Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression and have been implicated in the pathogenesis of cancer. In this study, we applied next generation sequencing techniques to comprehensively assess miRNA expression, identify genetic variants of miRNA genes, and screen for alterations in miRNA binding sites in a patient with acute myeloid leukemia. RNA sequencing of leukemic myeloblasts or CD34+ cells pooled from healthy donors showed that 472 miRNAs were expressed, including 7 novel miRNAs, some of which displayed differential expression. Sequencing of all known miRNA genes revealed several novel germline polymorphisms but no acquired mutations in the leukemia genome. Analysis of the sequence of the 3′-untranslated regions (UTRs) of all coding genes identified a single somatic mutation in the 3′-UTR of TNFAIP2, a known target of the PML-RARα oncogene. This mutation resulted in translational repression of a reporter gene in a Dicer-dependent fashion. This study represents the first complete characterization of the “miRNAome” in a primary human cancer and suggests that generation of miRNA binding sites in the UTR regions of genes is another potential mechanism by which somatic mutations can affect gene expression.
Introduction
MicroRNAs (miRNAs) are endogenous small noncoding RNAs that regulate gene expression posttranscriptionally by binding of target mRNAs to regulate their stability or translation.1 miRNAs are transcribed as long primary miRNA (pri-miRNA) transcripts that are processed into precursor hairpin intermediates (pre-miRNA) and then to 19-27 nucleotide mature miRNAs through a complex and highly regulated biogenesis process. The mature miRNA binds to semicomplementary bases in the target mRNA, usually in the 3′-untranslated region (3′-UTR) but also occasionally in 5′ untranslated region (5′-UTR)2,3 or coding regions of target mRNAs.4 Each miRNA is believed to target multiple mRNAs, thereby providing a critical posttranscriptional mechanism to regulate diverse cellular functions including development, proliferation, and apoptosis.5
Accumulating evidence suggests that mutation of miRNAs may contribute to the pathogenesis of cancer. Genes encoding miRNAs are often located in fragile sites and regions of chromosomal aberrations in cancer.6 A survey of miRNA genes by array-based comparative genome hybridization in breast, ovarian, and melanoma samples showed that regions containing miRNAs were often targeted for amplification, loss of heterozygosity, and structural breakpoints in tumors.7 In chronic lymphocytic leukemia (CLL), for example, deletion of chromosome 13q14, the most common cytogenetic abnormality in CLL, results in loss of miR-16-1 and miR-15a, which are negative regulators of bcl2.8 Interestingly, no somatic single nucleotide mutation of miRNA genes in primary human cancer has been reported to date.
miRNA expression profiling has identified deregulated miRNAs in many different types of cancer.9 In leukemia, expression profiles of just 4 miRNAs can reliably distinguish between acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL).10 In AML, miRNA signatures can also distinguish between cytogenetic subtypes11-14 and even molecular subtypes (presence of NPM1, CEBPA, or FLT3 mutations).12 miRNA expression patterns may have prognostic significance in cancer. For example, in cytogenetically normal high-risk AML patients, the expression pattern of 12 specific miRNAs is associated with improved survival.15
miRNA expression profiling, while providing important information, does not interrogate all known (or predicted) miRNAs and is unable to detect mutations in miRNAs. Herein, we used next generation sequencing technologies to comprehensively assess miRNA expression, miRNA mutations, and miRNA binding site mutations in a patient with AML. RNA sequencing showed that 472 miRNAs were expressed in the AML sample (including 7 novel miRNAs), some of which were differentially expressed compared with normal CD34+ cells. Massive parallel sequencing of all known miRNA genes revealed several novel germline polymorphisms but no acquired mutations. Finally, we analyzed the previously generated whole-genome sequencing data for this AML genome16 to detect somatic mutations in the UTR of all coding genes. A single mutation in the putative tumor suppressor gene TNFAIP2 was detected. We provide evidence that this mutation generates a new miRNA binding site that leads to translational repression of TNFAIP2. To the best of our knowledge, this represents the first description of somatic mutation affecting gene expression through the generation of a new miRNA binding site.
Methods
Human subjects
All AML samples were obtained from a study at Washington University to identify genetic factors contributing to AML initiation and progression. In addition, bone marrow samples were obtained from healthy volunteers. Approval was obtained from the Washington University Institutional Review Board for these studies. After obtaining written informed consent in accordance with the Declaration of Helsinki, a bone marrow sample was obtained. In addition, for patients with AML, a 6-mm punch biopsy of skin was obtained for analysis of unaffected somatic cells.
miRNA sequencing
RNA was prepared from bone marrow sample from patient 933124 (AML1)16 (> 95% blasts) using Trizol reagent (Invitrogen). CD34+ cells were isolated from the bone marrow of 5 healthy donors using MACS according to the manufacturer's instructions (Miltenyi Biotec), and RNA was prepared using Trizol reagent. Small RNA libraries were constructed from 508 ng CD34+ cell and 789 ng AML1 bone marrow RNA using the SOLiD small RNA expression kit according to the manufacturer's instructions (Applied Biosystems). The amplified cDNAs were resolved on a 6% DNA Retardation Gel (Invitrogen), and cDNA fragments between 104-120 bp, corresponding to miRNAs between 19 and 26 nucleotides in length were excised. DNA was eluted from the gel slices by incubation overnight in 400mM NaCl and precipitated by adding 2.5 volumes of 100% ethanol and 15 μg GlycoBlue (Ambion), incubating at −20°C for 4 hours, and centrifuging at 12 000g for 30 minutes at 4°C. The resulting libraries were amplified onto beads using emulsion polymerase chain reaction (PCR), deposited on slides, and sequenced using the SOLiD v 2 sequencing system (Applied Biosystems). Post run filters found 27.9 and 20.7 million AML1 and CD34 pooled beads, respectively. All sequence data were deposited in the National Center for Biotechnology Information (NCBI)'s short read archive (study accession no. SRP002249).
Novel miRNA discovery
SOLiD reads were mapped to the Hs36 reference sequence using Maq17 in colorspace mode. The resulting read alignments were clustered and assessed for coverage using RefCov, which provides a topologic representation of alignments at each nucleotide of a cluster and statistics to represent the depth of maximal alignment. Some 72 433 clusters in AML1 and 34 320 clusters in CD34 cells had at least 10 supporting reads in the SOLiD data; the read depth per cluster ranged from 302 to 626 334 for AML1 clusters and from 161 to 500 220 for CD34 clusters. The top 1000 clusters (by coverage) in both AML1 and CD34 cells were combined together (yielding 1488 unique clusters), and their RNA secondary structure analyzed using an in-house validation tool (miRNAViewer). In brief, this tool simultaneously provides cluster information (cluster genome location, sequence, zenith depth, annotation of genome location), MFold-based secondary structure predictions,18 and RefCov alignment coverage maps. In addition, the tool allows for flexible adjustment of the candidate precursor location to allow for manual changes to the cluster sequence genomic window based on the initial folding and read alignment results. Criteria used to identify novel miRNAs are detailed in the Results section. Conservation scores for individual bases were determined using phylogenetic-hidden Markov model and the University of California Santa Cruz Genome Browser Database, as described previously.19 For miRNA clusters, conservation scores were calculated as the mean of positional scores across the cluster region.
Expression profiling of known miRNAs
The hairpin, mature, and mature* sequences for known human miRNA genes were downloaded from miRBase release 13.0.20-22 To determine relative expression of known miRNAs, SOLiD reads were mapped to the human hairpin sequences using SHRiMP v1.2.23 Alignments were filtered to remove multiple/ambiguous read placements and hits with probcalc P > .000001. The filtered alignments were compared with the hairpin-based coordinates of known mature, mature*, mature 5P, and mature 3P sequences to obtain digital read counts of miRNA expression for each sequence.
Expression of known miRNAs in the AML1 and CD34 cells samples was also assessed using Agilent miRNA microarrays. Total RNA (100 ng) was labeled using the miRNA Microarray System version 1.5 according to the manufacturer's instructions (Agilent) and hybridized to Agilent prerelease human miRNA arrays (array design ID 018077), containing probes to all miRNAs in miRBase v10.0. Microarrays were scanned with an Agilent DNA Microarray Scanner. Images were gridded and analyzed using Agilent feature extraction software version 9.5.3.1. Data were normalized to the 75% median intensity of each array, and values are reported as a total gene signal of multiple probes for each miRNA. The microarray expression data were submitted to Gene Expression Omnibus; series entry GSE24222.
Real-time reverse transcription PCR of miRNAs
Total RNA was reverse transcribed using the TaqMan microRNA Reverse Transcript Kit per manufacturer's instructions (Applied Biosystems). Real-time PCR for the indicated miRNA and RNU48 (as a control) was performed using the relevant TaqMan MicroRNAassay (Applied Biosystems).
454 sequencing of miRNA genes
Genomic coordinates for all 695 known human miRNA genes at the time this study was initiated were obtained from miRBase (release 12.0). PCR amplicons were designed to sequence approximately 200 bp of sequence flanking each miRNA precursor. These were later supplemented with 18 amplicons from low-coverage regions, bringing the total to 1396 amplicons (average size: 315 bp). PCRs were performed separately to produce 1396 products from whole genome amplified tumor genomic DNA of patient AML1. All PCR products were pooled and sequenced using the 454-XLR platform (Roche). Because sequence coverage was variable, the sequencing was repeated, this time after normalizing amplicon input. The 454 sequence data were combined into a single SFF file (using the sfffile utility) that contained 288 862 reads totaling 72.3 Mbp, with an average read length of 250 bp.
454 sequence analysis and variant detection
Read sequences and quality values were extracted using the sffinfo program (Roche) and aligned to the Hs36 reference sequence using Blat (v32 × 1). We provided a soft-masked reference sequence and the mask = lower parameter, so that repetitive regions were excluded during seeding of alignments but included when extending alignments. The resulting BLAT alignments were screened for single nucleotide variants (SNVs) and insertion/deletions (indels) using VarScan.24 Reads with alignment identity of < 95%, alignment scores of < 50, or 2 possible locations with the highest alignment score were discarded. To avoid artifacts from regions homologous to multiplex identifier primer tails, we excluded variants called within 10 bp of the start or end of the read. To call a variant, we required > 10 × sequence coverage, with > 25% of reads showing the variant allele, and an average base quality of 15.
Variant cross-validation with AML1 whole-genome sequencing data
We examined Illumina SNV data from the previously published whole-genome sequencing of AML1.16 Depth-filtered SNV calls from the tumor sample and unfiltered SNV calls from the matched skin sample were obtained from Maq v0.6.5.17 The read counts supporting each allele in tumor (approximately 40 × coverage) and skin (matched control; approximately 18 × coverage) samples were extracted from Maq map files using the internally developed snp-metrics program. Known human sequence variants were downloaded from public database dbSNP build 129.25 These were parsed to remove entries that were ambiguously mapped, spanned multiple bases, had 3 or more alleles, or were classified as a variant type other than single nucleotide polymorphisms (SNPs).
Variant detection and validation in the UTRs of coding genes in AML1
The previously published whole genome sequencing data were analyzed for sequence variants in the UTRs of all coding genes. A total of 29 166 coding genes were analyzed based on the Ensembl and NCBI/GenBank databases as of August 27, 2007. All sequence variants identified in the whole genome sequencing were independently validated by 454-based sequencing as described previously.16 The TNFAIP2 3′-UTR mutation was screened in 187 other AML patient samples using the same approach.
Bioinformatic prediction of miRNA binding sites
Two miRNA target prediction algorithms, miRanda26 and RNAhybrid,27 were used to scan the wild-type and mutant 3′-UTR of TNFAIP2 for potential miRNA binding sites. Both miRanda and RNAhybrid were run using the default parameters, which includes a minimal folding energy cutoff of −20 kcal. miRNA binding sites identified by these 2 algorithms were further filtered to remove sites with more than one internal loop or more than 2 G-U pairs within the seed region.
Generation of the Dicer knockdown cell line
KLE endometrial cells were maintained in 1:1 Dulbecco modified Eagle medium/Ham F-12 mixture (Invitrogen) containing 10% fetal bovine serum (Atlas), and 1% penicillin/streptomycin (Invitrogen). Lentiviral constructs expressing short hairpin RNAs (shRNAs) against the red fluorescent protein (shRFP: 5′-TGCTAAGGAGTTTGGAGACAA-3′) or against DICER1 (shDCRA: 5′-GCTCGAAATCTTACGCAAATA-3′) were generated as previously described.28,29 Virus production and infections were carried out according to established methods.29 After infection, cells were selected with 1 μg/mL puromycin, and DICER1 protein expression assessed by immunoblotting using rabbit polyclonal anti-DICER1 H212 (sc-30 226; Santa Cruz Biotechnology).30 Immunoblotting for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using a mouse monoclonal anti-GAPDH (NB615; Novus Biologicals) was used as a protein loading control.
miR-223 and miR-181b overexpression
A miR-223 mini-gene containing the pre-miR-223 hairpin sequence and approximately 200 bp of flanking sequence (total 527 bp) was PCR-amplified using the following primers: 5′-CTTTACCTGCTTATCTTCAGGATCTCT-3′ and 5′-CGTACGCGCCCCCATCAGCACTCT-3′. The resulting amplicons were cloned downstream of the green fluorescent protein (GFP) gene in the pMND-GFP lentiviral vector.31 The resultant pMND-GFP-miR-223 construct was transfected into 293T cells using DharmaFECT Duo (Dharmacon). Transduction efficiency, as assessed by GFP expression ranged from 50%-85% at 48 hours. The pMND-GFP-miR-181b expression vector was generated similar to pMND-GFP-miR-223 using the following primers: 5′-AAGGCGCGCCGTCTCCCATCCCCTTCAGAT-3′ and 5′-AAGCATGCTTTGCCTTTTCTAAAACATGCTC-3′. The resulting pMND-GFP-miR-181b construct was transiently transfected into K562 cells using the Amaxa Nucleofector II (program T16; Amaxa). Transduction efficiency, as assessed by GFP expression ranged from 30%-35% at 48 hours.
Luciferase assays using psiCheck2 TNFAIP2 3′-UTR sensor plasmids
Oligonucleotides corresponding to the region of the 3′-UTR of TNFAIP2 shown in Figure 6A were synthesized containing the germline or mutated sequence, and the annealed oligonucleotides were cloned into the psiCheck2 vector (Promega) downstream of Renilla luciferase. The psiCheck2 vector also contains Firefly luciferase for normalization. For luciferase assays, the indicated psiCheck2-3′-UTR vectors were transfected into 293T cells, HS-5 and KLE cell lines using DharmaFect Duo, or into K562 cells by nucleofection. Luciferase activity was determined after 48 hours using the Dual-Glo substrate system (Promega) and a Beckman Coulter LD400 luminometer. Data are presented as the ratio of experimental (Renilla) luciferase to control (Firefly) luciferase.
Statistical analyses
Significance was determined using Prism software Version 5.0c (GraphPad) to perform 2-tailed Student t tests assuming equal variance. All data are presented as the mean plus or minus SEM.
Results
Case presentation
We previously reported the sequence of the cancer genome for a patient with AML (AML1).16 In brief, a female in her 50s with no significant past medical history presented with FAB M1 AML. Routine cytogenetics revealed a normal 46 XX karyotype, and high-resolution comparative genomic hybridization (CGH) studies revealed no somatic copy number alterations at a resolution of approximately 5 kb. The patient achieved a complete remission with standard chemotherapy but relapsed at 11 months and expired 23 months after the initial diagnosis. Whole genome sequencing identified a total of 10 genic, nonsynonymous somatic mutations, including mutations in NPM1 and FLT3.
Identification of novel miRNAs
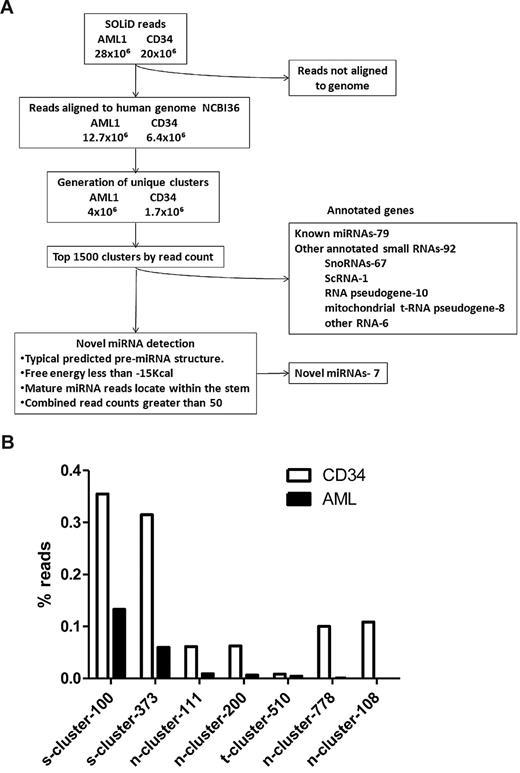
Massively-parallel sequencing of small RNAs isolated from the myeloblasts of AML1 was performed using the ABI SOLiD sequencing platform. As a control, we also analyzed pooled RNA isolated from CD34+ bone marrow cells of 5 healthy volunteers (CD34). In each case, cDNA was size-fractionated (corresponding to RNAs of 19-26 nucleotides in length) to enrich for miRNAs before sequencing. A total of 28 million sequence reads from AML1 and 20 million reads from CD34 were obtained. These sequences were aligned in color space to the human genome reference sequence (NCBI36) using the Maq aligner. Reads that did not map to the human genome were excluded from further analysis. In the AML1 sample, we identified 4 million unique genomic clusters from 12.7 million mapped reads. In the CD34 sample, we identified 1.7 million unique clusters from 6.4 million mapped reads (Figure 1A).
Identification of novel miRNAs. (A) Algorithm used to identify novel miRNAs. (B) Expression of novel miRNAs in the AML1 and normal CD34 samples. The percent contribution of each miRNA sequence to the total pool of known miRNA sequences was calculated by dividing individual miRNA read counts by the total number of miRNA sequence reads.
Identification of novel miRNAs. (A) Algorithm used to identify novel miRNAs. (B) Expression of novel miRNAs in the AML1 and normal CD34 samples. The percent contribution of each miRNA sequence to the total pool of known miRNA sequences was calculated by dividing individual miRNA read counts by the total number of miRNA sequence reads.
We selected the top 1000 clusters from AML1 and CD34 (a combined total of 1488) to screen for potential novel miRNAs. Only 171 of the clusters represented known genes, including 79 named miRNAs and 67 snoRNAs (Figure 1A). The remaining unannotated clusters were manually reviewed to identify novel miRNAs. An in-house program (miRNAViewer) was developed to view the RNA secondary structure of cluster sequences in 50, 100, 150, and 200 bp windows. The following criteria were used to identify miRNAs: (1) predicted RNA secondary structure with a free energy (ΔG) of < -15 kcal; (2) alignment of sequencing reads to one of the hairpin arms; and (3) a combined read count of 50 or more between AML1 and CD34. Applying these criteria in a blinded fashion, 78 of 79 known miRNAs in the top 1488 clusters were correctly identified as miRNAs. Among the top 1488 clusters, a total of 7 novel putative miRNAs were identified (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Consistent with the rate of star species reported for known miRNAs, a star species was detected for 2 of 7 (29%) novel miRNAs (supplemental Table 1). The highest expressed novel miRNA in both AML1 and CD34 cells was s-cluster-100, representing 0.15% and 0.35% of all miRNA sequences, respectively (Figure 1B). Several novel miRNAs, were differentially expressed in AML1 compared with CD34.
Assessment of miRNA expression using deep sequencing
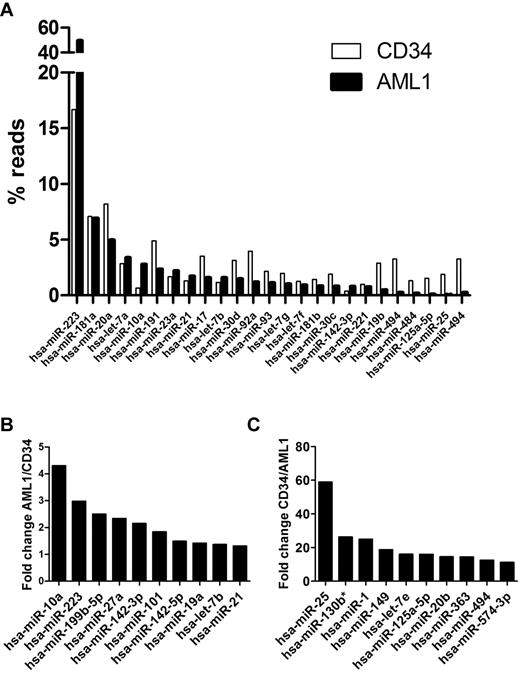
To assess miRNA expression, the sequence reads were aligned to hairpin sequences present in the miRNA Sanger database version 13 (795 known human miRNAs) and hairpin sequences from the 7 novel miRNAs identified in this study. Expression of 421 and 384 known miRNAs were detected in AML1 and CD34, respectively (supplemental Table 2). miR-233 was the most highly expressed miRNA in both AML1 and CD34 (Figure 2A). Remarkably, it represented 49.66% of all miRNA reads in AML1. We compared relative expression levels between AML1 and CD34 cells among miRNAs with read counts of 50 or higher (Figure 2B and supplemental Table 2). The most overexpressed miRNA in AML1 was miR-362-3p, which was increased 4.92-fold, compared with CD34 cells. The most underexpressed miRNA in AML1 was miR-25, which was decreased nearly 60-fold compared with CD34.
miRNA expression. (A) Top 20 expressed miRNAs in AML1 or normal CD34 cells (total 25 miRNAs); the percentage of reads for each miRNA relative to the total number of miRNA reads is shown. (B) Top 10 miRNAs with the most increased expression in AML1 compared with normal CD34. (C) Top 10 miRNAs with the most reduced expression in AML1 compared with CD34.
miRNA expression. (A) Top 20 expressed miRNAs in AML1 or normal CD34 cells (total 25 miRNAs); the percentage of reads for each miRNA relative to the total number of miRNA reads is shown. (B) Top 10 miRNAs with the most increased expression in AML1 compared with normal CD34. (C) Top 10 miRNAs with the most reduced expression in AML1 compared with CD34.
To compare our deep sequencing approach for miRNA expression with traditional array-based methods, we hybridized the AML1 and CD34 RNAs on Agilent miRNA arrays. In general, the correlation of expression levels between platforms was highly significant (r2 = 0.4372 for AML1 and r2 = 0.4143 for CD34; supplemental Figure 1). However, a few discrepancies were observed. For example, miR-720 was the 10th highest expressed miRNA in AML1 based on the Agilent array data, but was not detected by RNA sequencing. Conversely, miR-191 (the 4th highest expressed miRNA in AML1) was sequenced more than 36 000 times but was barely detected on the Agilent array.
miR-223 is expressed at high levels in a subset of AML
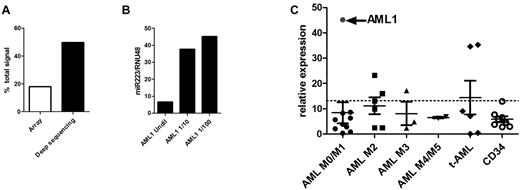
miR-223 was sequenced 765 476 times in AML1, representing 49.66% of all miRNA sequences obtained. Interestingly, although still the top expressed miRNA based on the Agilent miRNA array data, miR-223 represented only 16.66% of the total miRNA signal (Figure 3A), suggesting that the array data underestimated miR-223 expression. To test this possibility, real-time RT-PCR for miR-223 was performed on AML1 RNA (Figure 3B). At the standard input RNA (4 ng), the observed cycle number (< 16) was outside the dynamic range of this assay. Indeed, serial dilution of the AML1 RNA sample was required to fully appreciate the very high expression of miR-223.
miR-223 expression in AML. (A) The relative expression of miR-223 in AML1 compared with the total expression of all miRNAs based on the Agilent array (array) and RNA sequencing (deep sequencing) data are shown. (B) The relative expression of miR-223 compared with RNU48 was determined using real time RT-PCR on undiluted RNA from AML1 and RNA diluted 10-fold (1/10) or 100-fold (1/100) with water. (C) miR-223 expression relative to RNU48 in 27 AML samples and 8 CD34 samples from healthy donors is shown. AML1 is indicated by the arrow. The dashed line indicates 2 SDs above the mean using data from the CD34 samples.
miR-223 expression in AML. (A) The relative expression of miR-223 in AML1 compared with the total expression of all miRNAs based on the Agilent array (array) and RNA sequencing (deep sequencing) data are shown. (B) The relative expression of miR-223 compared with RNU48 was determined using real time RT-PCR on undiluted RNA from AML1 and RNA diluted 10-fold (1/10) or 100-fold (1/100) with water. (C) miR-223 expression relative to RNU48 in 27 AML samples and 8 CD34 samples from healthy donors is shown. AML1 is indicated by the arrow. The dashed line indicates 2 SDs above the mean using data from the CD34 samples.
To determine whether extremely high miR-223 expression is a consistent feature of AML, we performed real-time RT-PCR for miR-223 (after appropriate sample dilution) in an additional 26 AML samples (Figure 3C) and in CD34+ bone marrow cells from 8 healthy donors. In each case, miR-223 expression was normalized to RNU48 (SNORD48); a previous study showed that expression of this snoRNAs varied minimally in human AML samples.12 Compared with normal CD34+ bone marrow cells, miR-223 expression in AML1 was increased 6.8-fold. However, increased miR-223 expression was not a common feature of AML. Only 5 of 26 additional AML samples had relative miR-223 expression that was 2 standard deviations above that seen in normal CD34+ bone marrow cells.
miRNA isomiR patterns are similar in AML1 and normal CD34 cells
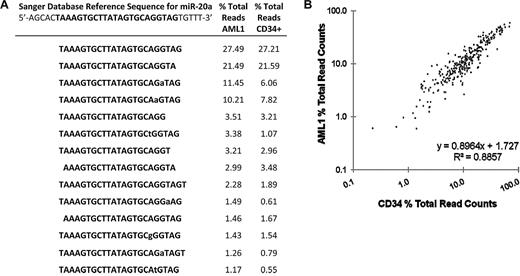
Previous miRNA sequencing studies have identified variants of miRNAs (termed isomiRs) that differ in mature miRNA length or less commonly in nucleotide sequence and are thought to arise from variable miRNA processing or RNA editing, respectively.32-35 isomiRs were detected for all abundantly expressed miRNAs. Consistent with previous reports,32,34,35 the majority of isomiRs represent truncations or extensions at the 5′- or 3′-ends of the mature miRNA (Figure 4A). Nucleotide substitutions were largely restricted to the last 10 nucleotides at the 3′ end of the miRNAs and do not conform to established RNA editing mechanisms in the majority of cases. Although the functional significance of isomiRs is unclear, we asked whether the pattern of isomiRs differed between AML1 and CD34 cells. We identified the isomiRs for all miRNAs that had at least 1000 read counts between the AML1 and CD34 samples and determined the percentage of each isomiR relative to the total sequence reads for that miRNA. Interestingly, the pattern of isomiR expression was nearly identical in the leukemic and normal CD34 cell samples (Figure 4B). These data suggest that, at least for the highest expressed miRNAs, the pattern of isomiRs is not altered during leukemic transformation.
isomiR pattern analysis. (A) miR-20a isomiRs. The reference sequence for miR-20a is shown with the mature miRNA bolded. Lower case letters represent nucleotide deviations from reference. The percentage of each miR-20a isomiR relative to the total sequence reads for miR-20a in the AML1 and CD34+ cells samples is shown. (B) The percentage of each isomiR relative to the total read counts for a specific miRNA were calculated for all miRNAs that had at least 1000 read counts between the AML1 and CD34 patient samples.
isomiR pattern analysis. (A) miR-20a isomiRs. The reference sequence for miR-20a is shown with the mature miRNA bolded. Lower case letters represent nucleotide deviations from reference. The percentage of each miR-20a isomiR relative to the total sequence reads for miR-20a in the AML1 and CD34+ cells samples is shown. (B) The percentage of each isomiR relative to the total read counts for a specific miRNA were calculated for all miRNAs that had at least 1000 read counts between the AML1 and CD34 patient samples.
Identification of genetic variants in miRNA genes in AML1 by next generation sequencing
We used the Roche/454 Titanium pyrosequencing platform to sequence all 695 miRNA genes in the Sanger miRNA database at the time this project was initiated (version 12.0). We sequenced a minimum of 200 bp flanking the miRNA precursor (total approximately 470 bp per miRNA gene) to detect mutations affecting the primary miRNA. The sequence coverage was excellent, with an average read depth per base position of 195.9 ± 17.3 (supplemental Figure 2). Of the 695 miRNA genes, 660 (95.0%) had at least one supporting sequence read and 90% of the bases had at least 40 × sequence coverage.
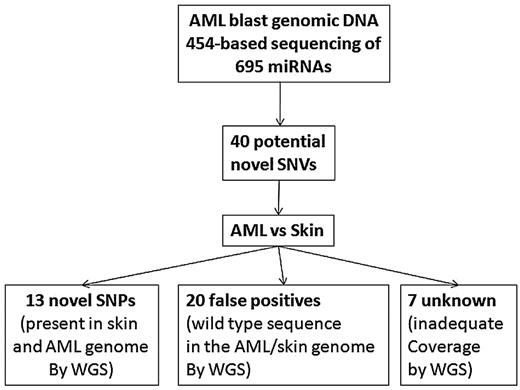
We applied the VarScan36 tool to identify sequence variants in the 454 data. A total of 148 high confidence SNVs were detected in AML1, of which 108 were previously annotated variants in dbSNPs (Figure 5). To validate the remaining 40 SNVs and determine their somatic status, we used the previously reported whole-genome sequencing data for the leukemic and normal (skin) genomes of this patient.16 Adequate sequence coverage of the genome was available for 33 of these SNVs. Twenty were not confirmed and likely represent sequencing artifacts. The remaining 13 SNVs were present in both the leukemic and normal genomes of this patient, suggesting that they represent novel germline SNPs. Interestingly, 3 of these novel SNPs were located in the precursor miRNA (Table 1). In addition to the SNVs, a single germline deletion in hsa-miR-620 was detected by both sequencing platforms (Table 1). Of note, no somatic mutations of miRNA genes were detected in the leukemic genome of this patient.
Deep sequencing of miRNA genes. All 695 miRNA genes present in the Sanger miRNA database at the time this study was initiated were PCR-amplified from leukemic genomic DNA from AML1 and subjected to 454-based sequencing. A total of 148 high confidence SNVs were identified, of which 108 were in dbSNPs. Analysis of previously generated whole genome sequence (WGS) data for the leukemic and skin (normal) genomes of AML1 showed that 13 novel germline SNPs were present in AML1. No somatic mutations were identified.
Deep sequencing of miRNA genes. All 695 miRNA genes present in the Sanger miRNA database at the time this study was initiated were PCR-amplified from leukemic genomic DNA from AML1 and subjected to 454-based sequencing. A total of 148 high confidence SNVs were identified, of which 108 were in dbSNPs. Analysis of previously generated whole genome sequence (WGS) data for the leukemic and skin (normal) genomes of AML1 showed that 13 novel germline SNPs were present in AML1. No somatic mutations were identified.
Genetic variants of miRNA genes in AML1
| miRNA gene . | Chr . | Position . | Ref . | Var . | Sequence Reads . | Location . | |
|---|---|---|---|---|---|---|---|
| Ref . | Var . | ||||||
| hsa-mir-1231 | 1 | 200044313 | C | T | 69 | 54 | pri-miRNA |
| hsa-mir-663b | 2 | 132731072 | G | C | 59 | 77 | pri-miRNA |
| hsa-mir-663b | 2 | 132731082 | A | C | 56 | 77 | pri-miRNA |
| hsa-mir-663b | 2 | 132731148 | A | G | 94 | 73 | pri-miRNA |
| hsa-mir-663b | 2 | 132731173 | T | C | 94 | 81 | pri-miRNA |
| hsa-mir-663b | 2 | 132731191 | T | C | 92 | 81 | pri-miRNA |
| hsa-mir-569 | 3 | 172307120 | T | C | 66 | 50 | pri-miRNA |
| hsa-mir-1324 | 3 | 75762663 | A | G | 132 | 94 | 1 nucleotide 5′ of mature miRNA |
| hsa-mir-1324 | 3 | 75762700 | T | C | 122 | 105 | 1 nucleotide 3′ of mature miRNA |
| hsa-mir-585 | 5 | 168623242 | C | G | 106 | 138 | pri-miRNA |
| hsa-mir-597 | 8 | 9636492 | G | A | 18 | 27 | pri-miRNA |
| hsa-mir-620 | 12 | 115070799-115070804 | ATATAT | deletion | 50 | 94 | pri-miRNA |
| hsa-mir-1283-2 | 19 | 58953361 | C | G | 131 | 107 | miRNA star |
| hsa-mir-133a-2 | 20 | 60572673 | C | T | 124 | 109 | pri-miRNA |
| miRNA gene . | Chr . | Position . | Ref . | Var . | Sequence Reads . | Location . | |
|---|---|---|---|---|---|---|---|
| Ref . | Var . | ||||||
| hsa-mir-1231 | 1 | 200044313 | C | T | 69 | 54 | pri-miRNA |
| hsa-mir-663b | 2 | 132731072 | G | C | 59 | 77 | pri-miRNA |
| hsa-mir-663b | 2 | 132731082 | A | C | 56 | 77 | pri-miRNA |
| hsa-mir-663b | 2 | 132731148 | A | G | 94 | 73 | pri-miRNA |
| hsa-mir-663b | 2 | 132731173 | T | C | 94 | 81 | pri-miRNA |
| hsa-mir-663b | 2 | 132731191 | T | C | 92 | 81 | pri-miRNA |
| hsa-mir-569 | 3 | 172307120 | T | C | 66 | 50 | pri-miRNA |
| hsa-mir-1324 | 3 | 75762663 | A | G | 132 | 94 | 1 nucleotide 5′ of mature miRNA |
| hsa-mir-1324 | 3 | 75762700 | T | C | 122 | 105 | 1 nucleotide 3′ of mature miRNA |
| hsa-mir-585 | 5 | 168623242 | C | G | 106 | 138 | pri-miRNA |
| hsa-mir-597 | 8 | 9636492 | G | A | 18 | 27 | pri-miRNA |
| hsa-mir-620 | 12 | 115070799-115070804 | ATATAT | deletion | 50 | 94 | pri-miRNA |
| hsa-mir-1283-2 | 19 | 58953361 | C | G | 131 | 107 | miRNA star |
| hsa-mir-133a-2 | 20 | 60572673 | C | T | 124 | 109 | pri-miRNA |
The number of sequence reads supporting the reference and variant alleles in the AML1 (and skin) genomes is shown.
Identification of a somatic mutation in the 3′-UTR region of TNFAIP2
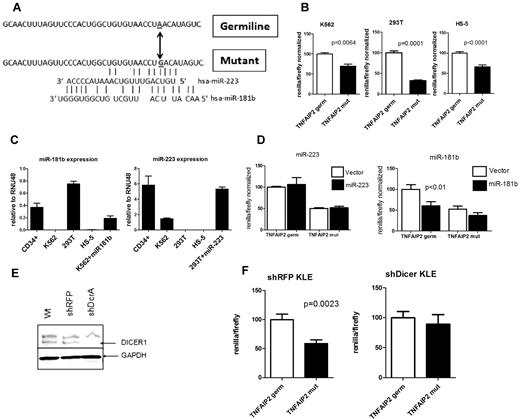
Mutations in coding genes might generate or destroy miRNA binding sites. To address this possibility, we analyzed the whole genome sequence data for somatic mutations in the untranslated regions of all 29 166 coding genes present in the ENSEMBL or NCBI/GenBank databases as of August 27, 2007. A total of 216 potential somatic SNVs were detected. Independent validation of all these SNVs was performed using 454-based sequencing of leukemic and skin genomic DNA from AML1. The majority of the variants (151) were not confirmed and likely represent sequencing artifacts. Forty-six of the SNVs were present in both the leukemic and normal genomes and are germline SNPs. Two somatic mutations were detected: a G- > A substitution in the 5′-UTR of ZSCAN10 (Chr 16, 3082843) and an A- > G substitution in the 3′-UTR of TNFAIP2 (Chr 14, 102673033; Figure 6A). Analysis of previously reported RNA expression profiling from this patient showed that TNFAIP2 but not ZSCAN10 was expressed in leukemic blasts (data not shown).16 Thus, we limited further study to TNFAIP2, a gene previously identified as a target gene for the PML-RARα oncogene.37,38 The variant allele frequency for the TNFAIP2 mutation was 76% (13 of a total 17 supporting sequence reads), suggesting that this heterozygous mutation is probably present in all cells in the tumor sample. Of note, no mutations of TNFAIP2 were identified by sequencing an additional 187 de novo AML samples.
The TNFAIP2 3′-UTR mutation confers translation repression in a miRNA dependent fashion. (A) The sequence of the 3′-UTR of TNFAIP2 (14:102673002-102673041) flanking the A→G mutation at 14:102673033 is shown, along with predicted binding sites for miR-223 and miR-181b. (B) The region of the TNFAIP2 3′-UTR shown in panel A containing germline (TNFAIP2 germ) or mutated sequence (TNFAIP2 mut) was cloned downstream of Renilla luciferase (psi-check2 vector) and transiently transfected into K562, 293T, and HS-5 cells. Expression of Renilla luciferase relative to firefly luciferase (an internal control present in the psi-check2 vector) after normalization to the germline TNAIP2 3′-UTR signal is shown. (C) Expression of miR-223 and miR-181b relative to RNU48 is shown. (D) 293T or K562 cells were cotransfected with the TNFAIP2 3′-UTR Renilla luciferase reporter constructs and a vector directing high level expression of either miR-223 or miR-181b. Expression of Renilla luciferase relative to firefly luciferase is shown. (E) Representative Western blot showing DICER1 protein expression in parent KLE cells (Wt) or KLE cells expressing shRNA directed against the red fluorescent protein (shRFP) or DICER1 (shDcrA). The upper band is nonspecific. GAPDH was used as a loading control. (F) The shRFP and shDicer KLE cells lines were transiently transfected with the TNFAIP2 3′-UTR Renilla luciferase reporter constructs. Expression of Renilla luciferase relative to firefly luciferase is shown. Data represent the mean ± SEM of 3-5 independent experiments.
The TNFAIP2 3′-UTR mutation confers translation repression in a miRNA dependent fashion. (A) The sequence of the 3′-UTR of TNFAIP2 (14:102673002-102673041) flanking the A→G mutation at 14:102673033 is shown, along with predicted binding sites for miR-223 and miR-181b. (B) The region of the TNFAIP2 3′-UTR shown in panel A containing germline (TNFAIP2 germ) or mutated sequence (TNFAIP2 mut) was cloned downstream of Renilla luciferase (psi-check2 vector) and transiently transfected into K562, 293T, and HS-5 cells. Expression of Renilla luciferase relative to firefly luciferase (an internal control present in the psi-check2 vector) after normalization to the germline TNAIP2 3′-UTR signal is shown. (C) Expression of miR-223 and miR-181b relative to RNU48 is shown. (D) 293T or K562 cells were cotransfected with the TNFAIP2 3′-UTR Renilla luciferase reporter constructs and a vector directing high level expression of either miR-223 or miR-181b. Expression of Renilla luciferase relative to firefly luciferase is shown. (E) Representative Western blot showing DICER1 protein expression in parent KLE cells (Wt) or KLE cells expressing shRNA directed against the red fluorescent protein (shRFP) or DICER1 (shDcrA). The upper band is nonspecific. GAPDH was used as a loading control. (F) The shRFP and shDicer KLE cells lines were transiently transfected with the TNFAIP2 3′-UTR Renilla luciferase reporter constructs. Expression of Renilla luciferase relative to firefly luciferase is shown. Data represent the mean ± SEM of 3-5 independent experiments.
The TNFAIP2 3′-UTR mutation results in decreased expression of a reporter gene in a miRNA-dependent fashion
A number of bioinformatic tools (see “Bioinformatic prediction of miRNA binding sites”) were used to determine whether the TNFAIP2 3′-UTR mutation altered an existing, or generated a new, miRNA binding site. No miRNA binding sites were identified near the mutated site in the germline TNFAIP2 3′-UTR. However, the TNFAIP2 mutation was predicted to generate imperfect binding sites for miR-223 and miR-181b (Figure 6A). To directly test the effect on expression, the germline or mutant TNFAIP2 3′-UTR (40 bp flanking the mutation) were cloned downstream of the Renilla luciferase reporter gene. In both K562 and 293T cells, the mutant TNFAIP2 sequence resulted in significant inhibition of reporter gene expression (Figure 6B). Of note, K562 cells have relatively high miR-223 expression and low miR-181b expression, whereas 293T cells have relatively high miR-181b and low miR-223 expression (Figure 6C). Surprisingly a similar decrease in Renilla luciferase expression was seen with the mutant TNFAIP2 in the HS-5 cell line (Figure 6B), a human bone marrow stromal cell line that has low levels of expression of both miR-223 and miR-181b (Figure 6C). We next assessed the effect of the enforced expression of miR-223 or miR-181b on translation repression by the mutant TNFAIP2 3′-UTR. We were unable to consistently achieve miR-223 or miR-181b overexpression in HS-5 cells. Instead, we transiently transfected 293T or K562 cells with expression vectors for miR-181b or miR-223, respectively; expression levels similar to that seen in normal CD34+ cells were achieved (Figure 6C). Enforced expression of miR-223 in 293T cells did not result in translation repression by the mutant or germline TNFAIP2 3′-UTR (Figure 6D). Interestingly, expression of miR-181b in K562 cells, though having no effect on the translation of constructs with the mutant TNFAIP2 3′-UTR, resulted in significant translational repression by the germline TNFAIP2 3′-UTR. Overall these results suggest that miR-223 and miR-181b do not mediate the translational repression of mutant TNFAIP2 3′-UTR.
To address whether the observed translational repression by the mutant TNFAIP2 3′-UTR was dependent on miRNAs, we developed a cell line with constitutive knockdown of Dicer. In brief, the human endometrial cancer cell line KLE was transduced with lentivirus expressing RNAi against Dicer (shDcrA) or, as a control, red fluorescent protein (shRFP). After puromycin selection, immunoblotting for Dicer protein was performed (Figure 6E). Densitometry showed that Dicer protein expression in shDcrA cells was reduced to 19.8% that of shRFP cells (data not shown). Moreover, expression of mature miR-181b and miR-223 in shDcrA cells was reduced 81.8% and 83.1%, respectively, compared with shRFP cells (supplemental Figure 3). Similar to K562 and 293T cells, the mutant TNFAIP2 3′-UTR sequence resulted in a significant inhibition of reporter gene expression in shRFP KLE cells (Figure 6F). In contrast, reporter gene expression was similar for the wild-type and mutant TNFAIP2 3′-UTR constructs in cells with constitutive Dicer knockdown. Collectively, these data suggest the possibility that the TNFAIP2 3′-UTR mutation generates a binding site for an, as yet unknown, miRNA that represses translation.
Discussion
In this study, we applied next generation sequencing technologies to comprehensively characterize miRNA expression, perform mutational profiling of miRNA genes, and screen for alterations in miRNA binding sites in a single patient with AML. To the best of our knowledge, this represents the first complete characterization of a microRNAome in a primary human cancer. We identified several novel miRNAs, some of which were differentially expressed in AML1 myeloblasts compared with normal CD34 cells. No somatic mutations of miRNA genes were identified in this leukemic genome, though several novel germline SNPs were detected. Sequencing of the untranslated regions of all coding genes identified a single somatic mutation in a expressed gene (TNFAIP2). We provide evidence that this mutation may result in translational repression of TNFAIP2 through generation of a new miRNA binding site.
There are 940 human miRNAs in the most recent version of the Sanger miRBase database (version 15, released April 2010). It is likely that additional miRNAs remain to be discovered, especially miRNAs with tissue-specific expression. High-throughput sequencing of small RNA libraries offers a powerful tool to discover novel miRNAs, because it uses an unbiased approach for miRNA detection.32-35 Indeed, in the present study, we identified 7 highly expressed novel miRNAs. Of note, because we limited our analysis to the top 1488 clusters, it is likely that additional novel miRNAs remain to be discovered that are expressed in AML or normal CD34+ cells.
A consistent finding of deep miRNA sequencing studies is the presence of isomiRs. isomiRs containing 5′- or 3′-extensions/deletions (1-2 nucleotides) are the most common, and likely arise from differential Drosha/Dicer processing. In addition, RNA editing enzymes, including adenosine deaminases, can generate isomiRs containing nucleotide substitutions. Importantly, some isomiRs have been shown to alter miRNA stability or target specificity. For example, RNA editing of miR-376 producing an adenine to inosine conversion within its seed sequence resulted in reduced suppression of its target mRNA PRPS1.39 In the present study, we identified isomiRs for all expressed miRNAs. In addition to sequence-length variants, we also detected frequent nucleotide substitutions, some of which are not consistent with known RNA editing mechanisms. Because some of the observed isomiRs are predicted to alter target specificity, we compared isomiR expression patterns between AML1 and CD34 cells. Interestingly, a nearly identical pattern was observed for the highest expressed isomiRs, suggesting that, at least for this case of AML, differential miRNA processing does not contribute to leukemic transformation.
A striking finding in our study is that miR-223 represents nearly 50% of all expressed miRNAs in AML1. Of note, both the Agilent arrays and real-time RT-PCR assays (using standard input RNA) underestimated miR-223 expression, emphasizing the wide dynamic range of deep sequencing to assess miRNA expression. MiR-223 expression in AML1 was increased 6.8-fold (by RT-PCR) compared with CD34. However, increased expression of miR-223 was only detected in 5 of 26 additional AML samples, suggesting that overexpression of miR-223 is not a common feature of AML. Indeed, a previous study showed that miR-223 expression is decreased in AML carrying t(8;21) generating AML1/ETO.40 miR-223 is thought to play an important role in myeloid development. It is expressed at low levels in pluripotent HSCs and common myeloid progenitors and increases with neutrophilic differentiation. miR-223–deficient mice display a myeloproliferative phenotype with increased myeloid progenitors, neutrophils, and neutrophil hyperactivity.41 Validated targets of miR-223 include transcription factors NFIA and MEF2C and transcriptional coregulator LMO2, all of which are involved in hematopoietic differentiation.41-43 Whether miR-223 overexpression contributes to leukemic transformation in some cases of AML will require further study.
There is accumulating evidence that genetic alterations in miRNA genes may contribute to malignant transformation. Approximately one-half of miRNA genes map to genomic fragile sites or regions of structural alterations in cancer.6 Indeed, recurrent copy number alterations (deletions or amplifications) of miRNA genes have been identified in many human cancers, including CLL, ovarian cancer, breast cancer, melanoma, and lymphoma.7,44 Interestingly, there have been no reports to date of somatic point mutations of miRNA genes in human cancer. In the present study, we identified no somatic copy number alterations (using genome-wide high-resolution aCGH) nor point mutations in any miRNA genes. However, our sequence was limited to approximately 200 bp flanking the mature miRNA; it is possible that mutations outside of this region might affect miRNA expression. Thirteen novel germline SNPs and a single Indel of miRNA genes were identified. Although functional studies were not performed, 3 of the novel SNPs were located in the pre-miRNA, raising the possibility that they may affect miRNA processing. In any case, the contribution of these and other germline SNPs to leukemic susceptibility will require the analysis of a much larger number of normal and leukemic samples.
There is emerging evidence that genetic alterations in miRNA binding sites in target genes may contribute to cancer susceptibility. A SNP in the let-7 miRNA binding site in the 3′-UTR of KRAS is associated with risk of lung cancer development.45 Polymorphisms in miRNA binding sites in the 3′-UTR of ITGB4 (integrin B4) are associated with estrogen negative breast cancer and predict improved survival.46 Finally, SNPs in miRNA binding sites in CD86 and INSR (the insulin receptor) are associated with colon cancer.47 In the present study, we provide evidence that a somatic mutation in the 3′-UTR of TNFAIP2 generates a new miRNA binding site that may result in translational repression of TNFAIP2. Specifically, we show that sequences containing the mutated region of the 3′-UTR of TNFAIP2 are able to inhibit reporter gene expression in a Dicer-dependent fashion. The identity of the miRNA that mediates this response is not yet clear, since overexpression of the 2 miRNAs (miR-223 and miR-181b) predicted to bind to the mutated site had no apparent effect on translational repression. Current bioinformatic approaches to identify miRNA target sites are imperfect, and biochemical approaches may be required to identify the miRNA(s) that bind to this mutated site. Of note, it is also possible that the TNFAIP2 3′-UTR mutation confers translational repression in a Dicer-dependent but miRNA-independent fashion.
TNFAIP2 encodes for tumor necrosis factor α-induced protein 2. It was originally identified as a tumor necrosis factorα-inducible immediate early gene in endothelial cells. TNFAIP2 is highly expressed in hematopoietic cells, and it appears to be a direct target for transcriptional repression by the PML-RARα and PZLF-RARα oncogenes.37,38 Accordingly, TNFAIP2 mRNA expression is repressed in M3-AML compared with M0-M2 AML. Interestingly, the TNFAIP2 gene is disrupted by human papilloma virus integration in a cervical cancer cell line.48 Although the function of TNFAIP2 is currently unknown, its transcriptional repression by PML-RARα and the potential translational repression by generation of a new miRNA binding site are consistent with a role in leukemic transformation. Of note, we did not identify TNFAIP2 3′-UTR mutations in an additional 187 de novo AML samples, indicating that this is a rare mutation in de novo AML.
These data demonstrate the feasibility of using next generation sequencing to comprehensively characterize the miRNAome in primary human samples. In addition to miRNAs, the RNA sequencing approach provides a wealth of information about other noncoding RNAs, such as snoRNAs. Interestingly, a number of the highly expressed small RNAs mapped to unannotated regions of the genome. Studies are underway to characterize these small RNAs and assess their contribution to leukemic transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Translational Research Program Award from the Leukemia & Lymphoma Society (to D.C.L.) and by grants from the National Institutes of Health (RC2 CA1455073, D.C.L.; PO1-CA101937, T.J.L.; U54-HG003079, R.K.W.).
National Institutes of Health
Authorship
Contribution: G.R. and D.C.K. designed, performed, and analyzed most of the experiments and wrote the manuscript; M.T. performed the isomiR analysis; T.W., S.K., L.-W.C., and R.N., provided bioinformatic support for this project; K.B.C. and P.G. developed and characterized the Dicer knockdown KLE cell line; T.A.F., V.M., R.K.W., L.D., T.J.L., and E.R.M. provided key expertise in the analysis of the sequence data; and D.C.L. was responsible for the overall design and analysis of all studies and edited the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, Division of Oncology, Washington University School of Medicine, Box 8007, 660 S Euclid Ave, St Louis, MO 63110; e-mail: dlink@dom.wustl.edu.
References
Author notes
G.R. and D.C.K. contributed equally to this work.