band intensity is shown in the right panel. (B) K562 cells were treated with pimozide (7.5μM), the MEK inhibitor U0126 (10μM), or both for 48 hours, after which viable cell number was quantitated by ATP-dependent bioluminescence. (C) Isobologram analysis was performed based on loss of cell viability for 9 different combinations of pimozide and UO126. Data points below the line indicate superadditive effects.
Pimozide is active against imatinib-resistant forms of BCR/ABL
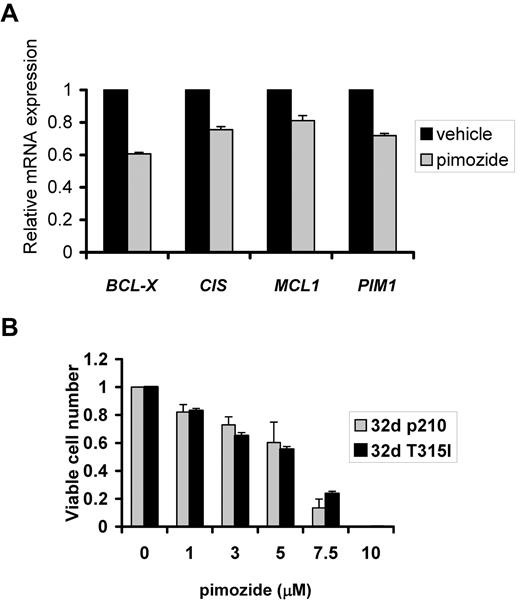
Because pimozide does not appear to target BCR/ABL directly and appears to be affecting STAT5 downstream of this kinase, we reasoned that pimozide might be effective against cells containing the imatinib-resistant T315I BCR/ABL mutation. Having shown that pimozide causes a significant reduction in expression of key STAT5 target genes in cells containing wild-type BCR/ABL (Figure 1C), we treated 32d cells reconstituted with the T315I BCR/ABL mutation with pimozide for 3 hours and harvested RNA. Quantitative RT-PCR analysis revealed a reduction in expression of STAT5 target genes by 20% to 40% (Figure 6A). We next evaluated the effect of pimozide on the viability of 32d cells transformed by wild-type BCR/ABL or the mutant T315I form. As predicted by its targeting of STAT5, pimozide was similarly effective at reducing viable cell number in isogenic cells transformed with either imatinib-sensitive or -resistant forms of BCR/ABL (Figure 6B). This effect is probably mediated through its inhibition of STAT5, as pimozide showed no direct inhibition of the kinase activity of either form of BCR/ABL (Table 1).
Pimozide is similarly effective against cells with unmutated BCR/ABL and kinase inhibitor-resistant T315I mutant BCR/ABL. (A) The 32d cells reconstituted with the T315I mutant form of BCR/ABL were treated for 3 hours with pimozide (5μM), after which RNA was harvested, and expression of the indicated genes was measured using quantitative RT-PCR and normalized to the expression of β-actin. (B) The 32d cells reconstituted with unmutated BCR/ABL (p210) or BCR/ABL with the kinase inhibitor-resistant T315I mutation were treated with pimozide at the indicated concentration for 48 hours after which viable cell number was measured by ATP dependent bioluminescence.
Pimozide is similarly effective against cells with unmutated BCR/ABL and kinase inhibitor-resistant T315I mutant BCR/ABL. (A) The 32d cells reconstituted with the T315I mutant form of BCR/ABL were treated for 3 hours with pimozide (5μM), after which RNA was harvested, and expression of the indicated genes was measured using quantitative RT-PCR and normalized to the expression of β-actin. (B) The 32d cells reconstituted with unmutated BCR/ABL (p210) or BCR/ABL with the kinase inhibitor-resistant T315I mutation were treated with pimozide at the indicated concentration for 48 hours after which viable cell number was measured by ATP dependent bioluminescence.
Discussion
STAT transcription factors are essential for the pathogenesis of many tumors by directly regulating the expression of genes that induce or maintain cancer cell growth and survival. STATs are attractive targets for cancer therapy because they sit at a convergence point of numerous tyrosine kinases. Because continued STAT activation is probably crucial for tumor survival, there is strong selective pressure to maintain STAT activation when an activated tyrosine kinase is inhibited. This can be achieved either by the acquisition of mutations rendering the inhibitor ineffective or by activating a kinase distinct from the one being inhibited.20-23 Thus, targeting STATs holds the promise of being useful against a wide spectrum of tumors, and for decreasing the emergence of resistance to kinase inhibitors.
Although STATs may be critical for tumor pathogenesis, they are largely dispensable for physiologic cellular function, probably because of redundancies in normal signaling. For example, activation of STAT3 is a common pathogenic event in a range of human tumors.2 However, the hyper-IgE syndrome, which is compatible with normal development in humans, is probably mediated by a dominant inhibitory form of STAT3 that is present since conception.24 Similarly, loss of STAT5 function may have limited consequences for hematopoiesis and other aspects of tissue homeostasis in adult animals.25,26 In particular, targeted STAT5 deletion in adult mice results in healthy animals with minimal defects in hematopoiesis, including fewer CD19+ cells and a slight impairment in erythroid differentiation.5 These studies suggest that STAT5 is largely dispensable in adult mice. Thus, targeting STATs has the potential for a very high therapeutic index in treating cancers driven by constitutive activation of these proteins.
Transcription factors have traditionally been thought to be suboptimal pharmacologic targets because their function relies on protein-protein and protein-DNA interactions that might not easily be disrupted by small molecules. Through a chemical biology approach focused on compounds already known to be safe in humans, we identified pimozide as a STAT5 inhibitor with significant activity in models of CML. Pimozide is Food and Drug Administration-approved in the United States for the treatment of Tourette syndrome and appears to work by blocking the D2 dopamine receptor.27 As with other dopamine antagonists, it can cause central nervous system toxicity and prolongation of cardiac repolarization. However, hematologic toxicity ascribable to pimozide has not been reported, consistent with our findings of minimal effects of this drug on CD34+ cells or peripheral blood mononuclear cells isolated from healthy donors (Table 2; Figure 3D).
Because STAT5 signaling is a critical mediator of the phenotype of CML,12,28,29 we used this leukemia to characterize the molecular and cellular effects of pimozide. Present therapy for CML includes the kinase inhibitors imatinib, nilotinib, and dasatinib,30 which lead to the reduction of BCR/ABL tyrosine kinase activity and a reduction in STAT5 activation. However, resistance can develop through mutations in BCR/ABL, which render the kinase refractory to inhibition. Therefore, targeting a downstream mediator such as STAT5 can be highly beneficial, and evidence from both RNA interference-based approaches31 and conditional deletions5 has provided validation for STAT5 as an excellent target in CML cells but not normal cells. In addition, a drug directly targeting STAT5 may be particularly useful in combination with a kinase inhibitor. The benefit of this multidrug approach probably arises through the enhanced inhibition of STAT5 activity (Figure 4A; supplemental Figures 7A, 8A). Such a combination may decrease the emergence of drug-resistant cells and may allow the use of less toxic doses of each drug. Finally, as shown by the inhibitory effects of pimozide on colony formation from primary CML cells (Table 2), a STAT5 inhibitor may be more effective at targeting the putative CML stem cell population.5,31,32
Like the kinase inhibitors, pimozide inhibits the activating tyrosine phosphorylation of STAT5. However, several lines of evidence suggest that pimozide is not functioning as a kinase inhibitor. Pimozide does not decrease the autophosphorylation of BCR/ABL, nor does it decrease the tyrosine phosphorylation of other cellular proteins mediated by BCR/ABL. In addition, pimozide does not decrease the activation of a distinct pathway downstream of BCR/ABL, MAP kinase. Finally, in vitro kinase assays showed no effect of pimozide on a spectrum of tyrosine kinases, including wild-type and mutant forms of BCR/ABL (Table 1). Thus, although pimozide functions as a STAT5 inhibitor through its ability to inhibit STAT5 tyrosine phosphorylation, it clearly does not act as a classic kinase inhibitor.
It is doubtful that the STAT5-inhibitory effect of pimozide is the result of inhibition of dopamine receptors, as the effects on STAT5 occur at a concentration of pimozide greater than that needed to inhibit dopamine signaling. Furthermore, other antipsychotics with dopamine antagonist activity in the Prestwick collection, including chlorpromazine and haloperidol, had no effect on STAT-dependent (or NF-κB-dependent) reporter gene expression in the initial screen (data not shown). It is possible that pimozide is interacting directly with STAT5 to inhibit its phosphorylation. Alternatively, pimozide may be activating a phosphatase to dephosphorylate STAT5, or altering its stability through interactions with chaperone proteins such as HSP90. Current studies are focused on elucidating the manner by which pimozide reduces STAT5 phosphorylation, which may reveal additional molecular strategies to target STAT5 selectively.
The finding that pimozide, in contrast to kinase inhibitors, leads to an increase in MAPK phosphorylation may provide insight into its cellular effects. It is known that STAT5 target genes encode negative regulators of kinases such as CIS.33 Thus, the increased phosphorylation level of BCR/ABL on treatment with pimozide may result from a loss of STAT5-dependent expression of CIS and other negative regulators. Consequently, non–STAT-signaling pathways, such as MAPK, may display greater activity. Because MAPK activation can promote enhanced survival and proliferation, it is not surprising that the combination of pimozide and an inhibitor of the MAP kinase pathway is particularly effective. This is similar to the effect of the AKT inhibitor perifosine, which also activates the MAPK pathway.34 Thus, understanding the role of feedback loops in response to targeted therapy may suggest particularly effective combinations of signal transduction inhibitors.
A major concern in the use of kinase inhibitors is the emergence of mutant forms of BCR/ABL that are resistant to multiple inhibitors. In particular, CML cells containing the T315I BCR/ABL mutation are resistant to all currently approved kinase inhibitors. Notably, however, cells containing this mutation are sensitive to pimozide (Figure 6), probably because pimozide is inhibiting STAT5 activity downstream of BCR/ABL. Furthermore, resistance to a variety of tyrosine kinase inhibitors in leukemias and carcinomas may involve the activation of alternative pathways that activate STATs. Thus, it is probable that pimozide or other STAT inhibitors will be effective at killing cells in which resistance to tyrosine kinase inhibitors has arisen through a variety of mechanisms.
Although pimozide levels in patients treated with this drug for neuropsychiatric disorders are generally lower than 1μM, the finding of the STAT5 inhibitory effect of pimozide may have several important implications. First, it may be possible to achieve the levels necessary for STAT5 inhibition, and only transient exposure to these levels may be sufficient to induce apoptosis. In addition, pimozide can now serve as a scaffold from which to identify compounds that have greater potency and efficacy. Because pimozide appears to decrease STAT5 phosphorylation through a unique mechanism, elucidation of the molecular target of the STAT5-inhibitory effect of pimozide may provide new opportunities for developing antileukemia drugs. Finally, because the efficacy of pimozide correlates with its inhibition of STAT5, it will allow the use of STAT5 phosphorylation, monitored by flow cytometry or immunoblot, as a pharmacodynamic marker of the effect of the drug on peripheral blood or bone marrow samples.
In conclusion, we have identified pimozide as a STAT5 inhibitor that is effective in models of CML. The efficacy of pimozide against resistant mutants of BCR/ABL, and its beneficial effects in combination with other agents, suggests that the strategy of targeting STATs and other oncogenic transcription factors may be highly beneficial.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Cancer Institute and the Initiative for Chemical Genetics (Bethesda, MD) and was assisted by the Chemical Biology Platform of the Broad Institute of Harvard and MIT (Boston, MA). Tissue samples were obtained from the Ted and Eileen Pasquarello Tissue Bank for Hematologic Malignancies (Dana-Farber Cancer Institute). This work was also supported by the Multiple Myeloma Research Foundation (Norwalk, CT), the Kittredge Foundation (Dana-Farber Cancer Institute), the Brent Leahey Fund (Dana-Farber Cancer Institute), Gabrielle's Angel Foundation (New York, NY), and the Claudia Adams Barr Program in Innovative Basic Cancer Research (Dana-Farber Cancer Institute).
National Institutes of Health
Authorship
Contribution: E.A.N. designed the research, performed experiments, and wrote the paper; S.R.W. and E.W. designed the research and performed experiments; M.B.-N., R.B., L.B.G., S.T., J.L.K., L.S., and M.R.A. performed experiments; J.D.G. and B.L.E. designed the research; and D.A.F. designed the research and wrote the paper.
Conflict-of-interest disclosure: J.D.G. has a financial interest with Novartis. The remaining authors declare no competing financial interests.
Correspondence: David A. Frank, Dana-Farber Cancer Institute, Department of Medical Oncology, Mayer 522B, 44 Binney St, Boston, MA 02115; e-mail: david_frank@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal