In this issue of Blood, Rossi and colleagues have identified a miR profile associated with 17p deletion in CLL, which may be used to further subdivide prognostic groups in CLL.1 Although the deletion of 17p is associated with poor outcome in different malignancies (acute myeloid leukemia, myeloma, CLL), its impact on outcome is particularly striking in CLL.2 It is therefore crucial to expand our understanding of the mechanisms underlying the impact of the 17p deletion in this subgroup of patients. Similarly, what other defect(s) could be a “phenocopy” of CLL with 17p deletion?
Despite the poor prognosis of most patients with 17p deletion, a small subgroup of chronic lymphocytic leukemia (CLL) cases with 17p deletion may be relatively static, not requiring treatment for years with relatively long survival.3,4
The authors have addressed the possibility of improving on current prognostic models in CLL by assessing the expression of a set of miRs in CLL.5,6 Similar approaches in a variety of solid tumors are under way using circulating miRs as the diagnostic (and potentially therapeutic) target. Because CLL is a disease of the blood, access to large quantities of sample should promote development, even if clonal heterogeneity could be problematic for expression-based assays. The authors have studied 2 cohorts that were quite disparate in that the initial cohort was selected for a high proportion of 17p deletion cases, while the validation cohort only included a few cases with 17p deletion. This suggests that the findings could be important independent of 17p deletions (see figure).
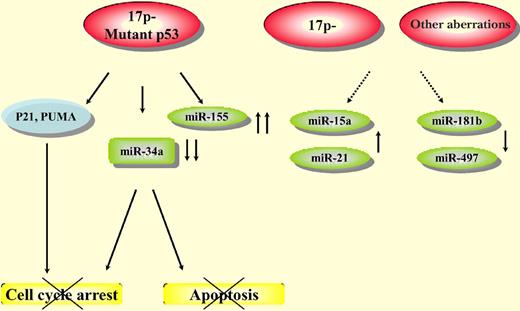
Model on miRs deregulated in CLL with 17p deletion. p53 targets are deregulated in cases with 17p deletion and TP53 mutation. Based on the work presented in this issue, additional miRs (miR-21, miR-181, miR-155, miR-497, and miR-15a) are differentially expressed in CLL with and without 17p deletion. Downstream targets potentially mediating clinical consequences of deregulation of these miRs in CLL are currently unknown.
Model on miRs deregulated in CLL with 17p deletion. p53 targets are deregulated in cases with 17p deletion and TP53 mutation. Based on the work presented in this issue, additional miRs (miR-21, miR-181, miR-155, miR-497, and miR-15a) are differentially expressed in CLL with and without 17p deletion. Downstream targets potentially mediating clinical consequences of deregulation of these miRs in CLL are currently unknown.
Rossi and colleagues were able to show that miRs correlated with 17p status and specifically were down-regulated (miR-34a, miR-181b, miR-497) or up-regulated (miR-15a, miR-21, miR-155) in CLL with 17p deletion. In addition, expression of miR-21 and miR-181b was shown to be associated with overall and progression-free survival, respectively. These data add important information to other recently published studies showing that miR-34a, miR-29c, miR-17-5p, and miR 151-3p are differentially expressed based on 17p status.7-9 To address the mechanisms underlying the deregulation of the miR, the authors have taken advantage of a cell line model where p53 was silenced to assess whether the miRs are direct p53 targets. In accordance with prior studies, the authors show evidence that both miR-34a and miR-155 are dependent on p53 as these were up-regulated (miR-34a) and down-regulated (miR-155) after silencing of p53. These findings pose the important question of whether the miRs not demonstrated to be directly p53 dependent are regulated by alternative genes on 17p or other important biologic pathways in CLL (immunoglobulin heavy-chain variable-region gene mutation status, B-cell receptor function). Although most 17p deletions target a large region on 17p, the TP53 locus is always affected and most cases with 17p deletion will have a TP53 mutation on the remaining allele. This observation, as well as the observation that CLL with TP53 mutation shows a similar outcome as cases with 17p deletion, suggests that p53 is central to the clinical course of CLL with 17p deletion.2,10
It is therefore intriguing that the model build based on miR-21 expression and 17p deletion also held up in a cohort with a low incidence of 17p. This finding raises the question of how miR-21 is regulated in cases with intact p53 status.
As with all important research there are many questions raised by the work of Rossi and colleagues. It will also be important to characterize the independent impact of miR-21 expression in models including 17p and TP53 mutation. Before translating the results into clinical practice, it will be important to confirm the impact of the deregulated miRs in a uniformly treated cohort; ideally, this should be done with purified CLL samples derived from a prospective trial. This is particularly important as the follow-up is relatively short (20 months) and the cohorts studied diverse.
Although the current work does not attempt to identify the basis for the association between miR-21 expression and outcome, the authors hint at some potential targets. It will be important to understand the function of these miRs and, particularly, how they relate and depend on functional p53. A first step toward this goal will be the detailed characterization of not only 17p status, but also TP53 mutation.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal