Abstract
Factor VIII binds to phosphatidylserine (PS)-containing membranes through its tandem, lectin-homology, C1 and C2 domains. However, the details of C1 domain membrane binding have not been delineated. We prepared 4 factor VIII C1 mutations localized to a hypothesized membrane-interactive surface (Arg2090Ala/Gln2091Ala, Lys2092Ala/Phe2093Ala, Gln2042Ala/Tyr2043Ala, and Arg2159Ala). Membrane binding and cofactor activity were measured using membranes with 15% PS, mimicking platelets stimulated by thrombin plus collagen, and 4% PS, mimicking platelets stimulated by thrombin. All mutants had at least 10-fold reduced affinities for membranes of 4% PS, and 3 mutants also had decreased apparent affinity for factor X. Monoclonal antibodies against the C2 domain produced different relative impairment of mutants compared with wild-type factor VIII. Monoclonal antibody ESH4 decreased the Vmax for all mutants but only the apparent membrane affinity for wild-type factor VIII. Monoclonal antibody BO2C11 decreased the Vmax of wild-type factor VIII by 90% but decreased the activity of 3 mutants more than 98%. These results identify a membrane-binding face of the factor VIII C1 domain, indicate an influence of the C1 domain on factor VIII binding to factor X, and indicate that cooperation between the C1 and C2 domains is necessary for full activity of the factor Xase complex.
Introduction
Factor VIII is a protein cofactor in the membrane-bound intrinsic factor Xase complex that cleaves the zymogen factor X to factor Xa.1 Deficiency or malfunction of factor VIII leads to hemophilia A, an X-linked bleeding disorder affecting one in 5000 males.2,3 The factor Xase complex consists of activated factor VIII, the serine protease factor IXa, and a phosphatidylserine (PS)-containing membrane.4 The binding to a PS-containing membrane increases the catalytic efficiency of the factor VIIIa-factor IXa complex approximately 100 000-fold.4 Despite the central importance of the membrane-binding function of factor VIII, the membrane-binding motifs of factor VIII and their functional implications remain poorly understood.
Factor VIII is a plasma glycoprotein containing 2332 amino acids and composed of repeating regions with A1-A2-B-A3-C1-C2 domain structure.5,6 Factor VIII is both structurally and functionally homologous with factor V, another blood coagulation cofactor.7 For both proteins, the 3 A domains share sequence homology and tertiary structure homology with ceruloplasmin,8 the C domains share sequence and tertiary structure homology with each other and with discoidin I as well as lactadherin,9-11 whereas the B domain is unique to each protein.12 Factor VIII circulates in plasma as a heterodimer in a noncovalent complex with von Willebrand factor (VWF).13 VWF binding competitively inhibits factor VIII from binding to PS-containing membranes14,15 and to activated platelets.16 On proteolytic cleavage by thrombin, activated factor VIII is released from VWF and is free to bind to PS-containing membranes where it assembles with factor IXa to form the factor Xase complex. In the factor Xase complex, the association of factor VIIIa with factor IXa and factor X is predominantly mediated by the A domains.17,18 Recent results indicate that the factor VIIIa C2 domain may interact with the Gla domain of factor IXa.19
Prior studies implicate the factor VIII C2 domain as the dominant membrane-binding structure.20-22 The lower end of the C2 domain displays 2 pairs of hydrophobic amino acids (Met 2199/Phe 2200, Leu 2251/Leu 2252) at the tips of protruding β-hairpin turns. Site-directed mutagenesis implicates these residues as playing a major role in membrane binding.23 The importance of the C2 domain in membrane binding is underscored by the fact that some anti–factor VIII antibodies that cause clinical bleeding are directed against the C2 domain and inhibit phospholipid binding.20,22,24,25 Consistent with their ability to block binding to phospholipid, these antibodies were potent inhibitors of factor VIIIa activity,26 underscoring the importance of membrane binding for normal function.
Recent evidence indicates that the C1 domain of factor VIII is also involved in membrane binding. Lipid-free factor VIII crystal structures indicate that the C1 domain is oriented nearly parallel to the C2 domain, positioned favorably for interaction with a membrane.27,28 A role for the C1 domain in platelet binding has recently been described.29 Moreover, we recently confirmed that Lys 2092 and Phe 2093, at the tip of the C1 domain, participate in binding to phospholipid vesicle and platelets.30 Factor VIII with the C2 domain deleted retains a moderately high phospholipid binding affinity and substantial residual activity when the phospholipid concentration is high.31 These considerations motivated studies to explore the participation of additional C1 domain amino acids and the way the corresponding motif(s) cooperate with motifs of the C2 domain for membrane binding.
We hypothesized that a surface on the C1 domain makes contact with the membrane and this membrane-binding motif cooperates with C2 domain motifs. We hypothesized, moreover, that the contribution of some membrane-binding amino acids would not be restricted to enhancing the affinity of factor VIII for membranes but would also alter the manner in which factor VIII interacts with factor IXa and/or factor X. To test the hypothesis, 7 residues encompassing the lower surface of the C1 domain (Figure 1) were changed into alanine, singly or in pairs. In addition, 2 prototype anti-C2 antibodies were chosen to block the membrane-interactive C2 domain epitopes, and the residual apparent membrane affinity and maximum activity of mutants were then evaluated. The results demonstrate that the mutated amino acids identify a membrane-interactive face of the factor VIII C1 domain. Engagement of this face not only affects the affinity of factor VIII for a membrane but also parameters of the factor Xase complex.
Methods
Materials
L-α-PS from bovine brain, L-α-phosphatidylethanolamine (PE) from egg yolk, L-α-phosphatidylcholine (PC) from egg yolk, and 1-oleoyl-2-[12-[(7-nitro-2–1,3-benzoxadiazol-4-yl) amino]lauroyl]-sn-glycero-3-phosphocholine (NBD-PC) were from Avanti Polar Lipids. Human α thrombin, human factor IXa, and human factor X were obtained from Enzyme Research Laboratories.
Human VWF was from Hematologic Technologies. Dulbecco modified Eagle medium and fetal bovine serum was purchased from Invitrogen. COAMATIC was purchased from DiaPharma Group Inc. Plasmid purification kits were purchased from QIAGEN. Superose-12 was obtained from GE Healthcare, and cyanogen bromide in 5M acetonitrile was obtained from Sigma-Aldrich.
Antibodies
Monoclonal antibody (mAb) ESH4 and horseradish peroxidase-labeled monoclonal antibody against human factor VIII:C (ESH-8HP) were obtained from American Diagnostica. Antibody GMA-8021 (anti–factor VIII A2 domain) was from Green Mount Antibodies. Factor VIII-specific human IgG4k monoclonal antibody, BO2C11, was produced by a cell line derived from a hemophilia A patient, as described.25
Preparation of factor VIII mutants
Factor VIII and mutants were constructed as described.23 Briefly, mutagenesis was performed within the mammalian expression vector pMT2. Oligonucleotide-directed mutagenesis was used to create a polymerase chain reaction template in which the codon targeted for substitution was mutated to either GCA or GCG, predicting an amino acid substitution of alanine. After the sequence of the coding regions of all mutants was verified by DNA sequence, plasmid DNA was purified and transfected into COS-1 cells. Conditioned medium was harvested at 64 hours after transfection in the presence of 10% fetal bovine serum. All proteins were purified from transiently transfected COS-1 cells by immunoaffinity chromatography. Proteins were concentrated 10- to 20-fold using Microcon Ultracel YM-100 centrifugal filter devices (Millipore) and stored at −80°C with Tris-buffered saline.
Measurement of specific activity
Specific activity of factor VIII was measured by an activated partial thromboplastin time clotting assay on an MLA Electra 750 fibrinometer by reconstitution of human factor VIII–deficient plasma. Factor VIII antigen was quantified by an antifactor light chain sandwich enzyme-linked immunosorbent assay (ELISA) method using the Asserachrom VIIIC:Ag commercial kit according to the manufacturer's instructions (Diagnostica Stago).
Phospholipid vesicles
Small unilamellar phospholipid vesicles were synthesized by sonicating large multilamellar vesicles in a bath sonicator (Laboratory Supplies) until the suspension was visually clear.32 Large multilamellar vesicles were prepared by evaporating chloroform from the desired phospholipid, resuspending the lipids in methylene chloride, and reevaporating 3 times under argon. Lipids were resuspended as vesicles by gently swirling Tris-buffered saline over the dried lipid.
Factor Xase assay
Factor Xase activity was measured with a 2-step amidolytic substrate assay.4 Small unilamellar vesicles containing 4% or 15% PS were mixed with 10nM factor IXa and 150nM factor X in 0.15M NaCl, 0.2% (weight/volume) bovine serum albumin, 50mM Tris-HCl, pH 7.8. Phospholipid vesicles had compositions of 15% PS and 4% PS with 20% PE and the balance as PC and were added at phospholipid concentrations of 2 and 0.15μM for the phospholipid-saturating and phospholipid-limiting assays, respectively. The reaction was started by adding Ca2+, factor VIII, and thrombin at final concentrations of 1.5mM, 1 unit/mL, and 0.1 unit/mL. After 5 minutes at 25°C, the reaction was stopped by diluting the mixture 1:0.8 with 16mM ethylenediaminetetraacetic acid, and factor Xa activity was determined immediately in a thermostatted kinetic microtiter plate reader (Molecular Devices) at 25°C using 0.1mM S-2765 (DiaPharma Group). Absorbance values were converted into molar concentrations using a standard curve of pure factor Xa. Apparent affinities were determined by fitting the factor Xase activity as a function of the factor X, factor IXa, or maximum velocity (Vmax) to the standard single-site binding model (GraphPad Prism Version 5.0). All values were evaluated by comparison with wild-type factor VIII produced in parallel with the mutants.
Fluorescein labeling of antibody BO2C11
BO2C11 was labeled with fluorescein isothiocyanate using FluoReporter fluorescein isothiocyanate protein labeling kit (Invitrogen) according to the instructions provided by the manufacturer. Protein concentration was determined using a Micro-BCA assay (Pierce Chemical).
Cyanogen bromide activation of Superose and preparation of antibody-coated Superose-12 microspheres
Superose-12 microspheres, 500 μL, were first equilibrated with 1M Na2CO3/NaHCO3 buffer, pH 11. Cyanogen bromide, 500 μL in 5M acetonitrile, was added to the microspheres and incubated for 10 minutes on an ice bath while swirling. The microspheres were extensively washed with distilled H2O, 95% acetone, and 0.2M Na2CO3/NaHCO3 buffer, pH 9.75, in ice bath. Antibody GMA-8021 (300 μL, 1.5 mg/mL) was added to the microsphere slurry and incubated overnight at 4°C on a rocker. The beads were washed twice with 0.2M Na2CO3/NaHCO3, pH 9.75, and once with 1M NaCl, 0.05M Na2CO3/NaHCO3. Nonspecific active sites were blocked by incubating the coated beads with 10 vol of 100mM Tris-HCl, pH 8.5, for 4 hours at room temperature with gentle mixing. The microspheres concentration was counted using a hemocytometer and beads were kept at 4°C until used.
Flow cytometric measurements of phospholipid vesicles binding to factor VIII
Factor VIII wild-type and mutants (the nominal concentration of factor VIII was 0.2nM) were incubated with various concentration of NBD-labeled phospholipid vesicles in 50 μL Tris-buffered saline, 1.5mM Ca2+, 0.2% (weight/volume) bovine serum albumin. The composition of the vesicles was 4% PS, 20% PE, 5% NBD-PC, with the balance as PC. After 30 minutes, aliquots containing approximately 4000 Superose-GMA-8021 beads were added and incubated for 30 minutes with constant vortex mixing at approximately 300 rpm. Samples were diluted to 300 μL in the same buffer within 30 seconds of reading the fluorescence/microsphere by flow cytometry with a BD Biosciences FACSCalibur. Data were acquired with all channels in log mode, and binding data were analyzed as the geometric mean fluorescence/microsphere. Microspheres were detected and distinguished from background noise based on the characteristic forward and side light scatter characteristics. Binding data were analyzed using the equation, F = F0 + Fmax(PL/(KD + PL)) (Eq. 1) where F is the measured fluorescence/Superpose bead, F0 is the fluorescence of free phospholipid vesicles resulting from unbound phospholipid and any nonspecific binding of phospholipid vesicles to control Superose beads lacking factor VIII. Because the fluorescence of Superose beads in the absence of NBD-labeled vesicles was very low, this background fluorescence was neglected. Fmax was the maximum signal from labeled vesicles, and KD is the dissociation constant for phospholipid (PL) binding to factor VIII. The variable KD was determined using nonlinear least squares analysis with the software GraphPad Prism Version 5.0. Fmax was determined by nonlinear least squares analysis for wild-type factor VIII, for which saturable binding was clearly evident. Fmax for each of the mutants was assigned based on the quantity of BO2C11 epitope/Superose-GMA-8021 microsphere relative to wild-type factor VIII.
Normalization of binding data
The quantity of immobilized factor VIII with exposed membrane-binding motif was measured by incubation with fluorescein-labeled BO2C11, a mAb that binds to a major membrane-interactive motif of the C2 domain.26,33 Antibody binding curves were generated as described in “Flow cytometric measurements of phospholipid vesicles binding to factor VIII.” The apparent affinities of BO2C11 for wild-type factor VIII and the mutants were equivalent, as predicted. The maximum fluorescence, with a BO2C11 concentration of 0.4nM, was used as the denominator to normalize binding curves for phospholipid vesicles to the quantity of immobilized factor VIII bound.
Sandwich ELISA
The affinity of factor VIII for VWF was measured in a competition sandwich ELISA. ELISA studies were performed in Nunc-Maxisorp 96-well plates, coated with BO2C11 capture antibody. Coating was achieved by incubating the plates with 50 μL/well of a 2.5-μg/mL BO2C11 antibody solution in carbonate buffer (Na2CO3, 200mM, pH 9.4) overnight at 4°C. The plates were washed 3 times with 100 μL of phosphate-buffered saline (10mM Na2HPO4,1.8mM KH2PO4, 14mM NaCl, 2.7mM KCl, pH 7.4) containing 0.05% Tween 20. The plates were incubated with 200 μL/well of 1% bovine serum albumin in phosphate-buffered saline for 2 hours at room temperature to eliminate nonspecific binding. Factor VIII at 0.4 unit/mL was incubated with various concentrations of VWF for 1 hour at room temperature. The mixtures of factor VIII with VWF were placed in microtiter wells coated with antibody BO2C11. After 1 hour of incubation, the cells were washed thoroughly, and bound factor VIII was detected with biotinylated mAb ESH8, developed with p-nitrophenyl-phosphate reagent (Sigma-Aldrich). The ELISA data were subtracted from a constant value equivalent to the maximum colorimetric intensity to generate upright binding curves. The data from 2 experiments were combined, and the combined curves were fitted by nonlinear, least squares analysis using GraphPad Prism Version 5.0 software.
Results
C1 domain mutations decrease specific activity and apparent affinity for phospholipid vesicles
Seven factor VIII C1 domain amino acids, encompassing a face on the lower half of the module, were changed. The C1 domain mutants (R2090A/Q2091A, K2092A/F2093A, Q2042A/Y2043A, and R2159A) had individual or paired amino acids mutated to alanine, as indicated (Figure 1). For 3 mutants, the amino acids were changed in pairs, rather than individually, to maximize the likelihood that properties would be measurably different from wild-type factor VIII. Mutants were expressed in COS-1 cells, together with wild-type factor VIII, and purified by immunoaffinity chromatography. The specific activities of all 4 mutants were compared with wild-type factor VIII in a commercial activated partial thromboplastin time assay containing excess phospholipid. All activities were diminished, with specific activity ranging from 24% to 61% of wild-type factor VIII (Table 1).
Factor VIII C1 domain mutations and the effects on apparent phospholipid binding affinity. (A) Space-filling display of the C1 and C2 domains of factor VIII (PDB ID: 3CDZ). Dark gray represents the C2 domain; and medium gray, the C1 domain. Mutated residues encompassing the lower surface of the C1 domain are highlighted in pink (Gln 2042/Tyr 2043), red (Arg 2159), blue (Arg 2090/Gln 2091), and green (Lys 2092/Phe 2093). The C2 domain epitopes of mAb ESH4 (magenta) and BO2C11 (cyan) are also shown. (B) Factor VIII mutants had diminished apparent affinity for phospholipid vesicles compared with wild-type factor VIII. Factor VIII wild-type or mutants (0.15 U/mL) were added to various concentrations of vesicles containing 4% or 15% PS, and factor Xase activity was measured as described in “Factor Xase assay.” Data are mean ± SEM for 3 experiments, each performed in duplicate.
Factor VIII C1 domain mutations and the effects on apparent phospholipid binding affinity. (A) Space-filling display of the C1 and C2 domains of factor VIII (PDB ID: 3CDZ). Dark gray represents the C2 domain; and medium gray, the C1 domain. Mutated residues encompassing the lower surface of the C1 domain are highlighted in pink (Gln 2042/Tyr 2043), red (Arg 2159), blue (Arg 2090/Gln 2091), and green (Lys 2092/Phe 2093). The C2 domain epitopes of mAb ESH4 (magenta) and BO2C11 (cyan) are also shown. (B) Factor VIII mutants had diminished apparent affinity for phospholipid vesicles compared with wild-type factor VIII. Factor VIII wild-type or mutants (0.15 U/mL) were added to various concentrations of vesicles containing 4% or 15% PS, and factor Xase activity was measured as described in “Factor Xase assay.” Data are mean ± SEM for 3 experiments, each performed in duplicate.
Specific activity of factor VIII mutants
| Mutant . | Amino acid(s) → alanine . | Relative specific activity* . |
|---|---|---|
| Wild-type | — | 1.00 |
| 2090/2091 | Arg/Gln | 0.61 |
| 2092/2093 | Lys/Phe | 0.42 |
| 2159 | Arg | 0.42 |
| 2042/2043 | Gln/Tyr | 0.24 |
| Mutant . | Amino acid(s) → alanine . | Relative specific activity* . |
|---|---|---|
| Wild-type | — | 1.00 |
| 2090/2091 | Arg/Gln | 0.61 |
| 2092/2093 | Lys/Phe | 0.42 |
| 2159 | Arg | 0.42 |
| 2042/2043 | Gln/Tyr | 0.24 |
— indicates not applicable.
Factor VIII activity was measured in an activated partial thromboplastin time assay with factor VIII-deficient plasma.
The concentration of phospholipid vesicles in a factor Xase assay was varied to determine the apparent affinities of the mutants for these vesicles (Figure 1B). The PS content was either 4% PS, simulating thrombin-stimulated platelets, or 15% PS, simulating platelets stimulated with thrombin plus collagen.34-36 All mutants had decreased apparent affinity for vesicles with 4% PS (Figure 1B closed bars). The reduction in affinity ranged from 33-fold for R2090A/Q2091A to 5-fold for Q2042A/Y2043A. Apparent affinities for phospholipid vesicles with 15% PS were reduced to a lesser degree with a range of approximately 8-fold for K2092A/F2093Ato 2-fold for R2159A. These results indicate that the face containing the amino acids that were selected for study participates in membrane binding and that the importance of this face is greater on membranes with lower PS content.
C1 domain amino acids participate in membrane binding
We have used a novel assay to determine the binding of C1 domain mutants to phospholipid membrane (Figure 2; Table 2). The assay is similar to an assay we previously described for measuring the affinity of vesicles for factor VIII mutants.23 Factor VIII mutants were first incubated with various concentrations of sonicated phospholipid vesicles of PS/PE/PC/NBD-PC (4:20:71:5) and then captured on mAb GMA-8021, against the A2 domain, coupled to Superose beads. Quasi-equilibrium binding of factor VIII-phospholipid vesicle complexes to the anti–factor VIII antibody on the Superose beads was evaluated by flow cytometry. To normalize the fluorescence signal to the quantity of immobilized factor VIII for each mutant, the fluorescence signals were divided by the fluorescence from the highest concentration of fluorescein-labeled BO2C11 that bound to factor VIII on the same beads. Compared with wild-type factor VIII, the fluorescence for the mutants was very low, although in all cases it exceeded that of control beads lacking factor VIII. This indicated that the mutants had markedly decreased affinity for phospholipid vesicles. The affinity of R2159A was deceased approximately 10-fold, and the other mutants did not exhibit clear evidence of saturation at phospholipid concentrations up to 200μM. In contrast, the saturable binding of phospholipid vesicles to wild-type factor VIII with half-maximal binding at a phospholipid concentration of approximately 25μM was consistent with our prior report.23 These results confirm that the affinity of factor VIII for phospholipid membranes is reduced when these amino acids encompassing the lower surface of the C1 domain were mutated. The relationship of these measurements to the apparent affinities in Figure 1B is discussed in “Discussion.”
Affinity of phospholipid vesicles for C1 domain mutants of factor VIII. Wild-type factor VIII (●) and mutants 2159 (■), 2042/2043 (▾), 2090/2091 (◇), and 2092/2093 (♦) were incubated with various concentrations of sonicated phospholipid vesicles of PS/PE/PC/NBD-PC (4:20:71:5) for 30 minutes. Aliquots of 1-μL Superose beads were added and incubated for another 30 minutes. Phospholipid vesicle-factor VIII complexes bound to mAb GMA-8021-Superose beads were measured by flow cytometry. Values were corrected for the quantity of fluorescence from phospholipid vesicles associated with Superose beads lacking factor VIII. The quantity of bound phospholipid vesicles was normalized to the quantity of mAb BO2C11-fluorescein isothiocyanate that bound to aliquots of the same quantity of factor VIII incubated with Superose beads. Curves indicate the best fit values for wild-type factor VIII (solid line), mutants 2159 (solid line), 2042/2043 (dashed line), 2090/2091 (dotted line), and 2092/2093 (dash dotted line) obtained with the GraphPad Prism Version 5.0. Data are from a single experiment representative of 2 experiments from 2 separate COS cell transfections.
Affinity of phospholipid vesicles for C1 domain mutants of factor VIII. Wild-type factor VIII (●) and mutants 2159 (■), 2042/2043 (▾), 2090/2091 (◇), and 2092/2093 (♦) were incubated with various concentrations of sonicated phospholipid vesicles of PS/PE/PC/NBD-PC (4:20:71:5) for 30 minutes. Aliquots of 1-μL Superose beads were added and incubated for another 30 minutes. Phospholipid vesicle-factor VIII complexes bound to mAb GMA-8021-Superose beads were measured by flow cytometry. Values were corrected for the quantity of fluorescence from phospholipid vesicles associated with Superose beads lacking factor VIII. The quantity of bound phospholipid vesicles was normalized to the quantity of mAb BO2C11-fluorescein isothiocyanate that bound to aliquots of the same quantity of factor VIII incubated with Superose beads. Curves indicate the best fit values for wild-type factor VIII (solid line), mutants 2159 (solid line), 2042/2043 (dashed line), 2090/2091 (dotted line), and 2092/2093 (dash dotted line) obtained with the GraphPad Prism Version 5.0. Data are from a single experiment representative of 2 experiments from 2 separate COS cell transfections.
Effect of mutants in C1 domain on affinity for phospholipid vesicles and VWF
| Factor VIII . | Dissociation constants . | |
|---|---|---|
| Phospholipid,* μM . | VWF,† nM . | |
| Wild-type | 15 ± 6 | 0.94 ± 0.17 |
| 2090/2091 | > 200 | 0.56 ± 0.15 |
| 2092/2093 | > 200 | 0.43 ± 0.10 |
| 2159 | 134 ± 53 | > 100 |
| 2042/2043 | > 200 | 2.3 ± 1.5 |
| Factor VIII . | Dissociation constants . | |
|---|---|---|
| Phospholipid,* μM . | VWF,† nM . | |
| Wild-type | 15 ± 6 | 0.94 ± 0.17 |
| 2090/2091 | > 200 | 0.56 ± 0.15 |
| 2092/2093 | > 200 | 0.43 ± 0.10 |
| 2159 | 134 ± 53 | > 100 |
| 2042/2043 | > 200 | 2.3 ± 1.5 |
KD ± SD of fit. Normalized binding curves from 2 experiments with vesicles of 15% PS were averaged and fitted by nonlinear, least squares analysis.
KD ± SD of fit. Displacement curves from 3 competition, solution phase VWF-binding assays were averaged and fitted by nonlinear, least squares analysis. The concentration of VWF was analyzed as the VWF subunit concentration, assuming an Mr of 220 000.
Amino acids in C1 domain contribute to VWF binding
To determine the extent to which the phospholipid and VWF-binding amino acids of the factor VIII C1 domain overlap, we evaluated the affinities of the mutants for VWF. We used a competition, solution-phase ELISA (Figure 3; Table 2) in which only free factor VIII was available to bind to immobilized BO2C11. Compared with wild-type factor VIII, the affinity of R2159A mutant was reduced more than 50-fold, consistent with a prior study.37 Q2042A/Y2043A had VWF affinity reduced 2.5-fold, whereas the other mutants bound with affinity at least as high as wild-type factor VIII. These results confirm that Arg 2159 and the adjacent amino acids, Gln 2042/Tyr 2043, also participate in VWF binding. However, the amino acids nearer the tip of the C1 domain (Figure 1A) do not influence affinity.
Affinity of factor VIII C1 domain mutants for plasma VWF. Wild-type factor VIII (●) and mutants 2159 (■), 2042/2043 (▾), 2090/2091 (◇), and 2092/2093 (♦) were incubated with various concentration of purified plasma VWF for 60 minutes before placement in microtiter wells coated with mAb BO2C11. Bound factor VIII was detected with biotinylated mAb ESH8. Because VWF competes for the BO2C11 epitope,25 it leads to decreased bound factor VIII. Displayed values are normalized to the maximum signals measured for each type of factor VIII in the absence of added VWF. The quantity of factor VIII used was 0.4 unit/mL for wild-type factor VIIII and mutants. Data are the average of 3 experiments. The molar concentration of VWF subunits, indicated on the abscissa, was obtained by dividing the VWF concentration by the molecular weight of a single VWF subunit.
Affinity of factor VIII C1 domain mutants for plasma VWF. Wild-type factor VIII (●) and mutants 2159 (■), 2042/2043 (▾), 2090/2091 (◇), and 2092/2093 (♦) were incubated with various concentration of purified plasma VWF for 60 minutes before placement in microtiter wells coated with mAb BO2C11. Bound factor VIII was detected with biotinylated mAb ESH8. Because VWF competes for the BO2C11 epitope,25 it leads to decreased bound factor VIII. Displayed values are normalized to the maximum signals measured for each type of factor VIII in the absence of added VWF. The quantity of factor VIII used was 0.4 unit/mL for wild-type factor VIIII and mutants. Data are the average of 3 experiments. The molar concentration of VWF subunits, indicated on the abscissa, was obtained by dividing the VWF concentration by the molecular weight of a single VWF subunit.
Role of the C1 domain in the assembly of factor Xase complex
To isolate the effects of mutations on assembly and function of the factor Xase complex from effects on phospholipid affinity, experiments were performed with saturating concentrations of phospholipid vesicles. (Figure 4A). Factor IXa and factor X concentrations were increased to saturating values for each mutant. In the presence of vesicles of 15% PS, only Q2042A/Y2043A had clearly reduced activity. However, in the presence of vesicles with 4% PS, only R2159A maintained a normal Vmax, with the other mutants having 70% to 90% reduced activity. Thus, engagement of the C1 domain tip with the phospholipid membrane affects activity of the factor Xase complex in addition to the effect on net affinity for phospholipid membranes.
Effect of C1 domain mutations on parameters of the factor Xase complex. (A) Vmax was obtained with saturating factor IXa and factor X. (B) Apparent factor IXa affinities were measured by various factor IXa concentrations in the presence of factor VIII (0.15 U/mL) and excess phospholipid containing 4% or 15% PS. (C) Apparent factor X affinities were determined by various factor X concentrations in the presence of excess phospholipid vesicles and factor IXa. The apparent KD values were determined using nonlinear least squares analysis with the software GraphPad Prism, Version 5.0. Data are mean ± SEM for 2 to 4 experiments, each performed in duplicate.
Effect of C1 domain mutations on parameters of the factor Xase complex. (A) Vmax was obtained with saturating factor IXa and factor X. (B) Apparent factor IXa affinities were measured by various factor IXa concentrations in the presence of factor VIII (0.15 U/mL) and excess phospholipid containing 4% or 15% PS. (C) Apparent factor X affinities were determined by various factor X concentrations in the presence of excess phospholipid vesicles and factor IXa. The apparent KD values were determined using nonlinear least squares analysis with the software GraphPad Prism, Version 5.0. Data are mean ± SEM for 2 to 4 experiments, each performed in duplicate.
The apparent affinities for factor IXa and factor X were measured by evaluating different concentrations of factor IXa and factor X (Figure 4B-C). Mutant R2090A/Q2091A and Q2042A/Y2043A had approximately 50% reduced affinity for factor IXa, although the experimental variation with 4% vesicles leaves a higher degree of uncertainty for Q2042A/Y2043A. R2159A and K2092A/F2093A had normal apparent affinity for factor IXa. In the presence of 4% PS vesicles, mutants R2090A/Q2091A, Q2042A/Y2043A, and R2159A had approximately 4-fold decreased apparent affinity for factor X. With vesicles of 15% PS, the apparent affinities for factor X were close to wild-type factor VIII. These results indicate that Arg 2090/Gln 2091, Gln 2042/Tyr 2043, and Arg 2159 affect the association between factor VIII and factor X.
Cooperativity of C1 and C2 domains for cofactor function
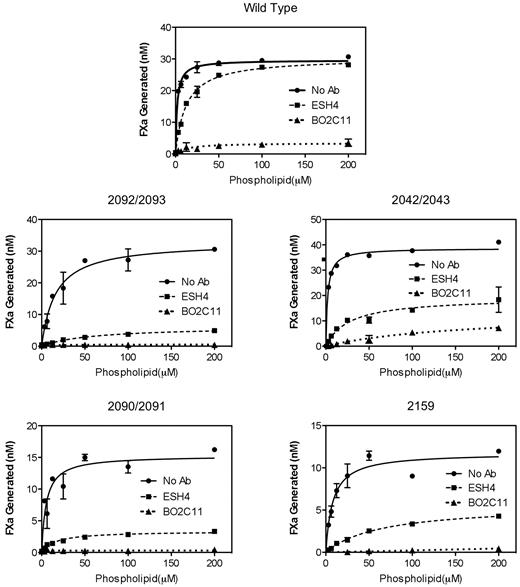
To determine the functional relationship between the membrane-interactive residues of the C1 domain and the membrane-interactive surfaces of the C2 domain, the activities of wild-type factor VIII were tested in the presence of mAbs BO2C11 and ESH4 (Figure 5). Both antibodies inhibit function of factor VIII by blocking membrane-interactive epitopes of the C2 domain26 (Figure 1A). Thus, the presumed mechanism of inhibition is reduction of membrane affinity. We asked whether the inhibitory function of these antibodies could be offset by addition of sufficient phospholipid vesicles so that the factor VIII-antibody complex would still bind to vesicles in the presence of factor IXa and factor X. Both antibodies substantially inhibited activity of wild-type factor VIII on vesicles with 4% PS (data not shown), making measurement of residual activity difficult. Thus, further experiments were performed with vesicles of 15% PS (Figure 5). ESH4 led to 7-fold reduction in apparent membrane affinity but left normal Vmax when phospholipid vesicles were at a saturating concentration. In contrast, BO2C11 led to a comparable decrease in apparent membrane affinity but 89% reduction of Vmax. Thus, availability of the ESH4 epitope affects membrane affinity but has little effect on function of the assembled factor Xase complex for membranes with 15% PS. In contrast, BO2C11decreases membrane affinity and also decreases activity of the assembled factor Xase complex.
Effect of mAbs ESH4 and BO2C11 on activities of factor VIII and mutants. ESH4 and BO2C11 inhibited activity of wild-type factor VIII and mutants on vesicles with 15% PS. Factor VIII (0.15 U/mL) in the absence (■) or presence of excess ESH4 (○) or BO2C11 (●) was added to various concentrations of phospholipid vesicles. Factor VIII was preincubated with ESH4 or BO2C11 for 30 minutes at room temperature. Data are mean ± SEM for 3 separate experiments, each performed in duplicate.
Effect of mAbs ESH4 and BO2C11 on activities of factor VIII and mutants. ESH4 and BO2C11 inhibited activity of wild-type factor VIII and mutants on vesicles with 15% PS. Factor VIII (0.15 U/mL) in the absence (■) or presence of excess ESH4 (○) or BO2C11 (●) was added to various concentrations of phospholipid vesicles. Factor VIII was preincubated with ESH4 or BO2C11 for 30 minutes at room temperature. Data are mean ± SEM for 3 separate experiments, each performed in duplicate.
BO2C11 inhibited the activity of R2090A/Q2091A, K2092A/F2093A, and R2159A more than 98%, whereas Q2042A/Y2043A retained approximately 15% activity, similar to wild-type factor VIII. However, the apparent membrane affinity was reduced 75-fold. These data indicate that most of the C1 domain membrane-interactive motif is essential for membrane binding and/or correct membrane alignment when the BO2C11 epitope is blocked. In contrast, ESH4 decreased apparent membrane affinity 3- to 12-fold for the 4 mutants, similar to the decrease for wild-type factor VIII. In contrast to wild-type factor VIII, ESH4 decreased the Vmax by 50% to 80% for all 4 mutants. These data indicate that membrane-interactive residues of the C1 domain tip function cooperatively with the ESH4 epitope to establish the effective membrane alignment for factor VIII. Together, these results indicate that the membrane-binding C1 domain face and the 2 C2 domain epitopes all participate to achieve normal membrane affinity and also normal membrane alignment that is required for full activity.
Discussion
This study demonstrates that 7 amino acids on a hydrophilic lower face of the factor VIII C1 domain affect interaction with phospholipid membranes. Amino acids Arg 2090/Gln 2091, Lys 2092/Phe 2093, Gln 2042/Tyr 2043, and Arg 2159 define this membrane-interactive face. Arg 2159 and the adjacent amino acids, Gln 2042/Tyr 2043 also participate in VWF binding. Further, the results indicate that membrane binding of the factor VIII C1 domain affects the assembly of factor Xase complex, modifying the affinity for factor X and the Vmax of the assembled complex. Our results also indicate that the C1 domain and the ESH4 epitope of the C2 domain function cooperatively to establish the optimal membrane alignment of factor VIII. Thus, the tip of the C1 domain contributes to the net membrane affinity of factor VIII as well as optimal alignment of factor VIII with the membrane and the substrate of the factor Xase complex.
Our results confirm and extend the conclusions from several prior studies. They are in agreement with the reports indicating that the C1 domain participates in platelet membrane binding29 and our prior report that amino acids Lys 2092 and Phe 2093 participate in binding to both synthetic phospholipid membranes and platelet membranes.30 Our results go beyond these prior studies in identifying additional amino acids, thus defining a C1 domain face that participates in membrane binding. In addition, our results shed light on the manner in which the C1 domain face cooperates with the 2 membrane-interactive epitopes of the C2 domain to influence assembly of the factor Xase complex.
Our results are readily interpretable in light of the lipid-free factor VIII crystal structures, placing the C1 domain beside and nearly parallel to the C2 domain.27,28 The results would be difficult to rationalize with an electron crystallography-based model of membrane-bound factor VIII, which places the C1 domain opposite the C2 domain and away from the membrane.38
Our findings demonstrate that a membrane-interactive face contains multiple amino acids that engage the phospholipid membrane. Five of the 7 mutated amino acids are predominantly hydrophilic in character, in contrast with the predominantly hydrophobic amino acids that were previously identified, suggesting that these residues interact with phospholipid head groups more than with the hydrophobic core of the membrane.
Our data indicate that the factor VIII C1 domain affects binding of factor X to the factor Xase complex (Figure 4C). Three of the 4 mutants had decreased apparent affinities for factor X on membranes with 4% PS. Only mutant K2092A/F2093A did not have a clearly decreased affinity. For vesicles with 15% PS, affinities approached wild-type factor VIII. This suggests that additional interactions of the C1 domain with these membranes can compensate for the loss of the mutated amino acids that participate in membrane binding. Our data suggest that factor X may contact the C1 domain, most probably via the Gla domain. Additional studies from our laboratory seem to confirm that contact of the factor X Gla domain with the C1 domain enhances activity of the factor Xase complex (X. L. Li, V. A. Novakovic, H. Wakabyashi, J. H. Lu, M.J., and G.E.G., manuscript in preparation). This suggests a need for refinement of the computational model of the factor Xase complex, which places the Gla domain of factor X domain in contact with the C2 domain and the Gla domain of factor IXa in contact with the C1 domain.27
Comparison of the directly measured affinities for phospholipid (Figure 2) with the apparent affinities implied by titrating phospholipid vesicles into the components of the factor Xase complex (Figure 1) suggests that factor IXa or factor X augments the membrane affinities of the mutants. Further experiments will be required to determine whether these effects are a consequence of direct contact between the C1 and/or C2 domain with factor IXa or factor X.
Prior reports indicate that an acidic peptide n-terminal to the A3 domain, the C2 domain, and the C1 domain contain VWF-binding amino acids.39,40 We have confirmed that Arg 2159 interacts with VWF37 and now demonstrated that Gln 2042/Tyr 2043 also participates in VWF binding. Our results are consistent with a prior report indicating a mild hemophilia phenotype associated with Arg2159 mutation.37 It should be noted that the amino acids at the tip of the C1 domain do not influence VWF affinity. These results demonstrate that the C1-binding sites for VWF and phospholipid overlap but are not identical.
The present study indicates that the structure-function relationships of factor VIII are analogous to those of factor V in that the factor VIII C1 domain also has a membrane-interactive motif. Residues Lys 2092, Phe 2093, and Arg 2159 of factor VIII C1 domain, homologous to Tyr 1956, Leu1957, and Arg 2023 of the factor V, are both involved in membrane binding.41 Both proteins were studied by substituting Ala for the membrane-interactive amino acids. Because Ala is less bulky than the substituted residues, it remains possible that some membrane-binding abnormalities could result from altered flexibility or minor changes in the conformation of the respective loops.
Model of factor VIII interacting with phospholipid membrane
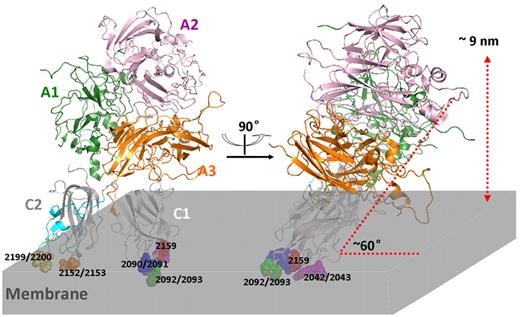
Our results enable refinement of a model for factor VIII interacting with phospholipid membranes (Figure 6). The model assumes that the lipid-free crystal structures of factor VIII provide the orientation of the C domains to one another and to the A domains when the protein is bound to a membrane. We assumed, further, that the previously identified membrane-binding hydrophobic amino acids, Tyr 2093, Met 2199, Phe 2200, Leu 2151, and Leu 2152 penetrate to the hydrophobic core of the membrane. Placing the hydrophilic membrane-binding interface of C1 domain in contact with the membrane then defines an angle of approximately 60 degrees for the long axis of factor VIII relative to the membrane. The right panel is rotated 90 degrees clockwise to better illustrate the membrane binding of amino acids in C1 domain. This model appears consistent with the cooperative membrane binding of the C1 domain and the C2 domain epitopes tested in this report as both the BO2C11 epitope and the ESH4 epitope of the C2 domain are in intimate contact with the membrane. This model also places His 2315 and Gln 2316, hydrophilic membrane-interactive amino acids tentatively identified by mutagenesis studies,42 adjacent to the membrane. It also places Trp 2313, another putative membrane-interactive residue,43,44 in intimate contact with the membrane. The distance from the top of the A2 domain to the membrane surface is approximately 9 nm, consistent with prior measurements.45 This orientation of factor VIII to the membrane is similar to the orientation recently proposed in a model of the factor VIIIa–factor IXa structure.27
Model of factor VIII interacting with phospholipid membrane. Front and side view of factor VIII crystal structure (PDB ID: 3CDZ) anchored into a phospholipid membrane (light gray). Green represents A1 domain (residues 1-336); pink, A2 domain (residues 373-721); gold-brown, A3 domain (residues 1690-2019); medium gray, C1 domain (residues 2020-2172), with mutated residues encompassing the lower surface highlighted by coloring; dark gray, C2 domain (residues 2172-2332); and orange spheres, 4 membrane-interactive hydrophobic residues. The epitopes of anti-C2 domain mAb ESH4 (blue and yellow-orange) and BO2C11 (orange and yellow-orange) are highlighted by coloring.
Model of factor VIII interacting with phospholipid membrane. Front and side view of factor VIII crystal structure (PDB ID: 3CDZ) anchored into a phospholipid membrane (light gray). Green represents A1 domain (residues 1-336); pink, A2 domain (residues 373-721); gold-brown, A3 domain (residues 1690-2019); medium gray, C1 domain (residues 2020-2172), with mutated residues encompassing the lower surface highlighted by coloring; dark gray, C2 domain (residues 2172-2332); and orange spheres, 4 membrane-interactive hydrophobic residues. The epitopes of anti-C2 domain mAb ESH4 (blue and yellow-orange) and BO2C11 (orange and yellow-orange) are highlighted by coloring.
Membrane-binding motifs in C1 and C2 domains for cofactor activity
ESH4 and BO2C11 represent 2 of the 3 classic anti-C2 inhibitory antibody types. Both block distinct epitopes that participate in phospholipid membrane binding.26 However, the quantitative effect on membrane affinity and the effect on residual activity had not been measured. We found that both antibodies decrease apparent affinity and activity on membranes with 4% PS to an extent that makes residual activity measurements difficult. On vesicles with 15% PS, both antibodies caused a comparable decrease in the apparent affinity of wild-type factor VIII, approximately 7-fold. However, they had a distinct effect on activity, with BO2C11 causing a major reduction in Vmax. This suggests that blockade of the BO2C11 epitope alters the alignment of factor VIII/VIIIa with the membrane and with factor IXa and factor X to a greater degree than the ESH4 epitope.
The effect of ESH4 on mutant factor VIII molecules was distinct from that on wild-type factor VIII. In contrast to wild-type factor VIII, ESH4 decreased the Vmax for all 4 mutants. However, the reduction in membrane affinity for the mutants was comparable with the effect on wild-type factor VIII. The extent of inhibition in Vmax was 55% to 90%.This pattern suggests that the C1 domain face containing the mutants and the ESH4 epitope cooperate to produce a correct alignment of factor VIII with a phospholipid membrane. Because the Vmax is modified more than the affinity suggests that interaction of the C1 residues causes a change in membrane orientation or a conformational change in factor VIII rather than only contributing additive binding energy.
The manner in which membrane engagement of the C1 domain face alters the orientation or conformation of factor VIII is an interesting topic for further investigation. Our data do not seem to require, or preclude, a conformational shift of the C2 domain.27 We have recently initiated studies of the dynamic response of the C2 domain and the C1 domain in response to membrane binding.46 These studies, using deuterium exchange, appear likely to provide insight into changes in conformation or rigidity that occur in response to membrane engagement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Department of Veterans Affairs and the National Hemophilia Foundation (G.E.G.).
Authorship
Contribution: J.L. designed and performed experiments; J.L. and G.E.G. collected and analyzed data and drafted the manuscript; S.W.P and M.J. participated in overall project planning and revised the manuscript; H.M. prepared factor VIII mutants; and M.J. provided BO2C11 antibody and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gary E. Gilbert, Veterans Administration Boston Healthcare System, 1400 VFW Pkwy, West Roxbury, MA 02132; e-mail: Gary_Gilbert@hms.harvard.edu or ggilbert@rics.bwh.harvard.edu.