Abstract
Follicular dendritic cells (FDCs), an essential component of the lymph node microenvironment, regulate and support B-lymphocyte differentiation, survival, and lymphoma progression. Here, we demonstrate that adhesion of mantle cell lymphoma and other non-Hodgkin lymphoma cells to FDCs reduces cell apoptosis and is associated with decreased levels of the proapoptotic protein, Bim. Bim down-regulation is posttranscriptionally regulated via up-regulation of microRNA-181a (miR-181a). miR-181a overexpression decreases, whereas miR-181a inhibition increases Bim levels by directly targeting Bim. Furthermore, we found that cell adhesion–up-regulated miR-181a contributes to FDC-mediated cell survival through Bim down-regulation, implicating miR-181a as an upstream effector of the Bim-apoptosis signaling pathway. miR-181a inhibition and Bim upregulation significantly suppressed FDC-mediated protection against apoptosis in lymphoma cell lines and primary lymphoma cells. Thus, FDCs protect B-cell lymphoma cells against apoptosis, in part through activation of a miR-181a–dependent mechanism involving down-regulation of Bim expression. We demonstrate, for the first time, that cell-cell contact controls tumor cell survival and apoptosis via microRNA in mantle cell and other non-Hodgkin lymphomas. Regulation of microRNAs by B-cell–FDC interaction may support B-cell survival, representing a novel molecular mechanism for cell adhesion–mediated drug resistance and a potential therapeutic target in B-cell lymphomas.
Introduction
Lymphomas comprise the fifth most common cancer type in the United States, with approximately 55 000 cases of non-Hodgkin lymphoma (NHL) and 7400 cases of Hodgkin lymphoma each year. An unexplained finding is the 80% rise in NHL in the last 3 decades.1,2 Despite intensive efforts in developing new therapies and recent observed improvements in overall survival likely due to routine incorporation of monoclonal antibody therapy,1 this disease remains essentially incurable with standard therapeutic approaches. The emergence of clinical drug resistance continues to be an obstacle to the successful treatment of lymphoma. Mounting evidence now suggests that a dynamic interaction occurs between the tumor (lymphoma) cell and its microenvironment (lymph node stroma), with each profoundly influencing the behavior of the other. This “tumor microenvironment” is a critical determinant for tumor initiation, progression, response to therapy, and drug resistance.3,4 Specific niches within the tumor microenvironment may provide a sanctuary for subpopulations of tumor cells and provide a survival advantage (due to stromal cell–tumor cell interaction. This survival advantage after initial drug exposure may facilitate the acquisition of drug resistance. Thus it is likely that lymph node stromal cells influence the survival and apoptosis of lymphoma cells. Furthermore, this interaction may play a role in the resistance of residual lymphoma cells to additional chemotherapeutic agents, a problem that remains a major challenge in the treatment of lymphoma. However, how the lymphoma microenvironment influences lymphoma cell survival and response to therapy, as well as the molecular mechanisms involved, remains unclear.
Follicular dendritic cells (FDCs) are stromal cells unique to primary and secondary lymphoid follicles. Recirculating resting B cells migrate through the FDC networks, whereas antigen-activated B cells undergo clonal expansion within the FDC network in a T cell–dependent fashion, thereby generating the germinal center (GC),5,6 Only B cells that bind to FDC survive in the GC and differentiate into high-affinity plasma cells and memory B cells.6 This led us to investigate the effects of FDC–B cell interaction on B-cell survival in normal B-cell differentiation and in lymphoma cell response to therapy. Mantle cell lymphoma (MCL) is a distinct aggressive subtype of B-cell NHL. It represents approximately 5%-10% of all NHLs.1 MCL patients have a poor prognosis due to the development of drug resistance, with a median survival of 3-4 years.2 Therefore, characterizing stroma-mediated drug resistance development in MCL has the potential to shed important new light on novel approaches to treat MCL as well as other lymphoma tumors.
Although the majority of MCL lymphoma cells exhibit characteristics consistent with pre-GC lymphocytes, it has been reported that FDC are ubiquitously present in all MCL either in a nodular or diffuse pattern.7 This suggests that although normal B cells residing in the mantle zone may not have extensive interactions with FDC, during MCL growth, the normal architecture is disrupted leading to abundant FDC-MCL colocalization and interaction. Furthermore, the diffuse distribution of FDC in MCL is associated with a worse clinical outcome. These observations suggest that interaction between FDC and MCL cells may contribute to drug resistance and MCL clinical survival. Similarly, the follicular dendritic meshwork present within neoplastic follicles of follicular lymphoma (FL) is consistent with a similar role for FDC in FL. Therefore, in this study, we have investigated the significance of FDC regulated B-cell lymphoma apoptosis and cell-mediated drug resistance with cell lines from both MCL and GC-derived lymphomas as well as primary tumor cells from MCL, FL, and diffuse large cell lymphomas.
MicroRNAs (miRNAs) are a newly discovered class of short (19-25 nucleotides) endogenous small RNAs that negatively regulate gene expression.8 miRNAs have been found to play key roles in a wide range of biological processes and to be aberrantly expressed in many types of cancer, including lymphomas.9,10 However, the role of miRNAs in B-lymphocyte development and B-cell lymphomagenesis is largely unknown. miRNA-155 has been shown to regulate GC response through modulation of cytokine production11 and regulation of the activation-induced cytidine deaminase.12 A more recent study showed that miRNA-155, by down-modulating Ship and C/enhancer binding proteinβ, initiates a chain of events that leads to the accumulation of large pre-B cells and acute lymphoblastic leukemia/high-grade lymphoma cells.13 miRNA-150 has been shown to play an important role in B-cell differentiation by targeting the transcription factor Myb.14 In B-cell lymphomas, 13q31 amplification has been shown to cause over-expression of the miRNA-17-92 cluster, playing a critical role in lymphoma progression.15 Furthermore, dysregulation of miRNA-15a and miRNA-16 has been implicated in the pathogenesis of B-cell chronic lymphocytic leukemia.16,17 A complete delineation of miRNAs and their targets in B-cell survival is essential to understanding their role in B-cell apoptosis and response to therapy. Because physical interactions between B cells and FDCs from the lymphoid tissue microenvironment are critical to the survival of normal and malignant B cells, we have identified the miRNAs induced by interactions between FDCs and B cells. We now show that FDC interaction induces expression of microRNA-181a (miR-181a), which in turn targets the proapoptotic protein Bim (B-cell lymphoma-2 [BCL-2]–interacting mediator of cell death) for silencing. These findings define a novel mechanism by which the tumor microenvironment alters specific microRNA levels leading to drug resistance and lymphoma survival.
Methods
Cell lines, cell cultures, and patient lymphoma specimens
The human lymphoma cell lines Jeko-1, Mino, SUDHL-4, and SUDHL-10 were provided and maintained as previously described.17 SUDHL-4 and SUDHL-10 are both GC type large B-cell lymphomas, while Jeko-1 and Mino are MCL cell lines. Ramos, a Burkitt lymphoma cell line, was obtained from K.L.W. The FDC line, HK, was obtained from Y.S.C.5 The cells were grown in suspension in RPMI 1640 (Cellgro, Fisher Scientific), supplemented with 5% fetal bovine serum (Omega Scientific), and 1% (vol/vol) penicillin (100 U/mL), streptomycin (100 U/mL), and 1% (vol/vol) l-glutamine (Gibco-BRL). HK cells were fed after 4 days by adding one-half volume of complete medium and then at weekly intervals by complete replacement of the medium and maintained at 37°C in 5% CO2-95% air.
Diffuse large B-cell lymphoma and MCL cells were obtained from fresh biopsy-derived lymphoma tissues (lymph nodes) after informed consent from patients and approval by the Institutional Review Board of the University of South Florida. The lymph nodes were cut and gently dispersed. For preparation of viable, sterile, single cell suspensions, the lymph node tissue was diced and forced through a metal sieve into RPMI 1640 tissue culture. Cells, obtained after low-speed centrifugation, were resuspended in media. After Ficoll-Paque purification, the CD19-positive cells were isolated using CD19 microbeads with the autoMACS magnetic cell sorter according to the manufacturer's instructions (Miltenyi Biotec). Either lymphoma cell lines or CD19-sorted lymphoma cells (from diffuse large cell lymphomas, 1.5 × 106 cells/mL) were adhered to a pre-established monolayer of HK or kept in suspension for 12 hours. MCL cells were used for experiments without CD19 sorting because all of our MCL samples had > 90% lymphoma cells in purity. Lymphoma cells were then carefully removed, with the monolayer of HK kept intact. The purity of the lymphoma cell population was greater than 95% by flow cytometry. Cells were then analyzed as described in the next paragraph. In transwell experiments, lymphoma cells were separated from the HK by transwell inserts.
Antibodies, Western blotting, and apoptosis detection assay
Monoclonal antibodies were purchased as follows: Bim from Cell Signaling Technology and actin from Sigma-Aldrich. Vorinostat (suberoylanilide hydroxamic acid [SAHA]) was provided by Merck. Mitoxantrone was purchased from Sigma-Aldrich. At the indicated times, cells were collected and washed; Western blot and apoptosis analyses were then performed as previously described.18,19
Quantitative analyses
RNA was isolated from samples in Trizol as recommended by the manufacturer (Invitrogen). The samples were further purified into total RNA fraction using RNeasy columns as described by the manufacturer (QIAGEN). For miRNA quantitation, miR-181a levels in cell lines and patient lymphoma cells were determined using TaqMan miRNA assays (Applied Biosystems), following manufacturer-recommended protocols; 750 ng of total RNA/15 μL of reaction volume was used during reverse transcription (RT), and 2 μL of RT product was used for subsequent real-time polymerase chain reactions (PCRs). For each sample, 3 independent RT reactions were performed, and each reaction was assayed in triplicate for real-time PCR. miRNA levels were normalized with U6 small nuclear RNA, and relative levels were calculated using the ΔΔCt method. The relative expression level of specific miRNA was presented by 2−ΔΔCt. For quantitative detection of Bim mRNAs, cDNAs were synthesized from 1 μg of total RNA using the SuperScript first-strand kit (Invitrogen). Real-time PCR was conducted using the ABI PRISM 7000 sequence detection system (Applied Biosystems) as described previously.18 Primer and probe sequences for real-time detection of Bim (assay ID: Hs00708019) and endogenous control gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA were purchased from Applied Biosystems. Real-time PCR was performed using human Taqman predeveloped assay reagents (Applied Biosystems).
Transfection of pre-miRNAs and anti-miRNAs, Bim overexpression by transient transfection, and Bim knockdown by small hairpin RNA
Optimized nucleofection protocols generated by Amaxa were followed for the transfection of Jeko-1 cells (Nucleofector Kit T, Program T-001) and SUDHL-4 (Nucleofector Kit V, Program X-001) with miRNA precursors (Ambion) or anti-miRNA inhibitors (Ambion), respectively. A concentration of 100 nmol/L was used for all transfections. Cells were collected at 24-48 hours after electroporation and subjected to Western blot analysis for determination of Bim expression.
For transient over-expression, the Jeko-1 cell line was transfected by electroporation using Nucleofector (Amaxa) according to the manufacturer's instructions and commercially available transfection instrument (Amaxa). Briefly, cells (5 × 106) from each lymphoma cell line were suspended in 100 μL of Nucleofector T solution with 1 μg of either the control empty vector (pCMV-Sport6) or a pCMV-Sport6-Bim expression plasmid (I.M.A.G.E. Clone ID: 5213713, ATCC) and then electroporated using program T-001. Cells were collected at 24-48 hours after electroporation and subjected to Western blot analysis for determination of Bim expression. Based on the expression of green fluorescent protein (GFP) analyzed by flow cytometry, the transfection efficiency was between 50% and 75%, with a median of 60% (5 independent experiments) as described previously.18
Bim small hairpin RNA (pSM2 shRNA vector) and control (pSM2-nonsilencing) shRNAs were obtained from Cold Spring Harbor Laboratory. For transient expression, cell lines were transfected by electroporation using Nucleofector (Amaxa) according to the manufacturer's instructions and using a commercially available transfection instrument (Amaxa). Briefly, Jeko-1 or SUDHL-4 cells (5 × 106 cells/mL) were suspended in 100 μL of Nucleofector T solution with shRNA duplex (final concentration 1 μg) and then electroporated using program T-001. On the basis of the expression of GFP analyzed by flow cytometry, the transfection efficiency was between 50% and 75%, with a median of 60% (3 independent experiments) as described previously.18,19
Computer prediction of miRNA target sites in the 3′ untranslated region of Bim mRNA
Putative target sites for miRNA in the 3′ untranslated region (UTR) of human, mouse, and rat Bim mRNA were identified using the MiRANDA Human miRNA Targets Web site (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl) and the TargetScan (Version 5.4) Web site (http://genes.mit.edu/targetscan) based on the target prediction algorithms.20,21
Bim gene 3′ UTR luciferase reporter assay
To create Bim gene 3′ UTR luciferase reporter constructs, 60-bp sequences from putative miR-181a binding sites were synthesized and ligated into the pmiR-Report vector (Ambion) at SpeI and HindIII sites. To create a mutant 3′ UTR, point mutations were introduced at the first 2 miR-181a–matching nucleotides within selected putative seeding sequence regions with the following rules: A was changed to T and vice versa, and G was changed to C and vice versa. Jeko-1 and SUDHL-4 cells in 24-well plates were transfected with 0.10 μg of the pmiR-Report-3′ UTR/Bim luciferase reporter, 0.05 μg of the normalization plasmid pCMV-β-galactosidase, and 0.6 μg of miR-181a or non-GFP–expressing control vector. Luciferase assays were performed using a luciferase assay system (Promega), and activities were normalized to β-galactosidase activity.22
Results
Cell adhesion to FDC induces down-regulation of Bim and protects B-lymphoma cell lines from drug-induced apoptosis
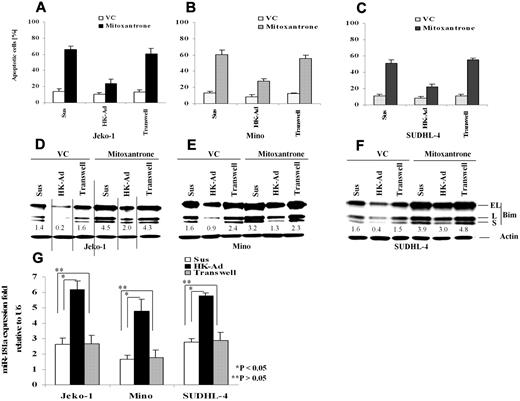
Our recent studies have shown that adhesion of lymphoma cells to bone marrow stromal cells significantly increases cellular resistance to a variety of cytotoxic drugs and induces cell cycle arrest.18,19,23 We have now extended this study to determine the role of the interaction between FDC and NHL B-cells in response to therapy and to further elucidate the molecular mechanisms by which stroma regulates lymphoma cell survival. We first performed immunohistochemical stain to evaluate the distribution of FDC in FLs (n = 10) and MCLs (n = 10) using CD21 antibody and confirmed that FDC is present in all lymphoma tested (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).7 We then asked whether adhesion of lymphoma cells to lymph node stromal cells (FDC) results in inhibition of cell apoptosis upon exposure to chemotherapeutic drugs. We initially profiled the response in lymphoma cell lines including the MCL lines Jeko-1 and Mino, the GC cell–derived large B-cell lymphoma cell lines SUDHL-4 and,SUDHL-10, as well as the GC-derived Burkitt lymphoma cell line, Ramos. We used FDC-derived cells (HK cells) as a source of the lymph node stromal cells.5,24 After 24 hours of adhesion, we observed that adhesion of lymphoma cells to HK cells protected the MCL cell lines (Jeko-1 and Mino) and a large B-lymphoma cell line (SUDHL-4) against mitoxantrone-induced apoptosis (Figure 1A-C). To examine the contribution of soluble factors independent of physical cell contact, we used microporous transwell inserts to prevent direct contact while allowing soluble factor diffusion across the membrane. Because soluble factors produced via dynamic interaction between lymph node stroma and lymphoma cells may be important in the protective effect, we incubated the lymphoma cells in one chamber of the transwell insert with a coculture of HK and lymphoma cells in the other chamber. As shown in Figure 1A through C, lymphoma cells that were in direct contact with the HK cell line, but not cells exposed to only soluble factors (in the transwell insert), have much higher protective effect on cell survival. In view of the central role of the BH-3–only protein, Bim, in apoptosis induced by various stimuli, we next investigated whether the observed increase in cell survival (anti-apoptosis) correlated with an expression change of the apoptotic regulator, Bim, in lymphoma cells. We examined the protein level of Bim in Jeko-1, Mino, and SUDHL-4 lymphoma cells adherent to HK cells cultured in the transwell system and in suspension. Western blot analysis showed that all 3 isoforms of Bim protein produced by alternative splicing were markedly decreased when the lymphoma cells were adherent to HK cells (Figure 1D-F). Given the close contact of FDC with follicular lymphocytes in GC in reactive lymph nodes and follicular lymphoma cells in FL, we repeated these experiments in 2 other GC-derived lymphoma cell lines, SUDHL-10 and Ramos. Similar effects of FDC on cell apoptosis and Bim expression were observed and are shown in supplemental Figure 2. Taken together, our data indicate that adhesion of lymphoma cells to the FDC cell line confer lymphoma cell drug resistance, a finding that is associated with decreased Bim expression. Furthermore, inhibition of drug response and Bim inhibition was not the result of soluble factors produced by the coculture system but rather required direct cell-to-cell contact.
Cell adhesion to follicular dendritic cells (HK cells) protects B-lymphoma cell lines from drug-induced apoptosis and induces down-regulation of Bim and up-regulation of miR-181a. Jeko-1 (A,D), Mino (B,E), or SUDHL-4 (C,F) lymphoma cells (105/mL) in suspension (Sus) adhered to the pre-established monolayer of HK cells (HK-Ad) or with a confluent HK monolayer but separated by transwell inserts were treated with and without mitoxantrone (2μM) or vehicle control (VC) for 12 hours, and the lymphoma cells were collected and analyzed for apoptosis with annexin V (A-C) and Bim protein (all Bim isoforms, EL, L, and S) by Western blot (D-F). Direct cell-cell contact but not soluble factor(s) decreased mitoxantrone-induced apoptosis and Bim expression in Jeko-1, Mino, and SUDHL-4 lymphoma cells. The relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane in panels D through F. (G) Jeko-1, Mino, or SUDHL-4 lymphoma cells (105/mL) were placed in suspension (Sus) or adhered to HK cells (HK-Ad) for 24 hours, and miR-181a expression was analyzed by TaqMan microRNA qRT-PCR assays. Results represent 4 independent experiments with mean ± SD.
Cell adhesion to follicular dendritic cells (HK cells) protects B-lymphoma cell lines from drug-induced apoptosis and induces down-regulation of Bim and up-regulation of miR-181a. Jeko-1 (A,D), Mino (B,E), or SUDHL-4 (C,F) lymphoma cells (105/mL) in suspension (Sus) adhered to the pre-established monolayer of HK cells (HK-Ad) or with a confluent HK monolayer but separated by transwell inserts were treated with and without mitoxantrone (2μM) or vehicle control (VC) for 12 hours, and the lymphoma cells were collected and analyzed for apoptosis with annexin V (A-C) and Bim protein (all Bim isoforms, EL, L, and S) by Western blot (D-F). Direct cell-cell contact but not soluble factor(s) decreased mitoxantrone-induced apoptosis and Bim expression in Jeko-1, Mino, and SUDHL-4 lymphoma cells. The relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane in panels D through F. (G) Jeko-1, Mino, or SUDHL-4 lymphoma cells (105/mL) were placed in suspension (Sus) or adhered to HK cells (HK-Ad) for 24 hours, and miR-181a expression was analyzed by TaqMan microRNA qRT-PCR assays. Results represent 4 independent experiments with mean ± SD.
Cell adhesion induces up-regulation of miR-181a and Bim is a direct target of miR-181a
Global miRNA expression profiling has indicated that the expression of multiple miRNAs is altered in NHL cells upon adhesion to FDC.25 Among these miRNAs, we identified that miR-181a was up-regulated by FDC interaction. Furthermore, we found that the 3′ UTR of Bim contains one sequence motif that perfectly matches with the “seed” sequence of miR-181a. This motif is well conserved among humans, mice, and rats, suggesting a potential regulation of Bim by miR-181a. We therefore first performed quantitative RT-PCR to validate that cell adhesion indeed induces miR-181a expression in MCL and other B-cell lymphoma cells. As shown in Figure 1G, lymphoma cells that were in direct contact with the HK cells increased miR-181a by 2-3-fold in Jeko-1, Mino, and SUDHL-4 lymphoma cells. In contrast, lymphoma cells exposed to soluble factors with no contact with HK cells (in the transwell insert) revealed no changes in miR-181a expression, supporting the role of lymphoma-HK cell-cell contact in miR-181a induction.
We next determined effect of miR-181a on Bim protein expression. We tested whether knockdown of miR-181a can induce Bim expression. The cells were transfected with anti–miR-181a or a scrambled oligonucleotide control. Quantitative (q)RT-PCR analysis revealed a significant reduction of miR-181a in the cells treated with anti–miR-181a (Figure 2A). Furthermore, the protein but not mRNA levels of Bim was increased in the anti–miR-181a–transfected Jeko-1 cells compared with the control transfected cells (Figure 2B-C). We then overexpressed miR-181a by ectopically transfecting pre–miR-181a into Jeko-1 cells and compared them to cells transfected with an empty pre-miR plasmid as a control. Twenty-four hours after transfection, overexpression of transfected miR-181a was verified by qRT-PCR (Figure 2D). As shown in Figure 2E, ectopic expression of miR-181a significantly reduced expression of all Bim isoforms (EL, L, and S) in Jeko-1 cells in suspension as well as after adhesion with HK cells. To further demonstrate that miR-181a can directly regulate Bim, we inserted the putative mir-181a binding site from the Bim 3′-UTR into the 3′-UTR of a constitutively active luciferase reporter (pmiR-Bim-3′UTR). To confirm specificity, a second reporter with a mutation in the seed sequence of the mir-181a binding site was created (Figure 2F). Both the wild-type and mutant reporters were introduced into Jeko-1 cells and SUDHL-4 cells. The luciferase activity of pmiR-Bim-3′UTR was significantly suppressed in miR-181a–transfected Jeko-1 cells (Figure 2G). To confirm Bim regulation by miR-181a in other types of B-cell lymphoma, we repeated the Figure 2G experiments in SUDHL-4, GC-derived lymphoma cells, and similar results were observed as shown in Figure 2H. Furthermore, in pmiR-Bim-3′UTR mutant transfected Jeko-1 cells as well as SUDHL-4, no changes in reporter activity were observed (Figure 2G-H). In addition, anti–miR-181a increased the luciferase activity in pmiR-Bim-3′UTR but not in pmiR-Bim-3′ UTR mutant–transfected Jeko-1 cells (Figure 2G-H). Taken collectively, these data indicate that Bim is a direct target of miR-181a.
miR-181a directly targets Bim and mediates cell adhesion–induced Bim down-regulation. (A). Knockdown of miR-181a using anti–miR-181a. The cells were transfected with anti–miR-181a or a scrambled oligonucleotide control, and miR-181a was analyzed by TaqMan microRNA qRT-PCR assays. Results in fold were obtained and expressed relative to small RNA U6 expression levels in Jeko-1 cells. The mean values and SDs from 4 independent experiments are shown. The Student t test was used for statistical analysis. *P < .05. (B-C). Knockdown of miR-181a increases Bim protein (B) but not mRNA (C) level with and without HK cell adhesion. Results for panel B are representative of 3 independent experiments. Results for panel E are means ± SD of 3 independent experiments. (D) Overexpression of miR-181a in pre–miR-181a–transfected Jeko-1 cells. Jeko-1 cells were transfected with pre–miR-control or pre–miR-181a (50 nmol/L) using Amaxa's Nucleofector system, and the miR-181a expression was analyzed by TaqMan microRNA qRT-PCR assays. (E) Overexpression of miR-181a down-regulated Bim (all Bim isoforms, EL, L, and S) expression. Pre–miR-control (pre–miR-Ctrl)– or pre–miR-181a–transfected (pre–miR-181a) Jeko-1 lymphoma cells (105/mL) were in suspension (Sus) or adhered to HK cells (HK-Ad) for 24 hours, and Bim expression was analyzed by Western blot. (D-E) Each are representative of at least 3 experiments with mean ± SD. Results (in fold) were obtained and expressed relative to small RNA U6 expression level. The Student t test was used for statistical analysis. *P < .05. (F) pmiR-Report.Bim 3′ UTR reporters were constructed using the miRANDA Human miRNA Targets Web site (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl) and the Targetscan (Version 5.4) Web site (http://genes.mit.edu/targetscan) based on the target prediction algorithms. Jeko-1 (G) or SUDHL-4 (H) cells were transfected with pmiR-Report control vector (Ctrl-vector) or pmiR-Report.Bim 3′ UTR wild-type (3′UTRw) or pmiR-Report.Bim 3′UTR mutant (3′UTRm) plasmids harboring point mutations in the target sites for miR-181a. These reporter plasmids were cotransfected with pre–miR-181a or anti–miR-181a (50 nmol/L) or control oligonucleotides or plasmid. The luciferase activities (in triplicate) of these transfected cells were measured 36 hours after transfection. Renilla luciferase activities were normalized against firefly luciferase activities, and mean normalized Renilla luciferase activities (± SD) from at least 3 independent experiments were determined and expressed relative to control values. In panels B and E, the relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane.
miR-181a directly targets Bim and mediates cell adhesion–induced Bim down-regulation. (A). Knockdown of miR-181a using anti–miR-181a. The cells were transfected with anti–miR-181a or a scrambled oligonucleotide control, and miR-181a was analyzed by TaqMan microRNA qRT-PCR assays. Results in fold were obtained and expressed relative to small RNA U6 expression levels in Jeko-1 cells. The mean values and SDs from 4 independent experiments are shown. The Student t test was used for statistical analysis. *P < .05. (B-C). Knockdown of miR-181a increases Bim protein (B) but not mRNA (C) level with and without HK cell adhesion. Results for panel B are representative of 3 independent experiments. Results for panel E are means ± SD of 3 independent experiments. (D) Overexpression of miR-181a in pre–miR-181a–transfected Jeko-1 cells. Jeko-1 cells were transfected with pre–miR-control or pre–miR-181a (50 nmol/L) using Amaxa's Nucleofector system, and the miR-181a expression was analyzed by TaqMan microRNA qRT-PCR assays. (E) Overexpression of miR-181a down-regulated Bim (all Bim isoforms, EL, L, and S) expression. Pre–miR-control (pre–miR-Ctrl)– or pre–miR-181a–transfected (pre–miR-181a) Jeko-1 lymphoma cells (105/mL) were in suspension (Sus) or adhered to HK cells (HK-Ad) for 24 hours, and Bim expression was analyzed by Western blot. (D-E) Each are representative of at least 3 experiments with mean ± SD. Results (in fold) were obtained and expressed relative to small RNA U6 expression level. The Student t test was used for statistical analysis. *P < .05. (F) pmiR-Report.Bim 3′ UTR reporters were constructed using the miRANDA Human miRNA Targets Web site (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl) and the Targetscan (Version 5.4) Web site (http://genes.mit.edu/targetscan) based on the target prediction algorithms. Jeko-1 (G) or SUDHL-4 (H) cells were transfected with pmiR-Report control vector (Ctrl-vector) or pmiR-Report.Bim 3′ UTR wild-type (3′UTRw) or pmiR-Report.Bim 3′UTR mutant (3′UTRm) plasmids harboring point mutations in the target sites for miR-181a. These reporter plasmids were cotransfected with pre–miR-181a or anti–miR-181a (50 nmol/L) or control oligonucleotides or plasmid. The luciferase activities (in triplicate) of these transfected cells were measured 36 hours after transfection. Renilla luciferase activities were normalized against firefly luciferase activities, and mean normalized Renilla luciferase activities (± SD) from at least 3 independent experiments were determined and expressed relative to control values. In panels B and E, the relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane.
Cell adhesion–mediated up-regulation of miR-181a and down-regulation of Bim are required for cell adhesion–mediated drug resistance
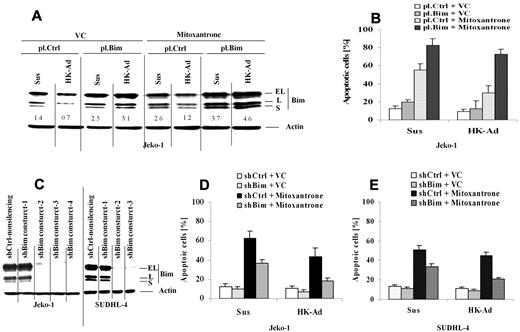
We next determined whether Bim down-regulation is required for cell adhesion–mediated drug resistance (CAM-DR) to occur upon cell adhesion to the lymph node stromal cells. We ectopically over-expressed Bim in Jeko-1 cells and applied these transfected cells to our coculture experiments. We examined the changes in Bim protein expression and drug-induced apoptosis upon adhesion to HK cells. Ectopic Bim expression suppressed the cell adhesion–mediated Bim down-regulation (Figure 3A). Importantly, maintaining Bim expression enhanced mitoxantrone drug-induced apoptosis specifically in the cells adhered to HK cells (Figure 3B). These results suggest that CAM-DR mediated by HK cell adhesion involves Bim down-regulation. We next ablated Bim expression using shRNAs and examined the effect on CAM-DR. Figure 3C reveals that 3 of 4 and 2 of 3 Bim shRNAs were highly effective in depleting Bim in both Jeko-1 cells and SUDHL-4, respectively. Reducing Bim levels conferred resistance to mitoxantrone-induced cell death in both Jeko-1 and SUDHL-4 cells as measured by annexin V staining (Figure 3D-E). These results support the key role of Bim in CAM-DR in MCL as well as other B-cell lymphomas.
Cell adhesion–mediated down-regulation of Bim is required for cell adhesion–mediated drug resistance. (A-B) Overexpression of Bim inhibits cell adhesion–induced Bim down-regulation, induces cell apoptosis, and overcomes CAM-DR. Jeko-1 lymphoma cells were transfected with pCMV-SPORT6 control (pl.Ctrl)- or pCMV-SPORT6-Bim (pl.Bim) for 24 hours. Cells, collected and washed in suspension versus in adhesion with HK, were treated with mitoxantrone (1μM) for another 12 hours; Bim protein levels (A) were then analyzed by Western blot, and apoptosis (B) was analyzed by annexin V with flow cytometry. Data are representative of 4 independent experiments. In panel A, the relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane. (C) Bim knockdown by shRNAs in Jeko-1 and SUDHL-4 cells. Bim expression levels in Jeko-1 cells were analyzed by Western blot after 48 hours of transfection with either Bim shRNA constructs 1 or 2 or 3 or 4 (shBim construct 1 or 2 or 3 or 4) or nonsilencing control shRNA (shCtrl-nonsilencing). Results are representative of 3 independent experiments. (D-E) Knockdown of Bim inhibits mitoxantrone-induced cell apoptosis in Jeko-1 and SUDHL-4 cells in the presence and absence of HK adhesion. Jeko-1 (D) or SUDHL-4 (E) cells transfected with Bim shRNA (shBim-3) or control shRNA (shCtrl) were incubated for 48 hours and then treated with vehicle control (VC) or mitoxantrone (0.2μM) for another 24 hours. Apoptosis was analyzed by flow cytometry using annexin V. Data are representative of 4 independent experiments.
Cell adhesion–mediated down-regulation of Bim is required for cell adhesion–mediated drug resistance. (A-B) Overexpression of Bim inhibits cell adhesion–induced Bim down-regulation, induces cell apoptosis, and overcomes CAM-DR. Jeko-1 lymphoma cells were transfected with pCMV-SPORT6 control (pl.Ctrl)- or pCMV-SPORT6-Bim (pl.Bim) for 24 hours. Cells, collected and washed in suspension versus in adhesion with HK, were treated with mitoxantrone (1μM) for another 12 hours; Bim protein levels (A) were then analyzed by Western blot, and apoptosis (B) was analyzed by annexin V with flow cytometry. Data are representative of 4 independent experiments. In panel A, the relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane. (C) Bim knockdown by shRNAs in Jeko-1 and SUDHL-4 cells. Bim expression levels in Jeko-1 cells were analyzed by Western blot after 48 hours of transfection with either Bim shRNA constructs 1 or 2 or 3 or 4 (shBim construct 1 or 2 or 3 or 4) or nonsilencing control shRNA (shCtrl-nonsilencing). Results are representative of 3 independent experiments. (D-E) Knockdown of Bim inhibits mitoxantrone-induced cell apoptosis in Jeko-1 and SUDHL-4 cells in the presence and absence of HK adhesion. Jeko-1 (D) or SUDHL-4 (E) cells transfected with Bim shRNA (shBim-3) or control shRNA (shCtrl) were incubated for 48 hours and then treated with vehicle control (VC) or mitoxantrone (0.2μM) for another 24 hours. Apoptosis was analyzed by flow cytometry using annexin V. Data are representative of 4 independent experiments.
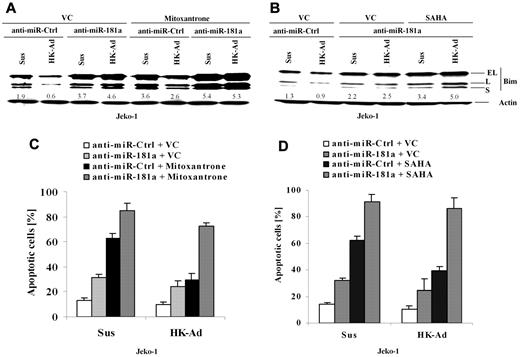
To determine whether miR-181a is involved in CAM-DR through cell adhesion–mediated Bim down-regulation, we blocked miR-181a induction using anti–miR-181a and examined the changes in Bim expression and drug-induced apoptosis. Cell extracts from Jeko-1 cells transfected with anti–miR-181a or a control anti-miR were prepared with or without cell adhesion to HK cells, followed by Western blot analysis. As shown in Figure 4A and B, anti–miR-181a increased Bim expression and blocked cell adhesion–induced Bim down-regulation, indicating that the miR-181a expression is required for cell adhesion–mediated Bim down-regulation. Corresponding apoptosis analysis (Figure 4C) demonstrated that miR-181a depletion not only increased cell apoptosis and sensitized the response to the cytotoxic drug Mitoxantrone but also abolished CAM-DR. Given that the histone deacetylase inhibitor SAHA has been shown as a potential new anti-MCL drug, we also tested whether inhibition of miR-181a by anti–miR-181a sensitized SAHA-induced cell apoptosis in Jeko-1 cells. Figure 4B and D reveal that anti–miR-181a enhanced SAHA-induced apoptosis in the absence and presence of HK adhesion and was associated with up-regulation of Bim protein. These results, for the first time, demonstrate that the level of Bim and cell apoptosis is tightly controlled by the interaction with surrounding FDCs via induction of miR-181a.
miR-181a is required for cell adhesion–mediated Bim down-regulation and drug resistance, and anti–miR-181a sensitizes lymphoma cells to mitoxantrone or SAHA-mediated apoptosis. (A-D) Jeko-1 cells transfected with anti–miR-181a or anti-miR-control (anti–miR-Ctrl) were treated with vehicle control (VC) or mitoxantrone (0.2μM) or SAHA (0.5μM) for another 24 hours with or without HK cell adhesion. Bim expression levels (A-B) were then analyzed by Western blot, and apoptosis results (C-D) were analyzed by flow cytometry using annexin V. Data are representative of 4 independent experiments with means ± SD. In panels A and B, the relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane.
miR-181a is required for cell adhesion–mediated Bim down-regulation and drug resistance, and anti–miR-181a sensitizes lymphoma cells to mitoxantrone or SAHA-mediated apoptosis. (A-D) Jeko-1 cells transfected with anti–miR-181a or anti-miR-control (anti–miR-Ctrl) were treated with vehicle control (VC) or mitoxantrone (0.2μM) or SAHA (0.5μM) for another 24 hours with or without HK cell adhesion. Bim expression levels (A-B) were then analyzed by Western blot, and apoptosis results (C-D) were analyzed by flow cytometry using annexin V. Data are representative of 4 independent experiments with means ± SD. In panels A and B, the relative change in Bim protein was measured by quantitative densotometry and is indicated below each lane.
Cell adhesion induces up-regulation of miR-181a and down-regulation of Bim in primary MCL and other non-Hodgkin B-cell lymphomas
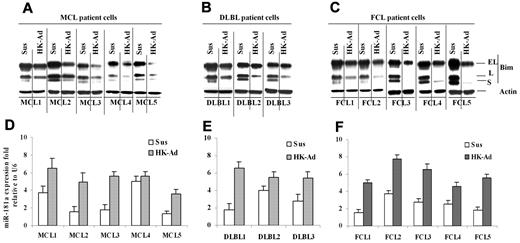
Finally, we determined whether the cell adhesion–mediated Bim down-regulation and miR-181a induction observed in lymphoma cell lines were operative in primary MCL and other NHL specimens. We examined the changes in Bim and miR-181a following adhesion of the primary lymphoma cells to HK cells. Figure 5A through C show that all 13 lymphoma samples (including 5 MCL samples, 3 GC-derived diffuse large B-cell lymphoma [DLBL], and 5 follicular center cell lymphoma) responded to stromal cell adhesion with the down-regulation of Bim expression similar to that observed in lymphoma cell lines. We also determined the effect of cell adhesion on miR-181a in primary lymphoma cells. As shown in Figure 5D through F, primary MCL, DLBL, and FL cells expressed increased levels of miR-181a upon adhesion to HK cells. Overall, this study implicates lymph node stromal cell control of tumor cell response via a miR-181a pathway in MCL and other NHLs.
Cell adhesion–induced down-regulation of Bim protein level and up-regulation of miR-181a expression in primary mantle cell and other non-Hodgkin B-cell lymphomas. (A-C) Bim levels expression in primary mantle cell lymphoma (MCL, A) and diffused large B-lymphoma and follicular cell lymphoma (DLBL and FCL, B-C) cells in suspension (Sus) versus HK adhesion (HK-Ad) were analyzed by Western blot. (D-F) miR-181a expression levels in primary MCL (D) and DLBL (E) and FCL (F) cells in suspension (Sus) versus HK adhesion (HK-Ad) were analyzed by TaqMan microRNA qRT-PCR assays. Error bars represent triplicate qRT-PCR analysis for each sample.
Cell adhesion–induced down-regulation of Bim protein level and up-regulation of miR-181a expression in primary mantle cell and other non-Hodgkin B-cell lymphomas. (A-C) Bim levels expression in primary mantle cell lymphoma (MCL, A) and diffused large B-lymphoma and follicular cell lymphoma (DLBL and FCL, B-C) cells in suspension (Sus) versus HK adhesion (HK-Ad) were analyzed by Western blot. (D-F) miR-181a expression levels in primary MCL (D) and DLBL (E) and FCL (F) cells in suspension (Sus) versus HK adhesion (HK-Ad) were analyzed by TaqMan microRNA qRT-PCR assays. Error bars represent triplicate qRT-PCR analysis for each sample.
Discussion
Mounting evidence now suggests that a dynamic interaction occurs between the tumor cell and its microenvironment, with each profoundly influencing the behavior of the other. This “tumor microenvironment” is a critical determinant for tumor initiation, progression, response to therapy, and drug resistance.3,4 Accumulating studies, including results from our own laboratory, have shown that cell adhesion of hematopoietic tumor cell lines to stromal cells confers a multidrug resistant phenotype and that disruption of cell adhesion–mediated signaling may increase the efficacy of currently used cytotoxic agents. To this end, our laboratory has focused on identifying targets that contribute to CAM-DR. In this study, we investigated the effects of lymph node stromal cells on B-cell lymphoma survival and apoptosis in cell lines and in primary B-lymphoma samples. We demonstrated a critical role of Bim down-regulation in the cell adhesion–mediated protection of lymphoma from drug-induced apoptosis. We characterized miR-181a signaling pathway by which cell adhesion regulates lymphoma cell survival and mediates drug resistance. This study, for the first time, demonstrates that the miRNA pathway is an important part of the mechanism by which cell adhesion regulates the level of Bim and cell survival in B-cell lymphomas. miR-181a serves as an important upstream effector in cell adhesion–mediated Bim down-regulation and hence confers lymphoma cell drug resistance when lymphoma cells are attached to their stroma. This study reveals a novel molecular signaling mechanism for interaction between lymphoma cells and the lymphoma microenvironment.
FDCs are restricted to the B-cell regions of secondary lymphoid tissue and to NHLs derived from the follicular center or the mantle zone. With their cytoplasmic ramifications, they form a dense network which contains the B lymphocytes. In normal physiological conditions, FDCs retain native antigens in the form of immune complexes and present these antigens to B cells during the secondary response. FDCs rescue bound B cells from apoptosis and induce the differentiation of B cells into long-term memory B cell clones. In pathologically lymphomatous conditions, lymphoma cells originate from their normal counterparts and also reside within lymph node. The monoclonal B cells invade new compartments and associate with FDCs to form a spherical network which contains neoplastic B cells. Thus, the adhesive interactions between FDC and benign or malignant B cells may directly influence B-cell survival and apoptosis both in physiological and pathological conditions. FDCs have been demonstrated to be indeed present in FL as well as other types of B-cell lymphoma.7,24 A recent study revealed that that FDCs are ubiquitously present in all 96 MCLs analyzed.7 This is in line with the observations of this study that FDCs confer drug resistance to B-cell lymphomas, regardless of type of lymphomas, as shown in Figure 5 (MCL vs DLBL vs follicular center cell lymphoma [FCL]). The FDC protection from lymphoma apoptosis was previously reported not only in FL but also in pregeminal center cell lymphoma such as chronic lymphocytic leukemia.24 The influence of lymph node stromal cells on lymphoma cell survival is likely to be clinically relevant following induction chemotherapy in B-cell lymphoma, when a small number of lymphoma cells survive in the lymph node due to CAM-DR, which ultimately leads to the recurrence of the disease. For instance, the diffuse distribution of FDCs in MCL is associated with a worse clinical outcome than the nodular distribution patter of FDC.7 Furthermore, recent gene expression profiling studies revealed that a signature, referred to as immune response-2, indicative of macrophage and/or FDC presence, conferred an inferior survival on patients with FL26 and shortened survival in patients with classic Hodgkin lymphoma.27 These findings suggest that FDC/lymphoma interaction has significant impact on most if not all lymphoma patient survival outcomes and contributes to poor survival. Therefore, a better understanding of the molecular mechanism involved in lymph node stroma/lymphoma cell interactions might lead to new therapeutic approaches for residual resistant lymphoma after chemotherapy.
Cell adhesion–mediated Bim down-regulation and its association with drug resistance have been less emphasized. The molecular mechanism of how stroma controls the level of Bim and hence the cell survival has not yet been understood. In the present study, we investigated the cell adhesion–mediated decrease in Bim in lymphoma cell lines and primary lymphoma cells. qRT-PCR analysis did not reveal any decrease in Bim mRNA levels as shown in Figure 2C, suggesting that posttranscriptional regulation was responsible for the down-regulation of Bim following cell adhesion to stroma. More importantly, our data reveal that levels of Bim were down-regulated by stroma and were dependent on the activity of the miR-181a pathway. First, we showed that miR-181a was markedly increased upon lymphoma cell adherence to the lymph node stromal cell line HK in both lymphoma cell lines and primary lymphoma cells. Increased miR-181a was correlated with reciprocal down-regulation of Bim. Second, inhibition of miR-181a expression by specific anti–miR-181a increased Bim levels. Third, overexpression of miR-181a by pre–miR-181a decreased Bim and induced lymphoma cell apoptosis. Finally, we showed that ectopic inhibition of miR-181a abolished cell adhesion–dependent Bim down-regulation, resulting in cell apoptosis and overcoming CAM-DR. Taken together, these data imply that regulation of miR-181a expression by stroma likely plays a significant role in the regulation of lymphoma survival and cell adhesion–regulated drug resistance.
Bim is one of the most potent proapoptotic BH3-only proteins: it binds to all prosurvival BCL-2 family members with high affinity, thereby releasing Bax or Bak proteins, the critical downstream effectors of the BCL-2–dependent pathway of apoptosis. Bim has been implicated in regulation of cell death in several cell types. Mice lacking Bim expression accumulate immune cells of various lineages associated with elevated levels of B cells (T cells), indicating that Bim is essential for regulating lymphocyte homeostasis.28 Moreover, T cells derived from Bim knockout mice are resistant to glucocorticoids, ionomycin, and γ radiation.28 Furthermore, reducing Bim levels with siRNA in K562 cells conferred resistance to imatinib-induced cell death.29 In our study, similar results were observed by reducing Bim levels with a shRNA expression vector in the MCL cell line Jeko-1. In addition, this study showed that reducing Bim levels confers resistance to the topoisomerase II inhibitor mitoxantrone. Together, our data indicate that reduced Bim levels can confer resistance to mechanistically distinct classes of drugs commonly used to treat lymphoma and that cell adhesion–mediated reduction in Bim levels may contribute to the CAM-DR phenotype. In recent years, advances in basic biology have provided a better understanding of how Bim kills cells; however, how Bim expression and activity are regulated has not been fully characterized. It has been reported that Bim is phosphorylated by c-Jun N-terminal kinase in response to various stresses, and this promotes apoptosis.30 In addition, particular attention has recently focused on the regulation of Bim by the prosurvival extracellular signal-regulated kinase 1/2 and protein kinase B pathways that act downstream of oncogenic protein kinases.31 In this study, we demonstrated that Bim is a direct target of miR-181a via translational inhibition, providing a novel mechanism for BIM regulation. Our findings indicate that miR-181a is a key factor inducing protection from drug-primed apoptosis in tumor microenvironment. Indeed, knockdown of miR-181a is sufficient to promote lymphoma cell apoptosis and overcome CAM-DR. This occurs through up-modulation of the proapoptotic factor Bim, unleashed from direct binding of miR-181a to BIM mRNA 3′ UTR. It has been shown that miR-181 is highly expressed in murine B-lymphoid cells, and its ectopic expression in progenitor cells causes an increase in the proportion of B cells.32 This effect is in agreement with our finding that miR-181a supports B lymphocyte survival through regulation of Bim. Thus, our work describes a novel oncogenic pathway underlying lymphoma development, whereby cell adhesion transactivates the miR-181a, which in turn down-modulates the tumor suppressor Bim. Recently, there has been considerable interest in understanding the roles of the miR-181 family of miRNAs in cancer. These studies have suggested that miR-181s function both as oncogene and tumor suppressors, depending on the cellular context. Accordingly, these miRNAs are elevated in breast,33 colon,34 and liver and pancreatic cancers35,36 but are reduced in gliomas37 and aggressive chronic lymphocytic leukemia.38 The role of miR-181s in multiple myeloma as oncomiRNAs has also been demonstrated and confirmed by in vivo studies.39 Pichiorri et al39 revealed overexpression of miR-181a in multiple myeloma and tumor regression of transplanted tumors after treatment with miR181 antagomiRs. These data suggest that miRNAs could have a therapeutic potential in antagonizing the survival of transformed lymphoma cells.
In addition, deletion at the Bim locus has been identified by array CGH.40 It was detected in approximately 17% of MCL samples. This finding demonstrates the biological importance of repressing Bim expression for the survival of MCL. This observation is consistent with our findings that adhesion to FDC can protect MCL by down-regulating Bim. Thus, MCL cells can control Bim expression through FDC interaction or through genomic deletion. Our study offers a molecular mechanism for the potential role of miR-181a in microenvironment-dependent tumor growth and response to therapy. Aberrant expression of miR-181a in the lymphoma results in down-regulation of Bim and enhances the survival of B-cell progenitors.32 The miR-181a–mediated microenvironment-dependent resistance of lymphoma cells to a widely used drug for lymphoma therapy is noteworthy. In this context, it would be of interest to examine therapeutic efficacy of anti–miR-181a in preclinical trials because it sensitizes lymphoma cells to cytotoxic treatment. It is logical to predict that anti–miR-181a alone or in conjunction with other anticancer agents could function as an effective, alternate therapeutic regimen against MCL and other types of B-cell malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to the Tissue Procurement, Molecular Core Laboratory, and Flow Cytometry Core Facilities at H. Lee Moffitt Cancer Center and Research Institute for providing specimens, molecular analysis, and cell apoptosis analysis. We thank Rasa Hamilton for editorial assistance.
This work was supported by grants from the National Cancer Institute (R01CA137123, to J.T.), Leukemia Research Foundation (to J.T.), Maher Fund (to J.T.), Moffitt Cancer Center Foundation (to J.T.), and Merck Inc (IISP32980, to J.T.).
National Institutes of Health
Authorship
Contribution: T.L., J.L., and X.Z. designed and did all the experiments and contributed to manuscript preparation; Y.S.C. and K.L.W provided essential reagents and intellectual support; E.M.S., L.C.M., W.S.D., and J.T. provided clinical samples and information and intellectual support; and J.T. and T.L. designed experiments, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jianguo Tao, MD, PhD, Department of Malignant Hematology and Experimental Therapeutics Program, H. Lee Moffitt Cancer Center and Research Institute at the University of South Florida, MCC-LAB 2071, 12902 Magnolia Dr, Tampa, FL 33613; e-mail: Jianguo.Tao@moffitt.org.