Abstract
Forodesine, a purine nucleoside phosphorylase inhibitor, displays in vitro activity in chronic lymphocytic leukemia (CLL) cells in presence of dGuo, which is the basis for an ongoing clinical trial in patients with fludarabine-refractory CLL. Initial clinical data indicate forodesine has significant activity on circulating CLL cells, but less activity in clearing CLL cells from tissues such as marrow. In tissue microenvironments, lymphocytes interact with accessory stromal cells that provide survival and drug-resistance signals, which may account for residual disease. Therefore, we investigated the impact of marrow stromal cells (MSCs) on forodesine-induced response in CLL lymphocytes. We demonstrate that spontaneous and forodesine-induced apoptosis of CLL cells was significantly inhibited by human and murine MSCs. Forodesine-promoted dGuo triphosphate (dGTP) accumulation and GTP and ATP depletion in CLL cells was inhibited by MSCs, providing a mechanism for resistance. Also, MSCs rescued CLL cells from forodesine-induced RNA- and protein-synthesis inhibition and stabilized and increased Mcl-1 transcript and protein levels. Conversely, MSC viability was not affected by forodesine and dGuo. Collectively, MSC-induced biochemical changes antagonized forodesine-induced CLL cell apoptosis. This provides a biochemical mechanism for MSC-derived resistance to forodesine and emphasizes the need to move toward combinations with agents that interfere with the microenvironment's protective role for improving current therapeutic efforts.
Introduction
The purine nucleoside phosphorylase (PNP) enzyme plays an important role in the purine salvage pathway. The function of PNP is to catalyze the phosphorolysis of 2′-ribonucleosides and 2′-deoxyribonucleosides of guanine and hypoxanthine. Genetic PNP deficiency syndrome leads to accumulation of dGuo in plasma1 and dGuo triphosphate (dGTP) in cells with a selective accumulation in T cells.2-4 Endogenous PNP deficiency leads to specific T-cell immunodeficiency, a genetic disease that has prompted the development of PNP inhibitors as potential therapeutics for T-cell–mediated diseases.5
Several PNP inhibitors have been developed and tested in cell-line model systems.6-11 Although their Ki values were in low micromolar, they inhibited PNP in whole cells and resulted in intracellular accumulation of dGTP; these agents were not effective in the animal model system because a high specific activity of PNP is present in large body organs such as liver, spleen, and thymus.12-14 These data suggested that a highly potent inhibitor would be needed for PNP inhibition in human system.
Forodesine (fodosine; immucilin H) was synthesized based on transition-state structure stabilized by target enzyme strategy.15,16 In contrast to the previous agents that inhibited PNP at micromolar or mid nanomolar concentration, forodesine suppressed the enzyme activity at low picomolar levels.17 Furthermore, a proof-of-principle phase 1 investigation demonstrated that this drug inhibited PNP, elevated the plasma dGuo levels to micromolar levels, and increased intracellular dGTP to millimolar levels in immature T cells when administered to patients.18 This proof-of-concept clinical study further elucidated the role of PNP inhibition in T-cell malignancies.
When PNP is blocked by forodesine, plasma dGuo is not cleaved to guanine but instead is intracellularly converted to dGTP by high deoxycytidine kinase (dCK) and deoxyguanosine kinase (dGK) in T cells, which leads to perturbation of dNTP pools and inhibition of DNA synthesis and cell replication, eventually resulting in apoptosis. Because chronic lymphocytic leukemia (CLL) cells have high dCK activity, have shown a positive response to a dGuo analog nelarabine, and could retain purine nucleoside analog triphosphates for a longer time, we hypothesized that these cells would respond to forodesine treatment. Our in vitro studies served as a proof-of-principle of this hypothesis.19 We demonstrated that when the CLL cells were incubated in vitro with forodesine and dGuo, the lymphocytes accumulated intracellular dGTP without any effect on other deoxynucleotides. This was associated with DNA damage-induced p53 stabilization, phosphorylation of p53 at Ser15, and activation of p21. The dGTP accumulation was related to induction of apoptosis measured by caspase activation, changes in mitochondrial membrane potential, and PARP cleavage.19
Based on these encouraging preclinical data, a clinical trial with forodesine was initiated for patients with fludarabine-refractory CLL at the University of Texas M. D. Anderson Cancer Center. This trial recruited 8 patients. The clinical response to forodesine in this heavily pretreated population was limited; 2 patients had a decrease in their peripheral blood absolute lymphocyte count to normal levels after the first cycle, but this was short-lasting, and the counts increased thereafter. In the other 5 patients, the white blood cell (WBC) counts increased progressively, and in 3 patients there was also progression in the lymph node.20 In all cases, the bone marrow typically displayed residual disease, indicating that CLL cells under the influence of microenvironment may be resistant to forodesine. Compelling evidence suggests that cross-talk with accessory stromal cells in specialized tissue microenvironments, such as the bone marrow and secondary lymphoid organs, promotes growth and primary drug resistance of CLL and various other cancer cells.21,22 Therefore, we hypothesized that forodesine, similar to more “conventional” drugs such as fludarabine, can eliminate the bulk of clonal cells, whereas residual malignant cells lurk in protective marrow niches, where they receive survival and drug-resistance signals from marrow stromal cells (MSCs).
Our previous and recent investigations using CLL-MSC coculture approaches showed that MSCs protected CLL cells from spontaneous and fludarabine-induced apoptosis.23,24 This was shown to be paralleled by the induction of the antiapoptotic protein Mcl-1, and phosphorylation of signaling molecules involved in the survival pathways (AKT and ERK) in CLL cells.24 In general, there was a tight correlation observed between the Mcl-1 levels, PARP cleavage, and apoptosis in the CLL cells. These findings prompted us to explore whether MSC-derived survival and drug-resistance signals may also interfere with forodesine-induced apoptosis in CLL.
Methods
Drugs and chemicals
Forodesine for in vitro use was provided by BioCryst Pharmaceuticals, and dGuo was purchased from Sigma-Aldrich. For quantitation of deoxynucleotides, dNTPs were obtained from Amersham Biosciences and were used as standards. [3H]dATP and [3H]dTTP were purchased from Perkin Elmer Life Sciences and MP Biomedicals. [3H]uridine and [3H]leucine (specific activity, 41.2 and 120 Ci/mmol, respectively) were purchased from Moravek Biochemicals.
Patients and methods
The present in vitro studies were carried out in leukemic lymphocytes obtained from patients with CLL. All patients signed written informed consent forms in accordance with the Declaration of Helsinki, and the laboratory protocol was approved by the institutional review board at the University of Texas M. D. Anderson Cancer Center.
Isolation of lymphocytes
Whole blood was collected in heparinized tubes and diluted 1:3 with cold phosphate-buffered saline (PBS; 0.135M NaCl, 2.7mM KCl, 1.5mM KH2PO4, 8mM Na2HPO4 [pH 7.4]) and layered onto Ficoll-Hypaque (specific gravity, 1.086; Life Technologies). The blood was then centrifuged at 433g for 20 minutes, and mononuclear cells were removed from the interphase. Cells were washed twice with cold PBS and resuspended in 10 mL of RPMI 1640, supplemented with 10% fetal bovine serum. A Coulter channelyzer (Coulter Electronics) was used to determine cell number and the mean cell volume. The lymphocytes were suspended at a concentration of 1 × 107 cells/mL, and fresh cells were used for all experiments.
Preparation of CLL-stromal cocultures
Isolated lymphocytes were either cultured in suspension or cocultured on confluent layers of murine MSCs (M210B4; ATCC) or human MSCs (NKtert; RIKEN Cell Bank25 ) at a ratio of 100 CLL cells to 1 MSC. To perform coculture experiments, we maintained continuous culture of stromal cells and plated them at a concentration of 1 × 105 cells/well 24 hours before adding CLL cells at a concentration of 1 × 107 cells/well. Cultures were incubated without any drugs or with forodesine and dGuo. Although CLL cells are constantly interacting with stroma, these lymphocytes do not adhere to plastic or to stroma cells. At the end of incubation, these free-floating CLL cells were carefully removed, leaving the adherent stromal layer undisturbed, and used for different pharmacologic, biochemical, and molecular end points.
Measurement of dNTP pools
The primary CLL cells were incubated with or without 2μM forodesine and 20μM dGuo for 24 or 48 hours. These concentrations were selected based on plasma pharmacology data during the phase 1 study of forodesine.18 Cultures were maintained and aliquots (1 × 107cells/mL) were removed at the end of incubation times. The nucleotides in the leukemia cells were extracted by 60% methanol as described,26 and the dNTPs were quantitated by DNA polymerase assay as modified by Sherman and Fyfe27 in these cell extracts.
Quantitation of cellular NTP pool
Before and after forodesine and dGuo incubations, the cells were processed using perchloric acid, and extracts were neutralized with KOH. The cellular NTP pool was determined using a high-pressure liquid chromatography (HPLC) procedure as described before.28 The nucleotides were expressed as micromolar, which assumes that they are equally distributed in cells.
Measurement of cell viability
Determination of CLL cell viability was based on the analysis of mitochondrial transmembrane potential by 3,3-dihexyloxocarbocyanine iodine (DiOC6) and cell membrane permeability to propidium iodide. For viability assays, 200 μL of cell suspension was collected at the indicated time points and transferred to fluorescence-activated cell sorter (FACS) tubes containing 200 μL of 60nM DiOC6 (Molecular Probes) and 10 μg/mL of propidium iodide (PI; Molecular Probes) in RPMI with 0.5% bovine serum albumin (BSA). Cells were then incubated at 37°C for 15 minutes and analyzed within 30 minutes by flow cytometry using a FACScalibur (Becton Dickinson). Fluorescence was recorded at 525 nm (FL-1) for DiOC6 and at 600 nm (FL-3) for PI.
An alternate apoptotic assay, annexin V binding, was carried out with a Detection Kit I (PharMingen,) according to the manufacturer's instructions. Briefly, cells were washed with PBS and resuspended in 200 μL of 1× annexin binding buffer obtained from BD Biosciences, at a concentration of 1 × 106 cells/mL. Annexin V–FITC (5 μL) was added, and the cells were incubated in the dark for 15 minutes at room temperature. A total of 10 μL of PI (50 μg/mL) was added to the labeled cells and analyzed immediately with a FACSCALIBUR cytometer (Becton Dickinson). Data from at least 10 000 events per sample were recorded and processed using Cell Quest software (Becton Dickinson).
Inhibition of RNA and protein synthesis
Primary CLL cells were either untreated or treated with 2μM forodesine and 20μM dGuo for 48 hours. Before removal of the aliquot, 10μCi/mL [3H]uridine or [3H]leucine was added to these cultures, and the incubation was continued for an additional 30 minutes. The labeled cells were then extracted using perchloric acid, the pellets were incubated overnight with KOH at 37°C to dissolve RNA or protein, and the radioactivity was measured by scintillation counting.
RNA isolation and real-time quantitative PCR
Total cellular RNA was isolated from CLL cells using the RNeasy mini kit (QIAGEN) with DNase digestion to completely remove the genomic DNA. Total RNA (20-50 ng) was used for the 1-step real-time reverse transcriptase–polymerase chain reaction (RT-PCR) in the TaqMan One-Step RT-PCR Master Mix (Applied Biosystems). Each PCR reaction was carried out in a 25 μL volume on a 96-well optical reaction plate for 30 minutes at 48°C for reverse transcription reaction, followed by 10 minutes at 95°C for initial denaturing, then followed by 40 cycles of 95°C for 15 seconds and 60°C for 2 minutes in the 7900HT Sequence Detection System (Applied Biosystems). The relative gene expression was analyzed by the Comparative Ct method using 18s ribosomal RNA as endogenous control, after confirming that the efficiencies of the target and the endogenous control amplifications were approximately equal. All the primers and probes and reverse-transcribed PCR buffers were purchased from Applied Biosystems.
Immunoblot analysis
Cells were lysed on ice for 20 minutes in lysis buffer containing 25mM HEPES (pH 7.5), 300mM NaCl, 1.5mM MgCl2, 0.5% sodium deoxycholate, 20mM glycerophosphate, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.2mM EDTA (pH 8), 0.5mM dithiothreitol, 1mM sodium orthovanadate (pH 10), and protease inhibitor. Cells were centrifuged at 14 000g for 10 minutes at 4°C, and the supernatant was removed and stored at −80°C until use. Protein content was determined using a DC protein assay kit (Bio-Rad Laboratories). Aliquots (30 μg) of total cell protein were boiled with Laemmli sample buffer and loaded onto 8% to 12% sodium dodecyl sulfate–polyacrylamide gels and transferred to nitrocellulose membranes (GE Osmonics Labstore). Membranes were blocked for 1 hour in PBS–Tween-20 containing 5% nonfat dried milk and then incubated with primary antibodies for 2 hours, followed by species-specific horseradish peroxidase–conjugated secondary antibody (diluted 1:5000) for 1 hour. The blots were visualized by enhanced chemiluminescence (Pierce Biotechnology Inc) and normalized to the actin (Sigma-Aldrich). Rabbit polyclonal antibody to Mcl-1 and mouse monoclonal antibody to Bcl-2 (Santa Cruz Biotechnology) was used to detect these proteins in each sample.
Results
Effect of forodesine and dGuo incubations on CLL cell apoptosis
Incubation of CLL primary cells for 24 hours with forodesine (2μM) and dGuo (20μM) induced modest but statistically significant amount of apoptosis (P < .001; n = 26; mean percentage of viability: control, 72%; treated, 59%; Figure 1A) in a time-dependent manner (48 hours; P < .001; n = 23; mean percentage viability: control, 67%; treated, 51%; Figure 1B). The apoptotic response to forodesine and dGuo was not related to patient characteristics such as Rai stage, and other prognostic markers such as ATM mutation, ZAP-70 expression, β2M level, or IgVH gene mutation (Table 1; data not shown). Our data demonstrate that from 49 patients analyzed, 14 patients exhibited less than 2% apoptosis, 13 patients showed 4% to 8% cytotoxicity, 9 patients showed 10% to 17% cytotoxicity, 10 patients showed 20% to 45% apoptosis, and 1 patient displayed 61% apoptosis (Table 1; Figure 1A-B), demonstrating heterogeneity in patient response to forodesine.
Forodesine with dGuo induces apoptosis in CLL cells in a time dependent manner and the apoptosis was abrogated by MSC (murine M210B4 and human NKtert). (A-B) Chronic lymphocytic leukemia (CLL) cells were incubated with 2μM forodesine and 20μM dGuo for 24 hours (n = 26, A) or 48 hours (n = 23, B), and the percentage viability was measured by DiOC6 in treated and untreated time-matched controls. (C-D) CLL cells were kept in suspension or cocultured with M210B4 or NKtert marrow stromal cells (MSCs) and incubated with or without forodesine and dGuo for 24 hours (n = 12, C) or 48 hours (n = 12, D), and the percentage viability was measured by DiOC6 in treated and untreated time-matched controls. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Forodesine with dGuo induces apoptosis in CLL cells in a time dependent manner and the apoptosis was abrogated by MSC (murine M210B4 and human NKtert). (A-B) Chronic lymphocytic leukemia (CLL) cells were incubated with 2μM forodesine and 20μM dGuo for 24 hours (n = 26, A) or 48 hours (n = 23, B), and the percentage viability was measured by DiOC6 in treated and untreated time-matched controls. (C-D) CLL cells were kept in suspension or cocultured with M210B4 or NKtert marrow stromal cells (MSCs) and incubated with or without forodesine and dGuo for 24 hours (n = 12, C) or 48 hours (n = 12, D), and the percentage viability was measured by DiOC6 in treated and untreated time-matched controls. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
CLL patient characteristics and forodesine plus dGuo–induced apoptosis
| Patient no. . | Age, y . | Sex . | No. prior therapies . | WBC, 109/L . | Lymphocyte, % . | β2M level . | Rai stage . | ZAP-70+ . | IgVH status, %* . | Chromosomal aberrations . | Percentage viability† . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11q22 . | del 13 . | 17p13 . | Untreated . | Treated . | ||||||||||
| 1 | 63 | M | 3 | 202.7 | 95.0 | 14.4 | 4 | POS | 100.0 | 94.5 | Y | NEG | 88.60 | 72.90 |
| 2 | 77 | M | 2 | 73.4 | 86.0 | 9.0 | 3 | POS | 100.0 | NEG | ND | NEG | 41.50 | 51.60 |
| 3 | 54 | M | 4 | 51.4 | 86.0 | 2.3 | 2 | 8.6 | 99.7 | NEG | N | 18 | 64.49 | 47.06 |
| 4 | 70 | M | 0 | 40.8 | 84.0 | 2.8 | 2 | NEG | 88.5 | NEG | ND | NEG | 37.30 | 30.30 |
| 5 | 68 | M | 0 | 18.1 | 77.0 | 2.4 | 1 | POS | 100.0 | 44.5 | N | NEG | 72.95 | 59.96 |
| 6 | 67 | F | 0 | 98.3 | 94.0 | 3.5 | 1 | NEG | 94.2 | NEG | N | NEG | 54.55 | 49.68 |
| 7 | 69 | F | 1 | 59.6 | 89.0 | ND | 1 | 9.3 | 98.9 | NEG | ND | NEG | 28.40 | 26.20 |
| 8 | 63 | F | 0 | 128.2 | 96.0 | 4.3 | 3 | 1.7 | 93.9 | NEG | N | NEG | 70.40 | 57.70 |
| 9 | 65 | M | 6 | 174.9 | 74.0 | 16.7 | 3 | POS | 98.3 | 86.5 | Y | NEG | 53.00 | 10.36 |
| 10 | 77 | M | 0 | 47.8 | 86.0 | 4.5 | 0 | 0.9 | 92.1 | ND | ND | ND | 54.73 | 50.68 |
| 11 | 63 | M | 2 | 52.2 | 83.0 | 2.7 | 2 | 0.9 | 96.6 | ND | ND | ND | 53.00 | 48.00 |
| 12 | 44 | F | 0 | 23.4 | 84.0 | 4.2 | 2 | 12.2 | 93.8 | NEG | ND | NEG | 42.96 | 32.79 |
| 13 | 79 | M | 1 | 47.3 | 94.0 | 4.4 | 1 | NEG | 100.0 | NEG | ND | NEG | 87.32 | 81.82 |
| 14 | 70 | M | 2 | 21.9 | 81.0 | 2.8 | 2 | 1.5 | 93.2 | NEG | ND | NEG | 84.98 | 80.87 |
| 15 | 68 | M | 3 | 105.9 | 91.0 | 8.0 | 4 | POS | 98.6 | NEG | N | 96.5 | 75.05 | 70.26 |
| 16 | 78 | M | 3 | 13.5 | 91.0 | 6.1 | 4 | POS | 100.0 | NEG | ND | NEG | 65.00 | 39.00 |
| 17 | 46 | M | 0 | 61.4 | 76.0 | 1.9 | 0 | 41.3 | 100.0 | NEG | N | NEG | 65.60 | 59.90 |
| 18 | 53 | F | 0 | 51.8 | 84.0 | 1.6 | 0 | NEG | 95.2 | ND | ND | ND | 76.60 | 76.80 |
| 19 | 67 | M | 1 | 142.9 | 92.0 | 7.7 | 2 | POS | 98.2 | 79% | N | NEG | 68.50 | 60.50 |
| 20 | 62 | F | 4 | 32.6 | 94.0 | 5.8 | 4 | 13.5 | 94.1 | NEG | N | 29.5 | 60.60 | 39.80 |
| 21 | 71 | M | 0 | 45.9 | 92.0 | 2.1 | 1 | NEG | 92.1 | NEG‡ | N | NEG‡ | 66.60 | 41.60 |
| 22 | 61 | F | 0 | 23.0 | 71.0 | 2.9 | 0 | 3.65 | 100.0 | NEG | ND | NEG | 78.60 | 78.10 |
| 23 | 61 | M | 0 | 33.8 | 74.0 | 2.8 | 1 | NEG | 99.6 | NEG | Y | NEG | 27.40 | 26.30 |
| 24 | 41 | M | 0 | 49.0 | 95.0 | 2.8 | 1 | NEG | 96.6 | NEG | N | NEG | 77.50 | 76.00 |
| 25 | 51 | M | 0 | 29.0 | 68.0 | 1.8 | 1 | ND | 99.7 | NEG | ND | NEG | 34.40 | 36.50 |
| 26 | 49 | F | 0 | 24.0 | 81.0 | 2.4 | 1 | POS | 95.3 | NEG | N | NEG | 61.00 | 57.30 |
| 27 | 51 | F | 0 | 10.0 | 68.0 | 1.6 | 0 | 20.5 | ND | NEG | ND | NEG | 85.70 | 85.90 |
| 28 | 85 | M | 1 | 10.4 | 51.0 | 7.2 | 4 | NEG | 93.1 | NEG | N | NEG | 85.30 | 78.00 |
| 29 | 65 | M | 0 | 91.9 | 90.0 | 5.2 | 4 | 0.91 | 96.3 | NEG | Y | 79 | 82.70 | 71.40 |
| 30 | 78 | F | 0 | 35.7 | 68.0 | 2.6 | 2 | 30.3 | 100.0 | NEG | ND | NEG | 46.80 | 44.80 |
| 31 | 59 | M | 0 | 94.7 | 84.0 | 5.3 | 1 | POS | 93.5 | NEG | N | NEG | 83.00 | 83.00 |
| 32 | 48 | M | 1 | 64.9 | 91.0 | 5.0 | 4 | NEG | 97.5 | NEG | N | NEG | 64.60 | 66.30 |
| 33 | 67 | M | 5 | 43.1 | 99.0 | 4.3 | 4 | POS | 100.0 | 88 | ND | NEG | 61.30 | 57.10 |
| 34 | 59 | M | 2 | 14.2 | 47.0 | 4.1 | 4 | 10.8 | 100.0 | NEG | ND | NEG | 43.20 | 14.80 |
| 35 | 66 | M | 0 | 145.5 | 89.0 | 5.8 | 1 | ND | 100.0 | 90.5 | ND | NEG | 62.00 | 17.90 |
| 36 | 50 | F | 0 | 22.7 | 86.0 | 2.5 | 2 | 29.7 | 100.0 | NEG | ND | NEG | 59.46 | ND |
| 37 | 78 | F | 0 | 53.7 | 91.0 | 2.6 | 2 | 30.3 | 99.6 | NEG | ND | NEG | 38.99 | ND |
| 38 | 52 | M | 0 | 40.7 | 84.0 | 4.1 | 2 | 25.1 | 100.0 | 13 | N | NEG | 58.68 | ND |
| 39 | 71 | F | 0 | 150.5 | 96.0 | 4.1 | 0 | 0.72 | 100.0 | DEL | ND | NEG | 54.40 | 54.10 |
| 40 | 60 | M | 0 | 37.5 | 75.0 | 2.7 | 2 | 8.88 | 91.5 | NEG | ND | NEG | 51.20 | 54.40 |
| 41 | 44 | F | 0 | 15.2 | 76.0 | 1.7 | 0 | 21.4 | ND | NEG | N | NEG | 60.30 | 50.10 |
| 42 | 70 | F | 0 | 22.6 | 84.0 | 2.1 | 0 | 18.2 | 100.0 | NEG | N | NEG | 83.80 | 47.10 |
| 43 | 65 | M | 3 | 110.7 | 96.0 | 3.2 | 2 | 0.9 | 96.6 | NEG | N | NEG | 75.30 | 70.40 |
| 44 | 48 | M | 0 | 72.2 | 86.0 | 2.2 | 2 | POS | 99.3 | NEG | N | NEG | 86.20 | 84.70 |
| 45 | 66 | M | 0 | 151.4 | 93 | 6.2 | 1 | NEG | 100.0 | 94 | N | NEG | 81.20 | 19.90 |
| 46 | 50 | M | 0 | 115.3 | 92.0 | 2.6 | 1 | POS | 100.0 | 93 | N | NEG | 89.00 | 61.50 |
| 47 | 88 | F | 0 | 47.9 | 81 | ND | 0 | ND | ND | N | Y | N | 75.00 | 44.00 |
| 48 | 77 | M | 1 | 73.4 | 98 | 4.6 | 4 | NEG | 94.9 | ND | ND | ND | 84.00 | 71.00 |
| 49 | 53 | F | 1 | 62.9 | 94 | ND | 3 | POS | 100.0 | ND | ND | ND | 56.00 | 40.00 |
| Patient no. . | Age, y . | Sex . | No. prior therapies . | WBC, 109/L . | Lymphocyte, % . | β2M level . | Rai stage . | ZAP-70+ . | IgVH status, %* . | Chromosomal aberrations . | Percentage viability† . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11q22 . | del 13 . | 17p13 . | Untreated . | Treated . | ||||||||||
| 1 | 63 | M | 3 | 202.7 | 95.0 | 14.4 | 4 | POS | 100.0 | 94.5 | Y | NEG | 88.60 | 72.90 |
| 2 | 77 | M | 2 | 73.4 | 86.0 | 9.0 | 3 | POS | 100.0 | NEG | ND | NEG | 41.50 | 51.60 |
| 3 | 54 | M | 4 | 51.4 | 86.0 | 2.3 | 2 | 8.6 | 99.7 | NEG | N | 18 | 64.49 | 47.06 |
| 4 | 70 | M | 0 | 40.8 | 84.0 | 2.8 | 2 | NEG | 88.5 | NEG | ND | NEG | 37.30 | 30.30 |
| 5 | 68 | M | 0 | 18.1 | 77.0 | 2.4 | 1 | POS | 100.0 | 44.5 | N | NEG | 72.95 | 59.96 |
| 6 | 67 | F | 0 | 98.3 | 94.0 | 3.5 | 1 | NEG | 94.2 | NEG | N | NEG | 54.55 | 49.68 |
| 7 | 69 | F | 1 | 59.6 | 89.0 | ND | 1 | 9.3 | 98.9 | NEG | ND | NEG | 28.40 | 26.20 |
| 8 | 63 | F | 0 | 128.2 | 96.0 | 4.3 | 3 | 1.7 | 93.9 | NEG | N | NEG | 70.40 | 57.70 |
| 9 | 65 | M | 6 | 174.9 | 74.0 | 16.7 | 3 | POS | 98.3 | 86.5 | Y | NEG | 53.00 | 10.36 |
| 10 | 77 | M | 0 | 47.8 | 86.0 | 4.5 | 0 | 0.9 | 92.1 | ND | ND | ND | 54.73 | 50.68 |
| 11 | 63 | M | 2 | 52.2 | 83.0 | 2.7 | 2 | 0.9 | 96.6 | ND | ND | ND | 53.00 | 48.00 |
| 12 | 44 | F | 0 | 23.4 | 84.0 | 4.2 | 2 | 12.2 | 93.8 | NEG | ND | NEG | 42.96 | 32.79 |
| 13 | 79 | M | 1 | 47.3 | 94.0 | 4.4 | 1 | NEG | 100.0 | NEG | ND | NEG | 87.32 | 81.82 |
| 14 | 70 | M | 2 | 21.9 | 81.0 | 2.8 | 2 | 1.5 | 93.2 | NEG | ND | NEG | 84.98 | 80.87 |
| 15 | 68 | M | 3 | 105.9 | 91.0 | 8.0 | 4 | POS | 98.6 | NEG | N | 96.5 | 75.05 | 70.26 |
| 16 | 78 | M | 3 | 13.5 | 91.0 | 6.1 | 4 | POS | 100.0 | NEG | ND | NEG | 65.00 | 39.00 |
| 17 | 46 | M | 0 | 61.4 | 76.0 | 1.9 | 0 | 41.3 | 100.0 | NEG | N | NEG | 65.60 | 59.90 |
| 18 | 53 | F | 0 | 51.8 | 84.0 | 1.6 | 0 | NEG | 95.2 | ND | ND | ND | 76.60 | 76.80 |
| 19 | 67 | M | 1 | 142.9 | 92.0 | 7.7 | 2 | POS | 98.2 | 79% | N | NEG | 68.50 | 60.50 |
| 20 | 62 | F | 4 | 32.6 | 94.0 | 5.8 | 4 | 13.5 | 94.1 | NEG | N | 29.5 | 60.60 | 39.80 |
| 21 | 71 | M | 0 | 45.9 | 92.0 | 2.1 | 1 | NEG | 92.1 | NEG‡ | N | NEG‡ | 66.60 | 41.60 |
| 22 | 61 | F | 0 | 23.0 | 71.0 | 2.9 | 0 | 3.65 | 100.0 | NEG | ND | NEG | 78.60 | 78.10 |
| 23 | 61 | M | 0 | 33.8 | 74.0 | 2.8 | 1 | NEG | 99.6 | NEG | Y | NEG | 27.40 | 26.30 |
| 24 | 41 | M | 0 | 49.0 | 95.0 | 2.8 | 1 | NEG | 96.6 | NEG | N | NEG | 77.50 | 76.00 |
| 25 | 51 | M | 0 | 29.0 | 68.0 | 1.8 | 1 | ND | 99.7 | NEG | ND | NEG | 34.40 | 36.50 |
| 26 | 49 | F | 0 | 24.0 | 81.0 | 2.4 | 1 | POS | 95.3 | NEG | N | NEG | 61.00 | 57.30 |
| 27 | 51 | F | 0 | 10.0 | 68.0 | 1.6 | 0 | 20.5 | ND | NEG | ND | NEG | 85.70 | 85.90 |
| 28 | 85 | M | 1 | 10.4 | 51.0 | 7.2 | 4 | NEG | 93.1 | NEG | N | NEG | 85.30 | 78.00 |
| 29 | 65 | M | 0 | 91.9 | 90.0 | 5.2 | 4 | 0.91 | 96.3 | NEG | Y | 79 | 82.70 | 71.40 |
| 30 | 78 | F | 0 | 35.7 | 68.0 | 2.6 | 2 | 30.3 | 100.0 | NEG | ND | NEG | 46.80 | 44.80 |
| 31 | 59 | M | 0 | 94.7 | 84.0 | 5.3 | 1 | POS | 93.5 | NEG | N | NEG | 83.00 | 83.00 |
| 32 | 48 | M | 1 | 64.9 | 91.0 | 5.0 | 4 | NEG | 97.5 | NEG | N | NEG | 64.60 | 66.30 |
| 33 | 67 | M | 5 | 43.1 | 99.0 | 4.3 | 4 | POS | 100.0 | 88 | ND | NEG | 61.30 | 57.10 |
| 34 | 59 | M | 2 | 14.2 | 47.0 | 4.1 | 4 | 10.8 | 100.0 | NEG | ND | NEG | 43.20 | 14.80 |
| 35 | 66 | M | 0 | 145.5 | 89.0 | 5.8 | 1 | ND | 100.0 | 90.5 | ND | NEG | 62.00 | 17.90 |
| 36 | 50 | F | 0 | 22.7 | 86.0 | 2.5 | 2 | 29.7 | 100.0 | NEG | ND | NEG | 59.46 | ND |
| 37 | 78 | F | 0 | 53.7 | 91.0 | 2.6 | 2 | 30.3 | 99.6 | NEG | ND | NEG | 38.99 | ND |
| 38 | 52 | M | 0 | 40.7 | 84.0 | 4.1 | 2 | 25.1 | 100.0 | 13 | N | NEG | 58.68 | ND |
| 39 | 71 | F | 0 | 150.5 | 96.0 | 4.1 | 0 | 0.72 | 100.0 | DEL | ND | NEG | 54.40 | 54.10 |
| 40 | 60 | M | 0 | 37.5 | 75.0 | 2.7 | 2 | 8.88 | 91.5 | NEG | ND | NEG | 51.20 | 54.40 |
| 41 | 44 | F | 0 | 15.2 | 76.0 | 1.7 | 0 | 21.4 | ND | NEG | N | NEG | 60.30 | 50.10 |
| 42 | 70 | F | 0 | 22.6 | 84.0 | 2.1 | 0 | 18.2 | 100.0 | NEG | N | NEG | 83.80 | 47.10 |
| 43 | 65 | M | 3 | 110.7 | 96.0 | 3.2 | 2 | 0.9 | 96.6 | NEG | N | NEG | 75.30 | 70.40 |
| 44 | 48 | M | 0 | 72.2 | 86.0 | 2.2 | 2 | POS | 99.3 | NEG | N | NEG | 86.20 | 84.70 |
| 45 | 66 | M | 0 | 151.4 | 93 | 6.2 | 1 | NEG | 100.0 | 94 | N | NEG | 81.20 | 19.90 |
| 46 | 50 | M | 0 | 115.3 | 92.0 | 2.6 | 1 | POS | 100.0 | 93 | N | NEG | 89.00 | 61.50 |
| 47 | 88 | F | 0 | 47.9 | 81 | ND | 0 | ND | ND | N | Y | N | 75.00 | 44.00 |
| 48 | 77 | M | 1 | 73.4 | 98 | 4.6 | 4 | NEG | 94.9 | ND | ND | ND | 84.00 | 71.00 |
| 49 | 53 | F | 1 | 62.9 | 94 | ND | 3 | POS | 100.0 | ND | ND | ND | 56.00 | 40.00 |
11q22 indicates percentage of cells positive for this chromosomal deletion; del 13, cytogenetic analyses for del of chromosome 13 or 13q14; and 17p13, percentage of cells positive for this chromosomal deletion.
β2M indicates beta-2-microglobulin; WBC, white blood cell; M, male; ND, not determined; F, female; POS, positive; NEG, negative; PB, peripheral blood; Y, yes; and N, no.
98% or greater homology indicates unmutated; less than 98% homology, mutated.
CLL cells were incubated in vitro without (untreated) or with 2μM forodesine and 20μM dGuo and percentage of viability was measured by DiOC6 assay.
Done in 2006.
Effect of MSCs on forodesine and dGuo-induced CLL cell apoptosis
To test if forodesine displays activity in the presence of MSCs, we cocultured CLL cells with murine or human MSCs (M210B4 and NKtert, respectively, which provide conditions representing bone marrow microenvironment) and compared the results to CLL controls cultured under identical conditions, but incubated without MSCs. There was no phenotypic change in the CLL cells grown on MSCs. We found that MSCs protected CLL cells from spontaneous and forodesine-induced apoptosis. At 24 hours, CLL cells in suspension culture had a mean percentage of viability of 69%, which increased to 73% when cocultured on M210B4 and 82% with NKtert (C vs CM, P = .034; C vs CN, P < .001). Likewise, the stromal cells also abrogated the forodesine-induced apoptosis in CLL cells. Forodesine-treated CLL suspension cells at 24 hours had a mean percentage of viability of 54%, which increased to 62% with M210B4 and 70% with NKtert (n = 15). Data from 12 individual patients is shown in Figure 1C (CF vs CMF, P < .005; CF vs CNF, P < .001). For the same patients with the extended incubation time (48 hours; n = 12), there was further decrease in the viability of CLL cells treated with forodesine and time-dependent abrogation of spontaneous and forodesine-induced cell death in the presence of MSCs. Forodesine-treated CLL suspension cells at 48 hours had a mean percentage of viability of 47%, which increased to 58% on M210B4 and 70% on NKtert (CF vs CMF, P < .001; CF vs CNF, P < .001; n = 12; Figure 1D).
Influence of microenvironment on the forodesine-induced accumulation of dGTP in CLL cells
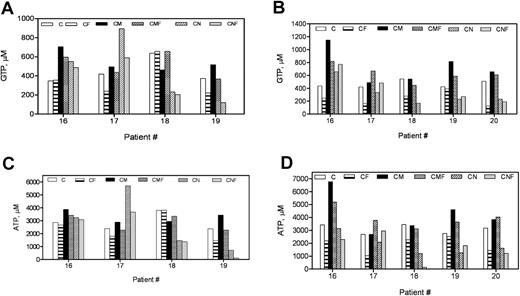
Our previous studies demonstrated that dGuo, which is added with forodesine in vitro, accumulates as dGTP in the leukemic cells and is proportional to the level of apoptosis.19 To test if MSCs can modulate the accumulation of dGTP in malignant B lymphocytes, CLL cells incubated with or without forodesine in the presence or absence of MSCs were analyzed for all 4 dNTPs by DNA polymerase assay. Consistent with our previous work, the endogenous dGTP levels in CLL lymphocytes were mean 5μM, which increased to mean 15μM with the incubation of forodesine and dGuo (24 hours; P = .023; n = 5; Figure 2A). There was heterogeneity among patients regarding increase in dGTP in CLL cells (Figure 2A-B). There was no significant change in the levels of other dNTPs such as dATP, dCTP, and TTP (supplemental Figure 1A-C, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). After 48 hours of drug incubation, the dGTP accumulation further increased to a mean 25μM (P = .009; n = 5; Figure 2B), but with no change in other dNTPs (supplemental Figure 1D-F). When the same parameters were tested in the presence of MSCs, the endogenous dGTP levels in CLL cells was not modulated by stromal cells; however, both stromal cells were able to attenuate the extent of dGTP accumulation promoted by forodesine and dGuo in a time-dependent fashion (CF vs CMF, P = .021; CF vs CNF, P = .039; 48 hours; n = 5; Figure 2B).
dGuo in presence of forodesine is accumulated as dGTP in CLL cells, and the accumulation decreased in presence of bone marrow stromal cell lines. CLL cells (n = 5) were incubated without or with 2μM forodesine and 20μM dGuo in suspension or on MSCs for 24 hours (A) or 48 hours (B), and the accumulation of dGTP was measured by DNA polymerase assay in treated and untreated time-matched controls. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
dGuo in presence of forodesine is accumulated as dGTP in CLL cells, and the accumulation decreased in presence of bone marrow stromal cell lines. CLL cells (n = 5) were incubated without or with 2μM forodesine and 20μM dGuo in suspension or on MSCs for 24 hours (A) or 48 hours (B), and the accumulation of dGTP was measured by DNA polymerase assay in treated and untreated time-matched controls. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Influence of MSCs on forodesine-induced perturbation of NTP pool
We next tested for NTP pools in CLL cells by measuring GTP, ATP, CTP, and UTP levels. Interestingly, in contrast to dGTP increase, there was a time-dependent decline in the purine nucleoside triphosphates. Of 4 patients tested, with treatment of forodesine, the GTP and ATP concentrations decreased in 2 patients (nos. 17 and 19; GTP at 24 hours, 57% and 59% of control; ATP at 24 hours, 75% and 62% of control; Figure 3A,C), but no change was observed in the other 2 patient samples. By 48 hours there was a significant decline in the GTP (C vs CF, P = .018) and ATP (C vs CF, P = .011) in all samples tested (48 hours; n = 5; Figure 3B,D). In the presence of M210B4 stromal cells, in 3 of 5 patients tested (nos. 16, 19, and 20) the endogenous GTP and ATP levels increased in a time-dependent manner and remained similar to control after the treatment with forodesine (48 hours: CF vs CMF, GTP, P = .01; ATP, P = .009). NKtert stromal cells did not modulate the NTP pool in these patients. Unlike GTP and ATP intracellular concentrations, there was no change in CTP or UTP levels (supplemental Figure 2A-D).
Forodesine in presence of dGuo lowered the GTP and ATP levels in CLL cells, while the same NTPs increased when cocultured with MSCs. CLL cells (n = 5) were incubated without or with 2μM forodesine and 20μM dGuo in suspension or on MSCs for 24 hours (A,C) or 48 hours (B,D), and the levels of GTP (A-B) and ATP (C-D) were measured by HPLC. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Forodesine in presence of dGuo lowered the GTP and ATP levels in CLL cells, while the same NTPs increased when cocultured with MSCs. CLL cells (n = 5) were incubated without or with 2μM forodesine and 20μM dGuo in suspension or on MSCs for 24 hours (A,C) or 48 hours (B,D), and the levels of GTP (A-B) and ATP (C-D) were measured by HPLC. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Effect of forodesine and dGuo on the global RNA and protein synthesis in CLL cells
CLL cells were incubated with forodesine and dGuo, and the total RNA and protein synthesis were measured by the [3H]uridine or [3H]leucine incorporation method, respectively (Figure 4A-B). Forodesine inhibited global RNA synthesis by a mean 68% of control (n = 9). Coculturing with MSCs generally induced an increase in RNA synthesis in CLL cells (C vs CM, P = .003, n = 9; C vs CN, P = .009, n = 10; Figure 4A). There was a trend for inhibition of protein synthesis by forodesine and dGuo, but this was not statistically significant (Figure 4B). These data suggest that forodesine actions on CLL cells are directed toward RNA synthesis inhibition, and the MSC-derived protection of CLL cells was associated with an increased transcriptional signaling, more than toward protein synthesis.
Forodesine in presence of dGuo inhibited the global RNA and protein synthesis in CLL cells, while MSCs augmented the macromolecule syntheses. (A-B) CLL cells (n = 10) were incubated with or without 2μM forodesine and 20μM dGuo in suspension or on MSCs for 48 hours. The global RNA synthesis was measured by uridine incorporation assay in treated and untreated time-matched controls (A), and the global protein synthesis was measured by leucine incorporation assay in treated and untreated (n = 5) time-matched controls (B). C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Forodesine in presence of dGuo inhibited the global RNA and protein synthesis in CLL cells, while MSCs augmented the macromolecule syntheses. (A-B) CLL cells (n = 10) were incubated with or without 2μM forodesine and 20μM dGuo in suspension or on MSCs for 48 hours. The global RNA synthesis was measured by uridine incorporation assay in treated and untreated time-matched controls (A), and the global protein synthesis was measured by leucine incorporation assay in treated and untreated (n = 5) time-matched controls (B). C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Effect of forodesine and dGuo on Mcl-1 mRNA and protein levels
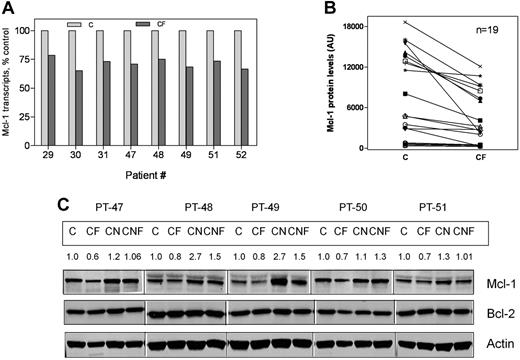
The observation that forodesine induced inhibition of total RNA and protein synthesis in CLL cells led us to postulate that this would likely affect short-lived transcripts and proteins such as Mcl-1. A 21% to 35% decrease in the Mcl-1 transcript was observed after 48 hours of treatment with forodesine and dGuo (n = 8; Figure 5A; P < .001). In concordance with the decline in transcript levels, there were consistently low Mcl-1 protein levels in all 19 CLL samples treated with forodesine (P < .001, n = 19; Figure 5B). In contrast, the CLL cells from 5 patients cocultured on NKtert stromal cells showed induction of Mcl-1 protein (n = 5; Figure 5C); however, in the presence of stromal cells, the decrease in Mcl-1 levels was compromised by MSCs.
Reduction in Mcl-1 transcript and protein levels in CLL cells after treatment with forodesine and dGuo. CLL cells were incubated with or without 2μM forodesine and 20μM dGuo in suspension or on MSCs. The levels of Mcl-1 transcript (n = 8, A), and Mcl-1 protein (n = 19, B), were measured by real time RT-PCR and immunoblotting analysis, respectively. The Western blot images from 5 representative patients for Mcl-1 and Bcl-2 proteins (C). Actin was used as loading control. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Reduction in Mcl-1 transcript and protein levels in CLL cells after treatment with forodesine and dGuo. CLL cells were incubated with or without 2μM forodesine and 20μM dGuo in suspension or on MSCs. The levels of Mcl-1 transcript (n = 8, A), and Mcl-1 protein (n = 19, B), were measured by real time RT-PCR and immunoblotting analysis, respectively. The Western blot images from 5 representative patients for Mcl-1 and Bcl-2 proteins (C). Actin was used as loading control. C indicates CLL; F, forodesine plus dGuo; M, M210B4; and N, NKtert.
Influence of ZVAD.fmk, a pancaspase inhibitor, on forodesine-induced apoptosis
Chemotherapeutic agents induce apoptosis through caspase-dependent or -independent processes. To determine whether forodesine-induced apoptosis involves activation of caspases, CLL cells (n = 5; 48 hours) were incubated with or without 50μM ZVAD.fmk, and the percentage of viable cells was measured by annexin assay for detection of apoptotic and dead cells (supplemental Figure 3A). ZVAD.fmk protected CLL cells from spontaneous and forodesine-induced apoptosis, suggesting that forodesine-induced apoptosis is caspase dependent (mean percentage viability: C, 52%; CZ, 60%; CF, 32%; CZF, 42%; CF vs CZF P = .047).
The percentage of viability after forodesine treatment measured by DiOC6 (supplemental Figure 3B; based on the analysis of mitochondrial transmembrane potential) did not differ significantly in the presence or absence of ZVAD.fmk, suggesting that the mitochondrial transmembrane potential is an earlier event in the cell death process and is not affected by ZVAD.fmk (mean percentage viability: C, 57%; CZ, 58%; CF, 43%; CZF, 40%; CF vs CZF P = .558). Similar to mitochrondrial outer membrane permeabilization (MOMP), forodesine-mediated inhibition of global RNA and protein synthesis (n = 5; P = .005) was not affected by caspase inhibitor (global RNA synthesis inhibition; F vs ZF, P = .056; protein synthesis inhibition; F vs ZF, P = .229; n = 5). Again, these events were upstream of the apoptosis (supplemental Figure 3C-D).
Effect of forodesine treatment on MSCs
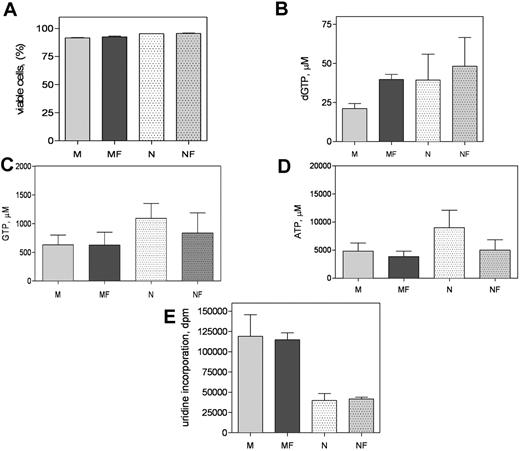
Because forodesine induced apoptosis in CLL cells, and this was abrogated by MSCs, we wanted to test if forodesine also affected the MSCs. Both M210B4 and NKtert MSCs were incubated with 2μM forodesine and 20μM dGuo; apoptosis was measured by DiOC6 assay. Interestingly, forodesine at the concentrations that induced apoptosis in CLL cells did not induce apoptosis in stromal cells (Figure 6A; M vs MF, P = .18; N vs NF, P = .51). In contrast to CLL cells, which are replicationally quiescent, M210B4 and NKtert are highly proliferating cells and have higher endogenous levels of dGTP (mean, 21μM in M210B4 and 39μM in NKtert). Similar to CLL cells, M210B4 cells accumulated significant amounts of dGTP (P < .001; Figure 6B), while NKtert cells had a modest increase in dGTP (P = .055; Figure 6B). In contrast to dGTP, the NTP levels such as GTP and ATP did not change in MSCs with forodesine treatment (Figure 6C; GTP, M vs MF, P = .725; N vs NF, P = .9; Figure 6D; ATP, M vs MF, P = .686; N vs NF, P = .673). Similarly, the global RNA synthesis in stromal cells was not affected after treatment with forodesine and dGuo (Figure 6E). These sets of experiments illustrate that the action of forodesine is largely restricted to CLL lymphocytes, whereas MSCs are not affected by this PNP inhibitor.
Murine and human MSC response to forodesine and dGuo. MSCs were treated without or with forodesine and dGuo for 48 hours. The apoptosis was measured by DIOC6 assay (A), dGTP accumulation was analyzed (B), GTP and ATP levels were quantitated (C-D), and the global RNA and protein synthesis were measured (E). Data in panel A represent mean + SD from 3 independent experiments and data in panels B through E represent mean + SEM from 5 independent experiments. M indicates M210B4; F, forodesine; and N, NKtert.
Murine and human MSC response to forodesine and dGuo. MSCs were treated without or with forodesine and dGuo for 48 hours. The apoptosis was measured by DIOC6 assay (A), dGTP accumulation was analyzed (B), GTP and ATP levels were quantitated (C-D), and the global RNA and protein synthesis were measured (E). Data in panel A represent mean + SD from 3 independent experiments and data in panels B through E represent mean + SEM from 5 independent experiments. M indicates M210B4; F, forodesine; and N, NKtert.
Discussion
Though many chemotherapeutic agents display efficacy in preclinical studies, their activity varies to a greater extent in the clinical settings. For leukemia cells and selected other cancer cells,22 this may be because the microenvironment in the marrow and secondary lymphoid tissues provides drug-resistance signals to the leukemia cells, a primary drug resistance mechanism that may account for residual disease after conventional treatments and disease persistence in the marrow. The marrow is invariably infiltrated with CLL cells, and the pattern and the extent of marrow involvement correlates with clinical stage and prognosis.29-32 We previously demonstrated that stromal cells protected CLL cells from fludarabine (a purine nucleoside analog)–induced apoptosis as an example of cell adhesion–mediated drug resistance.23,24 In contrast, CLL cells were equally sensitive to a BH3 mimetic (AT-101) when grown alone or in cocultures with MSCs.24
Previously, we demonstrated that forodesine (2μM), a PNP inhibitor, in the presence of dGuo (10μM) induces apoptosis in CLL cells.19 Based on the encouraging preclinical data, a phase 2 study of forodesine in patients with fludarabine refractory CLL was initiated. Because the mechanism of action of forodesine (with dGuo) is different from fludarabine or AT-101, and because preliminary clinical data indicated that CLL cells persisted in the marrow, after treatment with forodesine,20 we initiated the current project to determine whether and how the microenvironment affects forodesine-mediated biochemical changes leading to CLL cell death. Using 2 established MSC lines, we investigated if forodesine with dGuo could overcome CAM-DR, or if there is MSC-dependent resistance. When CLL cells were cocultured on murine (M210B4) or human (NKtert) MSCs, forodesine-promoted apoptosis was significantly inhibited (Figure 1C-D).
Previous studies with forodesine in T-cell leukemia lines demonstrated that accumulation of dGTP is a necessary step for its activity.17,33,34 In addition, the association between dGTP increase in T-leukemia cells and cytoreduction was observed during clinical trial.18 During in vitro incubations in T-leukemia cell lines as well as in primary T cells obtained from patients with acute lymphoblastic leukemia (ALL), there was an increase in dATP levels.18,34 This was similar in primary T cells from patients with ALL and also during therapy. Similar to T-leukemia cells, our current study also elucidated that there was an increase in the level of dGTP in CLL B cells, which was proportional to hallmark features of apoptosis.19 However, in these quiescent cells there was no increase in dATP levels (supplemental Figure 1). When dGTP levels were measured after coculturing on stromal cells, MSCs were able to reverse the increase of dGTP in CLL cells (Figure 2A-B). Although the mechanism is not known, this observation further confirms that the accumulation of dGTP is a necessary step in the forodesine-induced apoptosis in CLL cells. As expected, other dNTPs did not show any significant changes with forodesine in solo cultures or in presence of MSCs (supplemental Figure 2A-F).
Because forodesine would also affect phosphorolysis of Guo, this should tip the balance between the NTP levels. Hence we analyzed the NTP levels in CLL cells treated with forodesine. Interestingly, instead of increase in intracellular GTP, we noticed that there was a 25% to 50% decrease in the ATP and GTP levels. This was a paradoxical observation without a known mechanism (Figure 3A-D). Because the ribonucleoside triphosphates (GTP and ATP), the building units of RNA, were declined, we expected that the RNA synthesis might also be inhibited with forodesine treatment. There was a significant drop in the RNA synthesis after forodesine and dGuo incubations (68%; n = 9; Figure 4A). CLL cells had an increase in global RNA synthesis when cocultured with MSCs. Although there was a subsequent decrease in RNA synthesis, when these cocultured cells were treated with forodesine and dGuo, the declined level was similar or higher than the value obtained in untreated (control) CLL cells (Figure 4A). These data suggest that MSC-mediated increase in RNA synthesis promotes CLL cell survival.
Investigations on intrinsic ability of CLL cells to escape from apoptosis35,36 and the assessment of individual antiapoptotic protein molecules37 have led to the notion that though 14;18 chromosomal translocation is rare in CLL, the Bcl-2 protein is consistently overexpressed in CLL cells when compared with CD19+ B-cell fractions purified from healthy donors. However, there was an inverse relationship between Bcl-2 protein expression and miR15-a and miR16-1. These miRs are deleted or down-regulated in most CLL, leading to overexpression of Bcl-2 protein.38 Thus, the overall pattern of the BCL-2 gene family expression is tuned to shield CLL cells from undergoing cell death.35,39 These observations suggested that the selected signals in the microenvironment that favor CLL survival may tip the balance between antiapoptotic and proapoptotic signals40 by up-regulating antiapoptotic proteins such as Mcl-1 and Bcl-2.41,42 Our previous studies also showed that Mcl-1 protein is induced significantly in CLL cells coincubated with stromal cells,24,43 and there is a relationship between Mcl-1 protein levels in CLL cells and in vivo and in vitro response to therapy.44,45 Other studies convincingly showed that physical contact between BM stromal cells and leukemic cells extends the survival of CLL cells.31,32,46 It would be of interest to test what roles other cells, such as cytotoxic T lymphocytes and dendritic cells, play in forodesine-induced apoptosis.
Consistently, with forodesine incubation, Mcl-1 transcript (Figure 5A) and protein (Figure 5B-C) levels dropped significantly in these leukemic lymphocytes. This was not the case when CLL cells treated with forodesine and dGuo in coculture conditions. Global transcription in CLL was robustly increased in presence of stromal cells, suggesting that this is a more important mechanism for CLL cell extended survival.47 Furthermore, transcription is shown to be a good target because transcriptional inhibitors such as C8-substituted adenosine analogs,28 flavopiridol,48 SNS-032,49 and roscovitine50 have been shown to be effective therapeutics for CLL cells in vitro and during therapy.
Finally, we analyzed the actions of forodesine on MSCs; interestingly, forodesine did not induce apoptosis in both MSC lines (Figure 6A). However, dGTP accumulation was to a similar extent in murine M210B4 stromal cells, but there was not much increase by human NKtert stromal cells as that of CLL cells, which suggests that dGTP accumulation is necessary for the forodesine-induced apoptosis, but not sufficient to induce apoptosis, and that there are additional mechanisms involved in this process. Moreover, we do not know the endogenous PNP levels in these stromal cells. Data with these stromal cells are consistent with recent results in normal lymphocytes.51 Collectively, the MSC data (from current work) and normal lymphocytes51 suggested that forodesine has some selectivity to CLL lymphocytes while sparing normal cells.
In conclusion, forodesine selectively kills CLL cells in suspension; however, primary drug resistance develops when CLL cells are cultured in the presence of MSCs. These observations may provide an explanation for the reduced clinical activity of forodesine in the marrow compartment compared with the peripheral blood. In addition, these results provide a rationale to further explore strategies that are aimed to disrupt the cross-talk between CLL cells and the marrow microenvironment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants CA136411 (Lymphoma SPORE) and P30-16672 Cancer Center Support Grant from the National Cancer Institute, Department of Health and Human Services, and a sponsored research agreement from MundiPharma International Limited to V.G. and J.A.B.
National Institutes of Health
Authorship
Contributions: K.B. designed and performed most experiments, analyzed data, and wrote the manuscript; J.A.B. provided expertise in stromal cell coculturing and participated in manuscript writing; M.P.Q. and M.H. performed the stromal coculture incubations and apoptosis experiments; M.L.A. performed the stromal coculture incubations; W.G.W. identified CLL patients for inclusion in the study and is principal investigator of the laboratory protocol to obtain blood samples; and V.G. conceptualized the research, directed K.B. in experiment design and data analysis, and finalized the text of the manuscript.
Conflict-of-interest disclosure: J.A.B. and V.G. have received research funding from MundiPharma International Limited. The remaining authors declare no competing financial interests.
Correspondence: Varsha Gandhi, Department of Experimental Therapeutics, Unit 71, The University of Texas M. D. Anderson Cancer Center, Houston, TX 77030; e-mail: vgandhi@mdanderson.org.