Abstract
FLIP is a well-established suppressor of death receptor-mediated apoptosis. To define its essential in vivo role in myeloid cells, we generated and characterized mice with Flip conditionally deleted in the myeloid lineage. Myeloid specific Flip-deficient mice exhibited growth retardation, premature death, and splenomegaly with altered architecture and extramedullary hematopoiesis. They also displayed a dramatic increase of circulating neutrophils and multiorgan neutrophil infiltration. In contrast, although circulating inflammatory monocytes were also significantly increased, macrophages in the spleen, lymph nodes, and the peritoneal cavity were reduced. In ex vivo cultures, bone marrow progenitor cells failed to differentiate into macrophages when Flip was deleted. Mixed bone marrow chimera experiments using cells from Flip-deficient and wild-type mice did not demonstrate an inflammatory phenotype. These observations demonstrate that FLIP is necessary for macrophage differentiation and the homeostatic regulation of granulopoiesis.
Introduction
Cell surface death receptors, including Fas, TRAIL and TNFR1, are capable of mediating programmed cell death through the activation of caspases 8 or 10. Cellular Flice-like inhibitory protein (FLIP) is critical in the protection against death receptor-mediated apoptosis.1-3 After death receptor ligation caspases 8/10 are recruited to the receptor through adaptor molecules, FADD or TRADD,4-6 resulting in homotypic interactions and autocatalytic caspase activation, which is suppressed by FLIP.7 FLIP is highly expressed in a variety of tumors, and the forced reduction of FLIP is capable of sensitizing certain tumors to death receptor-mediated apoptosis.8-10 Our earlier studies with human monocytes demonstrated that FLIP was induced during monocyte to macrophage differentiation, which protected macrophages from Fas-FasL mediated apoptosis.11 In addition, FLIP is highly expressed in the macrophages within the synovial tissue of patients with rheumatoid arthritis, and the forced reduction of FLIP sensitizes macrophages to Fas-mediated apoptosis.11,12
In addition to protecting against apoptosis, a variety of other functions of FLIP have been characterized. At low levels compared with caspase 8, FLIP may be proapoptotic by contributing to caspase 8 activation, promoting Fas-mediated apoptosis.13 The role of FLIP in T lymphocytes is complex, because it is necessary for the maturation of thymocytes by suppressing apoptosis14 and is capable of protecting against the extrinsic cell death of mature lymphocytes, but may also promote apoptosis and the activation of NF-κB (reviewed in Krammer et al15 ). In dendritic cells an N-terminal p22 fragment of FLIP induces NF-κB activation.16 The deletion of FLIP in B lymphocytes results in decreased B lymphocytes and increased sensitivity to Fas-mediated apoptosis and the increased activation of JNK and p38.17 Other nonapoptotic functions of FLIP include the suppression of TNFα-induced JNK activation18 and the impairment of the ubiquitin-proteasome system, resulting in the increased expression of genes regulated by β-catenin.19 However, the potential nonapoptotic functions of FLIP in myeloid cells are unknown.
To elucidate the in vivo role of FLIP in macrophages and neutrophils, we developed mice deficient in Flip in myeloid cells. These mice had significantly reduced body weight and died prematurely. There was a dramatic increase in circulating neutrophils and monocytes (> 10-fold), which was accompanied by multiorgan neutrophil infiltration. Isolated neutrophils deficient in Flip demonstrated a mature morphology and underwent normal apoptosis, oxidative burst, and degranulation. In contrast to monocytes, macrophages were greatly reduced in the spleen, lymph nodes, and peritoneal cavity. In ex vivo cultures, the deletion of Flip prevented macrophage differentiation, and mixed bone marrow chimeric mice failed to recapitulate the phenotype of the mice deficient in Flip in myeloid cells. Together, these observations identify a novel role for FLIP in myeloid homeostasis.
Methods
Generation of Flip conditional knockout in myeloid lineage
The Flip targeting vector, containing 3 loxP sequences inserted in tandem orientation and a neomycin resistance (neoR) cassette, was transfected into the B6/Blu ES cells (ES Cell Core at Washington University, St Louis, MO). After removing the neoR cassette, chimeras were generated by blastocyst injection and embryos reimplantation in C57BL/6 mouse establishing floxed Flip (Flipf/+) mice, which were bred with the Zp3-cre mice to generate mice with one allele of Flip deleted (Flipd/+). After further crosses, myeloid cells deficient in FLIP were generated by crossing with LysMc/+ mice (Jackson Laboratory,20 ), resulting in the genotypes identified in Table 1. Mating Flipf/+, LysMc/+ and Flip+/d, LysMc/+ mice resulted in Flipf/d, LysMc/+ mice, which were deficient in FLIP in myeloid cells.
Complete blood counts of Flipf/d, LysMc/+ and the littermate control mice
| Genotypes . | No. of Flip alleles in whole genome . | No. of Flip alleles in myeloid lineage . | ||||
|---|---|---|---|---|---|---|
| 2 alleles (Flip+/+, f/+, or f/f)LysM+/+ . | 1 allele (Flipd/+or f/d) >LysM+/+ . | 2 alleles (Flip+/+) LysMc/+ . | 1 allele (Flipd/+ or f/+) LysMc/+ . | 1-2 alleles (Flip+/+, f/+, or +/d) LysMc/c . | 0 alleles (Flipf/d) LysMc/+ . | |
| Number | 11 | 8 | 5 | 25 | 9 | 56 |
| Leukocytes | ||||||
| White blood cells (103/μL) | 9.3 ± 1.1‡ | 11.7 ± 1.3† | 9.9 ± 2.2‡ | 11.8 ± 1.2† | 12.6 ± 2.3‡ | 58.8 ± 5.9 |
| Neutrophils (103/μL) | 2.6 ± 0.60‡ | 2.8 ± 0.78† | 2.2 ± 0.50* | 2.6 ± 0.36‡ | 2.6 ± 0.58‡ | 33.9 ± 4.1 |
| Lymphocytes (103/μL) | 6.1 ± 0.8 | 7.9 ± 0.9 | 7.1 ± 1.8 | 7.4 ± 0.80 | 9.0 ± 1.5 | 13.9 ± 1.7 |
| Monocytes (103/μL) | 0.5 ± 0.18‡ | 0.8 ± 0.2† | 0.5 ± 0.2 | 0.7 ± 0.1‡ | 0.80 ± 0.15† | 8.9 ± 1.2 |
| Eosinophila (103/μL) | 0.2 ± 0.04* | 0.2 ± 0.07 | 0.1 ± 0.07 | 0.2 ± 0.04‡ | 0.19 ± 0.09 | 1.9 ± 0.3 |
| Basophila (103/μL) | 0.06 ± 0.02 | 0.05 ± 0.03 | 0.04 ± 0.02 | 0.06 ± 0.01 | 0.02 ± 0.01 | 0.2 ± 0.03 |
| Erythrocytes | ||||||
| Red blood cell (106/μL) | 9.8 ± 0.3‡ | 9.3 ± 0.3* | 9.7 ± 0.1* | 9.5 ± 0.3‡ | 9.52 ± 0.55† | 7.4 ± 0.3 |
| Hemoglobin (g/dL) | 13.5 ± 0.3‡ | 13.2 ± 0.4† | 13.7 ± 0.3* | 15.1 ± 1.5‡ | 15.5 ± 3.3‡ | 9.4 ± 0.3 |
| Hematocrit (%) | 40.8 ± 0.9‡ | 38.8 ± 1.5† | 41.4 ± 0.7† | 40.5 ± 1.2‡ | 41.0 ± 2.4‡ | 30 ± 0.8 |
| Thrombocytes | ||||||
| Platelet (103/μL) | 974 ± 78 | 941 ± 62 | 825 ± 45 | 902 ± 72* | 1026 ± 131 | 1389 ± 76 |
| Mean platelet volume (fL) | 4.6 ± 0.1 | 4.5 ± 0.07 | 4.5 ± 0.07 | 4.8 ± 0.1 | 4.3 ± 0.04† | 5.0 ± 0.06 |
| Genotypes . | No. of Flip alleles in whole genome . | No. of Flip alleles in myeloid lineage . | ||||
|---|---|---|---|---|---|---|
| 2 alleles (Flip+/+, f/+, or f/f)LysM+/+ . | 1 allele (Flipd/+or f/d) >LysM+/+ . | 2 alleles (Flip+/+) LysMc/+ . | 1 allele (Flipd/+ or f/+) LysMc/+ . | 1-2 alleles (Flip+/+, f/+, or +/d) LysMc/c . | 0 alleles (Flipf/d) LysMc/+ . | |
| Number | 11 | 8 | 5 | 25 | 9 | 56 |
| Leukocytes | ||||||
| White blood cells (103/μL) | 9.3 ± 1.1‡ | 11.7 ± 1.3† | 9.9 ± 2.2‡ | 11.8 ± 1.2† | 12.6 ± 2.3‡ | 58.8 ± 5.9 |
| Neutrophils (103/μL) | 2.6 ± 0.60‡ | 2.8 ± 0.78† | 2.2 ± 0.50* | 2.6 ± 0.36‡ | 2.6 ± 0.58‡ | 33.9 ± 4.1 |
| Lymphocytes (103/μL) | 6.1 ± 0.8 | 7.9 ± 0.9 | 7.1 ± 1.8 | 7.4 ± 0.80 | 9.0 ± 1.5 | 13.9 ± 1.7 |
| Monocytes (103/μL) | 0.5 ± 0.18‡ | 0.8 ± 0.2† | 0.5 ± 0.2 | 0.7 ± 0.1‡ | 0.80 ± 0.15† | 8.9 ± 1.2 |
| Eosinophila (103/μL) | 0.2 ± 0.04* | 0.2 ± 0.07 | 0.1 ± 0.07 | 0.2 ± 0.04‡ | 0.19 ± 0.09 | 1.9 ± 0.3 |
| Basophila (103/μL) | 0.06 ± 0.02 | 0.05 ± 0.03 | 0.04 ± 0.02 | 0.06 ± 0.01 | 0.02 ± 0.01 | 0.2 ± 0.03 |
| Erythrocytes | ||||||
| Red blood cell (106/μL) | 9.8 ± 0.3‡ | 9.3 ± 0.3* | 9.7 ± 0.1* | 9.5 ± 0.3‡ | 9.52 ± 0.55† | 7.4 ± 0.3 |
| Hemoglobin (g/dL) | 13.5 ± 0.3‡ | 13.2 ± 0.4† | 13.7 ± 0.3* | 15.1 ± 1.5‡ | 15.5 ± 3.3‡ | 9.4 ± 0.3 |
| Hematocrit (%) | 40.8 ± 0.9‡ | 38.8 ± 1.5† | 41.4 ± 0.7† | 40.5 ± 1.2‡ | 41.0 ± 2.4‡ | 30 ± 0.8 |
| Thrombocytes | ||||||
| Platelet (103/μL) | 974 ± 78 | 941 ± 62 | 825 ± 45 | 902 ± 72* | 1026 ± 131 | 1389 ± 76 |
| Mean platelet volume (fL) | 4.6 ± 0.1 | 4.5 ± 0.07 | 4.5 ± 0.07 | 4.8 ± 0.1 | 4.3 ± 0.04† | 5.0 ± 0.06 |
No difference among groups with 1 or 2 Flip alleles either in whole genome or in myeloid linage was identified. Significant differences were noted between the identified values compared with the Flip f/d, LyzM c/+ group, indicated by *P < .05,
P < .01, and
P < .001.
Phenotypic analysis
Complete blood counts and differentials were performed using a Hemavet 950 (Drew Scientific).21 FLIP expression was determined by immunoblotting with an antibody to the N-terminal peptide of FLIP (Cell Signaling). For cultivation of microorganisms, tissues was collected under sterile conditions and cultured in Brain Heart Infusion Broth (Remel) at 37°C overnight. Circulating cytokines/chemokines were quantified by the Premixed 32 Plex kit (Millipore), and the data were acquired by LUMINEX 200.
Histology and immunohistochemistry
Organs were dissected and fixed in 10% neutral formalin, embedded in paraffin, and then 5-μm sections were stained with hematoxylin and eosin. Cryosections were processed for immunofluorescence staining of macrophages as described,22 then incubated with primary antibodies to mouse F4/80 (B8; eBioscience), CD169 (AbD Serotec), or isotype control immunoglobulin G at 4°C overnight, and subsequently with AlexaFluor 594 conjugated anti-rat secondary antibody (Invitrogen) for 2 hours and counterstained with DAPI (4′,6-diamidino-2-phenylindole).
Immunophenotying
Cells were preincubated with anti-mouse CD16/CD32 antibody (BD Phamingen) to block cell surface Fc III/II receptors before antibody staining. Immunophenotyping was performed by multicolor fluorochrome-conjugated antibody cocktails, including antibodies to CD 45, CD11b, F4/80, Gr 1, CD115, CD62L, CD4, CD8, CD3, CD19, B220, and CD11c (eBioscience or BD Pharmingen). Data were acquired on a BD LSR II flow cytometer (BD FACSDIVA software) and analyzed by FlowJo Version 8.8.4 (TreeStar). Single cell populations were sorted by Moflo High Speed Sorter.
Bone marrow reconstitution
C57BL/6 recipients CD45.1+ congenic (B6.SJL-Ptprca Pepcb/BoyJ, 6-week-old females; The Jackson Laboratory) were lethally irradiated (1000 rads), followed by the retroorbital administration of 5 × 106 donor whole bone marrow cells. The donor bone marrow was collected from CD45.1+ (wild-type) and CD45.2+ (Flipf/d, LysMc/+) mice. The bone marrow was transferred to the recipients containing 0%, 25%, 50%, 75%, and 100% of cells from Flipf/d, LysMc/+ mice. Recipients received sulfamethoxazole (50 mg/mL) and trimethoprim (8 mg/mL) in the drinking water and were subjected to phenotypic analysis 8 weeks posttransplantation.
Programmed cell death
Assessment of neutrophil function
Mouse peripheral blood cells were incubated with dihydrorhodamine 123 (0.5μM), with or without phorbol 12-myristate 13-acetate (PMA; 10nM) at 37°C for 30 minutes. Thereafter, the samples were chilled on ice and incubated with fluorochrome-conjugated anti-CD11b and -Gr1, followed by erythrocyte lysis. Oxidative burst was determined by measuring the conversion of nonfluorescent dihydrorhodamine 123 into fluorescent rhodamine12324 and degranulation by the increased expression of cell surface CD11b.24 Myeloperoxidase staining was performed by incubating slides in a solution of 3.3′-diaminobenzidine and hydrogen peroxide.
Colony formation
Spleen (1 × 104) or bone marrow cells (0.5 × 105), combined from femurs and tibias, were seeded in methylcellulose media (Methocult: StemCell Technologies) in the presence of interleukin-3 (IL-3; 10 ng/mL), IL-6 (10 ng/mL), stem cell factor (50 ng/mL), and granulocyte macrophage colony-stimulating factor (GM-CSF; 10 ng/mL) for 7 days. Colony-forming unit of granulocyte and macrophage (CFU-GM) colonies were counted under light microscropy. For megakaryocyte-CFUs (CFU-Mk), cells (1 × 105) were seed in a collagen-based media (Megacult-C; StemCell Technologies) supplemented with IL-3 (10 ng/mL), IL-6 (10 ng/mL), IL-11 (10 ng/mL), thrombopoietin (10 ng/mL) for 7 days. The Megacult-C cultures were dehydrated and stained for acetylcholinesterase activity to determine the CFU-MK colonies.25
Macrophage survival during differentiation
CD117+ (c-Kit) hematopoietic stem cells from the bone marrow of 8- to 12-week-old Flipf/+ or Flipf/d mice were isolated using antibody to the stem cell receptor CD117 (StemCell Technologies) according to the manufacturer's instructions. These freshly isolated cKit+ cells were cultured in the presence of stem cell factor (10 ng/mL), IL-3 (10 ng/mL), and IL-6 (10 ng/mL) overnight, followed by culture with recombinant retroviral vectors expressing green fluorescent protein (GFP) alone or GFP plus Cre for an additional 24 hours.25 The cells were then incubated with 20% L929 conditioned medium containing macrophage colony-stimulating factor (M-CSF) to allow in vitro macrophage differentiation for 5 days.
Data analysis
All quantitative data are presented as mean ± SEM. Statistical analysis between groups was done with 2-tailed Student t test. Nonparametric data were analyzed by the Mann-Whitney rank sum test. One-way analysis of variance (Tukey pairwise mean comparison) was performed for multigroup analysis. Significance levels were set at .05.
Results
Flip conditional knockout in myeloid lineage
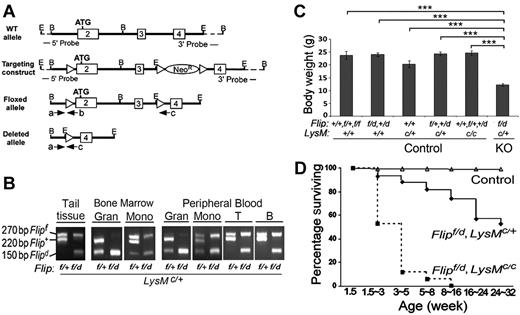
To generate mice with Flip deleted in myeloid cells, loxP sequences were inserted flanking exons 2 and 3 of Flip (Figure 1A), which was used to generate Flip floxed (Flipf) embryonic stem cells (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Deletion of one floxed allele in the whole genome (Flipd/+) was obtained by crossing heterozygous Flipf/+ mice with Zp3-Cre mice that express Cre in the female germ line (supplemental Figure 1B). Myeloid linage-specific deletion of the flipf was carried out by crossing Flipf/+ or Flipf/d mice with those expressing Cre driven by the LysM promoter (LysMc/+; Figure 1B). When Flipf/+ mice were crossed with the LysMc/+ mice, the floxed allele was deleted in granulocytes and monocytes, demonstrating the efficiency of Cre in these cells. Crossing Flipf/d mice with LysMc/+ mice resulted in the deletion of both alleles of Flip in bone marrow and peripheral blood granulocytes (Figure 1B). However, complete deletion of the floxed Flip allele was not observed in monocyte bone marrow precursors or peripheral blood monocytes (Figure 1B). As expected, neither allele of Flip was deleted in circulating T or B lymphocytes when Flipf/d or Flipf/+ mice were crossed with LysMc/+ mice (Figure 1B).
Deletion of Flip in myeloid linage results in postnatal growth retardation and premature death. (A) Schematic of the Flip targeting strategy. A 9.5-kb Flip genomic DNA segment (exon 2 to 4 from a C57BL/6J mouse BAC library) was used to engineer the Flip conditional mutagenesis construct. Exons are indicated by rectangular boxes and E and B represent the restriction cleavage sites of EcoRI and BamHI. Three loxP sites were inserted as indicated by triangles. The dashed lines represent the flanking region for Southern blot probes. The location of 3 PCR primers (a, b, c) and orientation are indicated by arrows. Amplification using primers a and b generates a 220-bp fragment for wild-type (Flip+) and a 270-bp fragment for the floxed (Flipf) allele. Cre-induced recombination generates a 150-bp fragment for depleted Flip allele (Flipd) using primers a and c. All recombinant DNA and animal procedures were approved by the Office of Research Safety and the Institutional Animal Care and Use Committee of Northwestern University. (B) PCR genotyping of the LysM-cre–induced cell type-specific deletion of Flipf. The representative Flipf/+, LysMc/+ littermate control and Flipf/d, LysMc/+ knockout (KO) mice were genotyped from tail biopsy, and the different cell types were isolated from bone marrow (BM), and peripheral blood were genotyped by 3 PCR primers indicated in panel A. Granulocytes (Gran) were 11b+/Gr1+ F4/80−; peripheral blood monocytes or bone marrow monocyte precursors (Mono) were 11b+, F4/80+; B cells were 11b−/CD19+, and T cells were 11b−CD3+. (C) Body weight of Flipf/d, LysMc/+ (n = 53) and littermate controls including: Flip+/+, Flipf/+ or Flipf/f, LysM+/+ (n = 9); Flipf/d or Flip+/d, LysM+/+ (n = 10); Flip+/+, LysMc/+ (n = 5); Flipf/+or Flip+/d, LysMc/+ (n = 24); and Flip+/+, Flipf/+or Flip+/d, LysMc/c (n = 12). All mice are between 6 to 24 weeks of age. ***P < .001, compared with indicated groups. (D) Postnatal viability of Flipf/d, LysMc/+ (n = 101) and the Flipf/d, LysMc/c (n = 17) and littermate controls (n = 475), which included: Flip+/+, Flipf/+ or Flipf/f, LysM+/+ (n = 112), Flipf/d or Flip+/d, LysM+/+ (n = 112), Flip+/+, LysMc/+ (n = 47), Flipf/+or Flip+/d, LysMc/+ (n = 127), and Flip+/+, Flipf/+or Flip+/d, LysMc/c (n = 77) mice.
Deletion of Flip in myeloid linage results in postnatal growth retardation and premature death. (A) Schematic of the Flip targeting strategy. A 9.5-kb Flip genomic DNA segment (exon 2 to 4 from a C57BL/6J mouse BAC library) was used to engineer the Flip conditional mutagenesis construct. Exons are indicated by rectangular boxes and E and B represent the restriction cleavage sites of EcoRI and BamHI. Three loxP sites were inserted as indicated by triangles. The dashed lines represent the flanking region for Southern blot probes. The location of 3 PCR primers (a, b, c) and orientation are indicated by arrows. Amplification using primers a and b generates a 220-bp fragment for wild-type (Flip+) and a 270-bp fragment for the floxed (Flipf) allele. Cre-induced recombination generates a 150-bp fragment for depleted Flip allele (Flipd) using primers a and c. All recombinant DNA and animal procedures were approved by the Office of Research Safety and the Institutional Animal Care and Use Committee of Northwestern University. (B) PCR genotyping of the LysM-cre–induced cell type-specific deletion of Flipf. The representative Flipf/+, LysMc/+ littermate control and Flipf/d, LysMc/+ knockout (KO) mice were genotyped from tail biopsy, and the different cell types were isolated from bone marrow (BM), and peripheral blood were genotyped by 3 PCR primers indicated in panel A. Granulocytes (Gran) were 11b+/Gr1+ F4/80−; peripheral blood monocytes or bone marrow monocyte precursors (Mono) were 11b+, F4/80+; B cells were 11b−/CD19+, and T cells were 11b−CD3+. (C) Body weight of Flipf/d, LysMc/+ (n = 53) and littermate controls including: Flip+/+, Flipf/+ or Flipf/f, LysM+/+ (n = 9); Flipf/d or Flip+/d, LysM+/+ (n = 10); Flip+/+, LysMc/+ (n = 5); Flipf/+or Flip+/d, LysMc/+ (n = 24); and Flip+/+, Flipf/+or Flip+/d, LysMc/c (n = 12). All mice are between 6 to 24 weeks of age. ***P < .001, compared with indicated groups. (D) Postnatal viability of Flipf/d, LysMc/+ (n = 101) and the Flipf/d, LysMc/c (n = 17) and littermate controls (n = 475), which included: Flip+/+, Flipf/+ or Flipf/f, LysM+/+ (n = 112), Flipf/d or Flip+/d, LysM+/+ (n = 112), Flip+/+, LysMc/+ (n = 47), Flipf/+or Flip+/d, LysMc/+ (n = 127), and Flip+/+, Flipf/+or Flip+/d, LysMc/c (n = 77) mice.
Disruption of Flip in myeloid lineage results in severe postnatal growth retardation and premature death
The deletion of Flip in myeloid cells resulted in the expected Mendalian birth rate with no indication of embryonic lethality (data not shown). However, the Flipf/d, LysMc/+ mice displayed a runted appearance and a 50% reduction in size (Figure 1C). None of the littermates of the Flipf/d, LysMc/+ mice (defined in Table 1) demonstrated reduced size (Figure 1C) or increased mortality (data not shown) and were therefore collectively used as the littermate controls in further experiments. Approximately 50% of the Flipf/d, LysMc/+ died before 32 weeks of age, whereas > 80% of Flipf/d, LysMc/c mice died by 3-5 weeks of age, most likely because of a more complete deletion of Flip (Figure 1D), and not because of the deletion of both LysM alleles, because there was no effect on phenotype, including body weight (Figure 1C) or blood count (Table 1), when one or both alleles of Flip were still present. These observations demonstrate that the reduction of FLIP in myeloid cells is critical for postnatal survival.
Deletion of Flip in the myeloid lineage results in leukocytosis, multiorgan neutrophil infiltration, and systemic infection
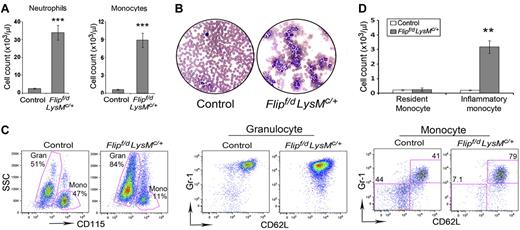
Examination of peripheral blood demonstrated a 5-fold increase of leukocytes in the Flipf/d, LysMc/+ mice compared with the littermate controls (58.8 ± 5.9 vs. 11.0 ± 0.7 × 103/μL, P < .001). Circulating neutrophils and monocytes were increased > 10-fold compared with the controls (P < .001; Figure 2A and Table 1). Eosinophila, thrombocytosis, and anemia were also present in the Flipf/d, LysMc/+ mice, but not in the littermate controls (Table 1). Examination of stained peripheral blood smears from the Flipf/d, LysMc/+ mice demonstrated mature neutrophils with appropriate nuclear morphology and cytoplasmic granularity. The neutrophils from the Flipf/d, LysMc/+ mice were CD45+, CD11b+, CD115−, Gr1hi, CD62Lhi, similar to the littermate controls (Figure 2C). The circulating monocytes appeared normal by morphology; however, the Flipf/d, LysMc/+ mice demonstrated an expanded population of monocytes that was CD45+, CD11b+, CD115+, Gr1+, CD62L+, identifying them as inflammatory monocytes (Figure 2C). In contrast, the expected distribution of resident (Gr-1−, CD62L−) and inflammatory (Gr-1+, CD62L+) monocytes were observed in the controls (Figure 2C). However, although the absolute number of inflammatory monocytes was increased in the Flipf/d, LysMc/+ mice, the number of resident monocytes was not different from the controls (Figure 2D). Analysis of peripheral blood also demonstrated a decreased percentage of B cells and CD4+ and CD8+ T cells, which was due to the marked increase of granulocytes. However, the absolute number of CD8+ cells was actually increased in the Flipf/d, LysMc/+ mice, whereas B cells and CD4+ lymphocytes were not different from the controls (supplemental Figure 2A-C).
Flip deletion in myeloid linage results in leukocytosis. (A) Peripheral blood from Flipf/d, LysMc/+ mice (n = 56) and their littermate controls (mixed genotypes, n = 50) were examined for completely blood count, and neutrophils and monocytes are presented. ***P < .001 compared with the controls. (B) Representative blood smears from a Flipf/d, LysMc/+ mice and controls stained with Hema-3. Data are representative of smears from 3-4 mice for each group. (C) Representative flow cytometric analysis of circulating monocytes and neutrophils of Flipf/d, LysMc/+ and sex-matched littermate controls. Cells are gated by side scatter (SSC) and the expression of CD115, CD62L, and Gr1. (D) Absolute cell count of resident and inflammatory monocytes calculated from the total number of monocytes and the percentage of each subset using 5 Flipf/d, LysMc/+ and sex-matched littermate controls.
Flip deletion in myeloid linage results in leukocytosis. (A) Peripheral blood from Flipf/d, LysMc/+ mice (n = 56) and their littermate controls (mixed genotypes, n = 50) were examined for completely blood count, and neutrophils and monocytes are presented. ***P < .001 compared with the controls. (B) Representative blood smears from a Flipf/d, LysMc/+ mice and controls stained with Hema-3. Data are representative of smears from 3-4 mice for each group. (C) Representative flow cytometric analysis of circulating monocytes and neutrophils of Flipf/d, LysMc/+ and sex-matched littermate controls. Cells are gated by side scatter (SSC) and the expression of CD115, CD62L, and Gr1. (D) Absolute cell count of resident and inflammatory monocytes calculated from the total number of monocytes and the percentage of each subset using 5 Flipf/d, LysMc/+ and sex-matched littermate controls.
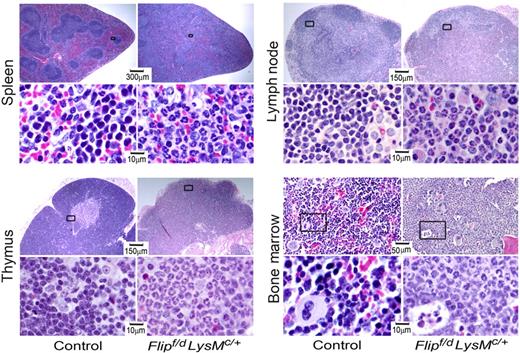
Because of the changes in leukocytes, lymphoid organs were subjected to pathologic analysis. A dramatic infiltration of neutrophils was observed not only in the bone marrow, but also in spleen, lymph nodes, and thymus (Figure 3). Within the spleen and lymph nodes, the architecture was disrupted, with loss of lymphoid follicles. The thymus demonstrated loss of a distinct medulla and cortex, an increase of neutrophils, and a reduction of thymocytes (Figure 3). Morphologically, the bone marrow revealed an expansion of neutrophils and a reduction of erythropoiesis, without morphologic evidence of myeloid dysplasia (Figure 3). No histologic abnormalities of bone were noted in the Flip-deficient mice. The spleen demonstrated a reduction in the percentage of B lymphocytes, and of CD8+ and CD4+ lymphocytes in the Flipf/d, LysMc/+ mice, although only the absolute number of B cells was reduced (P < .01; supplemental Figure 3). By immunophenotyping, a significant (P < .05-.01) increase in the absolute number of Gr-1+ neutrophils was identified in the spleen and lymph nodes, but not the bone marrow or thymus, even though the percentage of neutrophils was increased in each of the organs, from the Flipf/d, LysMc/+ mice (supplemental Figure 4). Nonlymphoid tissues were also affected. Accumulation of neutrophils was demonstrated in the lungs in the interalveolar spaces and around large blood vessels and in the small intestine, especially the jejunum (supplemental Figure 5). Neutrophilic infiltration was less severe in the liver and heart (supplemental Figure 5), as well as the stomach, large intestine, kidney, and brain (data not shown). Overall, pathologic analysis demonstrated an expansion and a multiorgan infiltration of neutrophils.
Flip deletion in myeloid linage results in multiorgan neutrophil infiltration. Hematoxylin and eosin staining of representative sections of tissues from the indicated organs from Flipf/d, LysMc/+ and control mice. The area in the box is enlarged in the panel below. Data are representative of sections from 3-4 mice for each group.
Flip deletion in myeloid linage results in multiorgan neutrophil infiltration. Hematoxylin and eosin staining of representative sections of tissues from the indicated organs from Flipf/d, LysMc/+ and control mice. The area in the box is enlarged in the panel below. Data are representative of sections from 3-4 mice for each group.
To determine whether the increased neutrophils might be due to an underlying infection, organs from Flipf/d, LysMc/+ mice were cultured for microorganisms. Cultures of the spleen, liver, lymph nodes, or peritoneal fluid were positive for bacteria in 30% of the mice, whereas none of the littermate controls was positive. There was no difference in the neutrophil counts between the Flipf/d, LysMc/+ mice that were culture positive and negative, although the culture positive mice were smaller (< 10 g) compared with those that were culture negative (supplemental Figure 6A-B). The cultures revealed several commensal intestinal organisms including Streptococcus sanguins, Enterococcus faecalis, Proteus mirabilis, and Escherichia coli. Based on susceptibility testing, Flipf/d, LysMc/+ mice were treated with antibiotics using ampicillin in drinking water and/or gentamicin subcutaneously. Treatment reduced the positive cultures by 50% but there was no reduction of circulating neutrophils (supplemental Figure 6C) and no increase of weight. These results suggest that infection was not the cause of the neutrophil expansion or the increased mortality.
Normal apoptosis, oxidative burst, and degranulation in Flip-deficient neutrophils
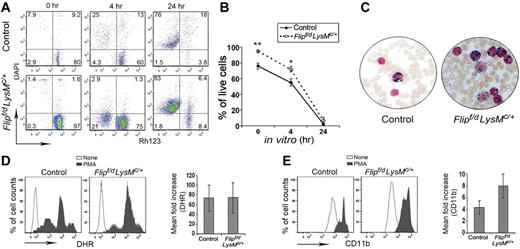
Because of the increased numbers of neutrophils and infections, studies were performed to determine whether there was an abnormality of survival or function of neutrophils. Control peripheral blood neutrophils exhibited spontaneous apoptosis with the loss of mitochondrial transmembrane potential and uptake of DAPI that was limited immediately after isolation and increased at 4 hours, with the marked loss of membrane integrity (DAPI+) by 24 hours (Figure 4A). The percentage of cells that had lost mitochondrial transmembrane potential or membrane integrity at 0 and 4 hours was somewhat less in the Flipf/d, LysMc/+ mice (Figure 4A). However, neutrophil cell death was essentially complete by 24 hours using circulating granulocytes from the Flipf/d, LysMc/+ mice, similar to the controls (Figure 4B). Because the Flipf/d, LysMc/+ mice experienced infections, studies were performed to examine neutrophil function. Peripheral blood neutrophils from Flipf/d, LysMc/+ mice demonstrated normal oxidative burst (Figure 4C) and degranulation after activation with PMA (Figure 4D). In response to the TLR4 ligand lipopolysaccharide, after 1 hour no neutrophil oxidative burst was detected, although degranulation was observed and was comparable in the Flip-deficient and control mice (data not shown). In addition, myeloperoxidase activity of peripheral blood neutrophils in the Flipf/d, LysMc/+ mice was comparable to the controls (Figure 4E). These observations do not suggest that an intrinsic abnormality of apoptosis or function was responsible for the neutrophilia or infection.
Normal apoptosis and function in Flip-deficient circulating neutrophils. (A) Time-dependent loss of mitochondrial transmembrane potential (Δ Ψm) was assessed by decreased Rh123 fluorescence (x-axis) and the loss of membrane integrity assessed by uptake of DAPI (y-axis). (B) The percentage of live cells is identified as Rh123+, DAPI− (n = 4 for each group); *P < .05 and **P < .01 between groups. (C) Representative myeloperoxidase staining of peripheral blood smears, observed by light microscopy (400×). Data are representative of 3 Flipf/d, LysMc/+ and sex-matched littermate controls. (D) The ability to oxidize nonfluoresent dihydrorhodamine 123 was accessed as increased mean florescence intensity after PMA activation. (E) Neutrophil degranulation was determined by increased mean florescence intensity of cell surface CD11b after PMA stimulation. The observations were obtained from 5 Flipf/d, LysMc/+ and sex-matched littermate controls.
Normal apoptosis and function in Flip-deficient circulating neutrophils. (A) Time-dependent loss of mitochondrial transmembrane potential (Δ Ψm) was assessed by decreased Rh123 fluorescence (x-axis) and the loss of membrane integrity assessed by uptake of DAPI (y-axis). (B) The percentage of live cells is identified as Rh123+, DAPI− (n = 4 for each group); *P < .05 and **P < .01 between groups. (C) Representative myeloperoxidase staining of peripheral blood smears, observed by light microscopy (400×). Data are representative of 3 Flipf/d, LysMc/+ and sex-matched littermate controls. (D) The ability to oxidize nonfluoresent dihydrorhodamine 123 was accessed as increased mean florescence intensity after PMA activation. (E) Neutrophil degranulation was determined by increased mean florescence intensity of cell surface CD11b after PMA stimulation. The observations were obtained from 5 Flipf/d, LysMc/+ and sex-matched littermate controls.
Flip deletion results in increased extramedullary myelopoiesis
Because there were increased granulocytes, monocytes, and platelets, studies were performed to determine the effect of Flip deletion on myelopoiesis. Examination of the bone marrow revealed no difference of GM-CFU between the Flipf/d, LysMc/+, and the littermate controls (Table 2). In contrast, using cells from the spleen, a significant (P < .01) increase of CFU-GM was observed in the Flipf/d, LysMc/+ mice (Table 2). The changes were not restricted to the GM, because the CFU-Mk were also increased (P < .001) in the spleens of the Flipf/d, LysMc/+ mice, but not the bone marrow (Table 2). Serum from the peripheral blood was used to determine whether growth factors might contribute to the extramedullary hematopoiesis. G-CSF, GM-CSF, M-CSF, IL-6, and IL-17 were all increased in the Flipf/d, LysMc/+ compared with the control mice (Table 3). These observations document a marked increase of splenic extramedullary hematopoiesis, which was associated with an increase of myeloid growth factors.
Extramedullary hematopoiesis in the Flipf/d, LysMc/+ mice
| Colony-forming units . | Littermate controls (no. of colonies) . | Flipf/d, LysMc/+ (no. of colonies) . | P . |
|---|---|---|---|
| Granulocyte monocyte macrophage | |||
| Spleen | 5.8 ± 0.8 | 270.2 ± 36.1 | < .002 |
| Bone marrow | 92.3 ± 12.7 | 74.2 ± 14.4 | NS* |
| Megakaryocyte | |||
| Spleen | 1.8 ± 0.4 | 42.1 ± 0.9 | < .0001 |
| Bone marrow | 31.1 ± 4.2 | 21.4 ± 1.6 | NS* |
| Colony-forming units . | Littermate controls (no. of colonies) . | Flipf/d, LysMc/+ (no. of colonies) . | P . |
|---|---|---|---|
| Granulocyte monocyte macrophage | |||
| Spleen | 5.8 ± 0.8 | 270.2 ± 36.1 | < .002 |
| Bone marrow | 92.3 ± 12.7 | 74.2 ± 14.4 | NS* |
| Megakaryocyte | |||
| Spleen | 1.8 ± 0.4 | 42.1 ± 0.9 | < .0001 |
| Bone marrow | 31.1 ± 4.2 | 21.4 ± 1.6 | NS* |
Values represented are mean of 3 independent experiments using gender-matched littermate controls performed in triplicate.
NS, not significant.
The Flipf/d, LysMc/+ mice exhibit increased myeloid cytokines
| Cytokines . | Littermate controls (pg/mL) . | Flipf/d, LysMc/+ (pg/mL) . | Mean fold increase . | P . |
|---|---|---|---|---|
| G-CSF | 598.4 ± 213.1 | 8648.2 ± 1690.9 | 14.45 | < .001 |
| IL-6 | 65.0 ± 39.4 | 1640.0 ± 464.4 | 25.23 | < .001 |
| M-CSF | 3.9 ± 0.54 | 11.4 ± 3.9 | 2.9 | < .03 |
| GM-CSF | 0.8 ± 0.8 | 21.7 ± 11.4 | 26.5 | NS* |
| IL-17 | 4.4 ± 1.0 | 14.9 ± 5.4 | 3.4 | < .003 |
| Cytokines . | Littermate controls (pg/mL) . | Flipf/d, LysMc/+ (pg/mL) . | Mean fold increase . | P . |
|---|---|---|---|---|
| G-CSF | 598.4 ± 213.1 | 8648.2 ± 1690.9 | 14.45 | < .001 |
| IL-6 | 65.0 ± 39.4 | 1640.0 ± 464.4 | 25.23 | < .001 |
| M-CSF | 3.9 ± 0.54 | 11.4 ± 3.9 | 2.9 | < .03 |
| GM-CSF | 0.8 ± 0.8 | 21.7 ± 11.4 | 26.5 | NS* |
| IL-17 | 4.4 ± 1.0 | 14.9 ± 5.4 | 3.4 | < .003 |
n = 14 in each group.
NS indicates not significant.
Deletion of Flip results in reduction of macrophages in the spleen and lymph nodes
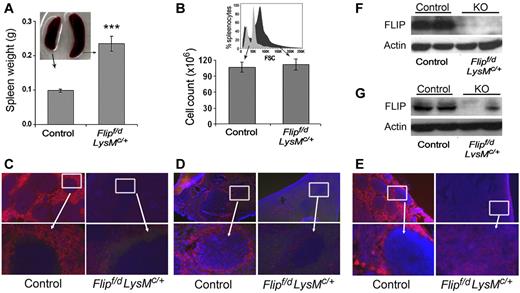
Splenomegaly (Figure 5A) with loss of lymphoid follicles and infiltration with granulocytes (Figure 3) was observed in the Flipf/d, LysMc/+ mice. However, the total number of cells in the Flipf/d, LysMc/+ and control spleens was not different (Figure 5B). This is most likely due to an increase of immature and myeloid cells that are larger than lymphoid cells, which were reduced. An increased size of cells in the spleens for the Flipf/d, LysMc/+ mice was documented by the right shift in forward scatter determined by flow cytometry (Figure 5B). No RBC sequestration was apparent (Figure 3). Immunofluorescence staining revealed a marked reduction of macrophages in both the red pulp (anti-F4/80; Figure 5C) and the marginal zone (anti-CD169; Figure 5D) in the Flipf/d, LysMc/+ mice. Examination of lymph nodes also showed a marked reduction of F4/80+ macrophages (Figure 5E). In contrast there was no reduction of macrophages in the liver (data not shown). By immunoblot analysis, FLIP was reduced but still present in lysates of the spleen (Figure 5F) and lymph nodes (Figure 5G) of the Flipf/d, LysMc/+ mice, most likely because of a marked increase of neutrophils that do not express FLIP and a marked reduction of B cells in spleen (supplemental Figure 3). Therefore, the deletion of Flip in myeloid cells results in a reduction of macrophages in the spleen and lymph nodes, with the loss of lymphoid follicles.
Decreased macrophages and FLIP expression in the spleen and lymph node. Spleen size (n = 31; A) and total number of spleen cells (n = 16; B) in Flipf/d, LysMc/+ and littermate controls. A representative flow histogram of forward scatter (FSC) from ungated splenocytes of Flipf/d, LysMc/+ and littermate control mice is presented in the inset of panel B. Immunofluorescence microscopy of spleen was performed to identify red pulp macrophages (anti-F4/80; C) or marginal zone macrophages (anti-CD169; D), and of lymph nodes with anti-F4/80 antibodies (E). The data are representative of sections from 3-4 mice of each group. The area in the box is enlarged in the bottom panel. Randomly selected spleens (F) and lymph nodes (G) from littermate controls and Flipf/d, LysMc/+ mice were used to examine the expression of FLIP determined by immunoblot analysis. The data are representative of > 4 mice for each group.
Decreased macrophages and FLIP expression in the spleen and lymph node. Spleen size (n = 31; A) and total number of spleen cells (n = 16; B) in Flipf/d, LysMc/+ and littermate controls. A representative flow histogram of forward scatter (FSC) from ungated splenocytes of Flipf/d, LysMc/+ and littermate control mice is presented in the inset of panel B. Immunofluorescence microscopy of spleen was performed to identify red pulp macrophages (anti-F4/80; C) or marginal zone macrophages (anti-CD169; D), and of lymph nodes with anti-F4/80 antibodies (E). The data are representative of sections from 3-4 mice of each group. The area in the box is enlarged in the bottom panel. Randomly selected spleens (F) and lymph nodes (G) from littermate controls and Flipf/d, LysMc/+ mice were used to examine the expression of FLIP determined by immunoblot analysis. The data are representative of > 4 mice for each group.
Deletion of Flip in the myeloid lineage results in increased neutrophils and the absence of mature macrophages in the peritoneal cavity
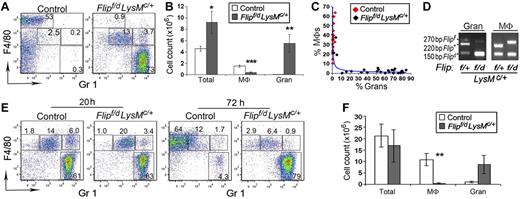
F4/80+, Gr1− resident macrophages were abundant in the peritoneal cavity of the littermate control mice expressing FLIP (Figure 6A). However, in the Flipf/d, LysMc/+ mice, the increased total cell count (Figure 6A-B) was due to a marked (P < .01) increase of Gr1+, F4/80− neutrophils, whereas F4/80+,Gr1− macrophages were significantly (P < .001) reduced (Figure 6A-B). There was a highly significant inverse relationship between the percentage of macrophages and granulocytes (R = -0.75, P < .001), that was more clearly described by the trend line (Figure 6C). Analysis of the peritoneal space of wild-type and the Flipf/d, LysMc/+ mice revealed that the percentage of granulocytes increased to > 10% only when the percentage of macrophages was reduced to < 10% of the total peritoneal cells (Figure 6C). The macrophages that could be isolated from the peritoneum of Flipf/d, LysMc/+ mice did not demonstrate deletion of Flip (Figure 6D). In addition, the Flipf/d, LysMc/+ mice demonstrated an increase percentage of cells expressing intermediate levels of F4/80 and Gr1 (F4/80mid, Gr1mid), which were also CD62L+ (data not shown) and may represent inflammatory monocytes. Further, there was a reduction of resident CD19+ B cells but not CD3+ T cells in the Flipf/d, LysMc/+ mice (supplemental Figure 7). To examine the role of FLIP in elicited macrophages, thioglycollate was injected into the peritoneum. At 20 hours, there was a marked increase of neutrophils in the controls, whereas neutrophil dominance persisted in the Flipf/d, LysMc/+ mice (Figure 6E). In both the Flipf/d, LysMc/+ mice and the controls there was a population of F4/80mid, Gr1mid cells at 24 hours (Figure 6E). At 72 hours, the controls demonstrated a marked increase of F4/80mid, Gr1− elicited macrophages. In contrast, in the Flipf/d, LysMc/+ mice, there was no increase of elicited macrophages, however, the marked increase of neutrophils persisted (Figure 6E-F). Therefore, the deletion of Flip in myeloid cells, not only results in an increase of peritoneal neutrophils, but also a marked reduction of resident and elicited macrophages.
Flip deletion results in increased neutrophils and decreased macrophages in the peritoneum. Resident (A-B) or thioglycollate elicited (E-F) cells from peritoneal cavities were analyzed by flow cytometry, gaiting on CD11b+ cells. The macrophages (MΦ) were identified as F4/80+, Gr1−, whereas granulocytes (Gran) were identified as F4/80−, Gr1+. (B) The total number of cells, resident macrophages and granulocytes from controls (n = 20) and Flipf/d, LysMc/+ mice (n = 26) are summarized. (C) There was an inverse relationship between the percentage of peritoneal macrophages and granulocytes in the wild-type and Flipf/d, LysMc/+ mice. (D) PCR genotyping of granulocytes and macrophages isolated from peritoneal cavities were performed using the 3 PCR primers indicated in Figure 1. (F) The total number of thioglycollate elicited macrophages and granulocytes at 72 hours (n = 7 for each group) are summarized. *P < .05, **P < .01, and ***P < .001 between groups.
Flip deletion results in increased neutrophils and decreased macrophages in the peritoneum. Resident (A-B) or thioglycollate elicited (E-F) cells from peritoneal cavities were analyzed by flow cytometry, gaiting on CD11b+ cells. The macrophages (MΦ) were identified as F4/80+, Gr1−, whereas granulocytes (Gran) were identified as F4/80−, Gr1+. (B) The total number of cells, resident macrophages and granulocytes from controls (n = 20) and Flipf/d, LysMc/+ mice (n = 26) are summarized. (C) There was an inverse relationship between the percentage of peritoneal macrophages and granulocytes in the wild-type and Flipf/d, LysMc/+ mice. (D) PCR genotyping of granulocytes and macrophages isolated from peritoneal cavities were performed using the 3 PCR primers indicated in Figure 1. (F) The total number of thioglycollate elicited macrophages and granulocytes at 72 hours (n = 7 for each group) are summarized. *P < .05, **P < .01, and ***P < .001 between groups.
FLIP is necessary for macrophage differentiation
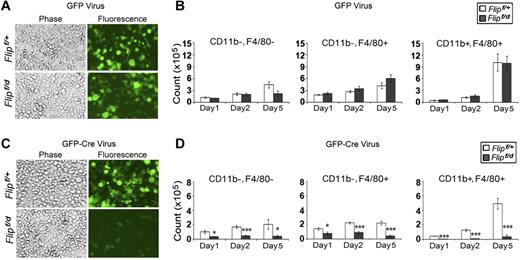
Experiments were performed to determine whether the reduction of macrophages upon Flip excision was due to a defect in differentiation or to cell death after differentiation. c-Kit+ hematopoietic stem cells from floxed Flip mice were infected with a recombinant GFP-Cre retrovirus to promote Flip excision in vitro. After infection, the cells were differentiated into macrophages for 5 days. There was no difference in the numbers of GFP+ cells (Figure 7A) or GFP+ cells that were CD11b−, F4/80−, or CD11b+, F4/80−, or CD11b+, F4/80+ mature macrophages (Figure 7B) when Flipf/+ and Flipf/d c-Kit+ cells were infected with a control GFP-expressing retrovirus. In contrast, there was a reduction of GFP+ cells (Figure 7C) and GFP+ cells that were CD11b−, F4/80−, or CD11b+, F4/80−, or CD11b+, F4/80+ mature macrophages (P < .01-.001; Figure 7D) on days 1, 2, and 5 after infection of Flipf/d c-Kit+ cells with the GFP, Cre-expressing retrovirus. This reduction does not appear to be due to increased apoptosis in response to Flip deletion, because there was no difference in the percentage of annexinV positivity for any of the cell types examined (data not shown). Further, when total bone marrow cells were used to generate macrophages, the caspase inhibitors IETD-fmk or zVAD.fmk did not rescue the GFP+, F4/80+ macrophages generated from the Flipf/d mice ex vivo after the deletion of Flip (data not shown). Together these observations suggest that FLIP is necessary for macrophage differentiation.
Deletion of Flip suppresses macrophage differentiation. cKit+ hematopoietic stem cells from Flipf/+ and Flipf/d were isolated from bone marrow and seeded at 2.5 × 105 cells/well followed by infection of retroviral vectors expressing GFP alone (A-B) or GFP and Cre (C-D). After the infection, the cells were differentiated in vitro to macrophages in 20% L929 medium. The total number of GFP+ cells (A,C) and the GFP+, CD11b−, F4/80−, the GFP+, CD11b+, F4/80−, and the GFP+, CD11b+, F4/80+ cells (B,D) were determined after 1, 2, and 5 days of differentiation. Data in panels B,D represents the mean ± SEM of 4 independent experiments.
Deletion of Flip suppresses macrophage differentiation. cKit+ hematopoietic stem cells from Flipf/+ and Flipf/d were isolated from bone marrow and seeded at 2.5 × 105 cells/well followed by infection of retroviral vectors expressing GFP alone (A-B) or GFP and Cre (C-D). After the infection, the cells were differentiated in vitro to macrophages in 20% L929 medium. The total number of GFP+ cells (A,C) and the GFP+, CD11b−, F4/80−, the GFP+, CD11b+, F4/80−, and the GFP+, CD11b+, F4/80+ cells (B,D) were determined after 1, 2, and 5 days of differentiation. Data in panels B,D represents the mean ± SEM of 4 independent experiments.
To determine whether the neutrophilia was intrinsic to the loss of Flip in myeloid cells, bone marrow reconstitution experiments were performed using CD45.1+ wild-type and CD45.2+Flipf/d, LysMc/+ bone marrow cells injected into CD45.1+ lethally irradiated hosts all on a C57BL/6 background. In mixed chimera experiments, when 25%, 50%, or 75% of the bone marrow cells injected into the lethally irradiated recipients were from the Flipf/d, LysMc/+ mice, no increase of circulating neutrophils or monocytes was observed (Table 4). Further, decreased body weight, increased spleen size, and decreased peritoneal macrophages were not observed in the mixed chimeras. However, the CD45.2+Flipf/d, LysMc/+ bone marrow cells were unable to compete with the wild-type CD45.1+ cells (supplemental Figure 8). It was only when 100% of the bone marrow cells were from the Flipf/d, LysMc/+ mice that the inflammatory phenotype, including increased circulating neutrophils (Table 4), was observed. All of the mice that received 100% Flipf/d, LysMc/+ bone marrow cells demonstrated an increase of CD45.2+ neutrophils compared with all the other groups (supplemental Figure 8C). Of interest, the one mouse with the greatest increase of CD45.2+ neutrophils also demonstrated an increase of recipient CD45.1+ neutrophils. G-CSF, GM-CSF, and IL-6 were increased only in this mouse and the 2 other mice with the greatest increase of neutrophils (data not shown), supporting the role of growth factors/cytokines in the neutrophilia. These observations suggest that the neutrophilia observed in the Flipf/d, LysMc/+ mice is due to a cell autonomous defect in the bone marrow progenitors which results in increased cytokines/growth factors that contribute to the granulopoiesis.
Mixed bone marrow chimeras do not develop an inflammatory phenotype in the peripheral blood
| . | Ratio of donor bone marrow cells implanted from control/KO mice . | ||||
|---|---|---|---|---|---|
| 100/0 . | 75/25 . | 50/50 . | 25/75 . | 0/100 . | |
| Granulocytes | 0.23 ± 0.04 | 0.25 ± 0.07 | 0.21 ± 0.03 | 0.31 ± 0.07 | 9.59 ± 6.3 |
| Inflammatory monocytes | 0.07 ± 0.02 | 0.06 ± 0.02 | 0.05 ± 0.02 | 0.09 ± 0.03 | 0.22 ± 0.14 |
| Resident monocytes | 0.20 ± 0.02 | 0.28 ± 0.09 | 0.20 ± 0.04 | 0.38 ± 0.14 | 0.1 ± 0.06 |
| B cells | 1.46 ± 0.28 | 2.22 ± 0.82 | 2.17 ± 0.60 | 5.21 ± 1.58 | 0.98 ± 0.43 |
| CD4+ T cells | 0.87 ± 0.13 | 1.02 ± 0.26 | 0.89 ± 0.15 | 1.66 ± 0.21 | 0.75 ± 0.14 |
| CD8+ T cells | 0.39 ± 0.04 | 0.45 ± 0.13 | 0.39 ± 0.07 | 0.82 ± 0.11 | 0.42 ± 0.05 |
| . | Ratio of donor bone marrow cells implanted from control/KO mice . | ||||
|---|---|---|---|---|---|
| 100/0 . | 75/25 . | 50/50 . | 25/75 . | 0/100 . | |
| Granulocytes | 0.23 ± 0.04 | 0.25 ± 0.07 | 0.21 ± 0.03 | 0.31 ± 0.07 | 9.59 ± 6.3 |
| Inflammatory monocytes | 0.07 ± 0.02 | 0.06 ± 0.02 | 0.05 ± 0.02 | 0.09 ± 0.03 | 0.22 ± 0.14 |
| Resident monocytes | 0.20 ± 0.02 | 0.28 ± 0.09 | 0.20 ± 0.04 | 0.38 ± 0.14 | 0.1 ± 0.06 |
| B cells | 1.46 ± 0.28 | 2.22 ± 0.82 | 2.17 ± 0.60 | 5.21 ± 1.58 | 0.98 ± 0.43 |
| CD4+ T cells | 0.87 ± 0.13 | 1.02 ± 0.26 | 0.89 ± 0.15 | 1.66 ± 0.21 | 0.75 ± 0.14 |
| CD8+ T cells | 0.39 ± 0.04 | 0.45 ± 0.13 | 0.39 ± 0.07 | 0.82 ± 0.11 | 0.42 ± 0.05 |
Data presented represent the number of cells × 103/μL determined by total cell count and flow cytometry, 8 weeks after bone marrow transplant. N = 5-6 in each group.
Discussion
The cellular mechanisms responsible for maintaining homeostatic balance in myeloid cells has not been fully characterized. Here, we demonstrate that the deletion of Flip in myeloid cells resulted in a marked increase of circulating neutrophils, associated with multiorgan neutrophil infiltration, including spleen, lymph nodes, thymus, intestine, and lung. There was no evidence of myeloid dysplasia in the bone marrow, spleen, or other organs. Increased granulopoiesis with an increase of CFU-GM was observed in the spleens of the Flipf/d, LysMc/+ mice, although not in the bone marrow. Under homeostatic conditions, the half life of circulating neutrophils is 8 hours or less, and neutrophilia may occur by increased survival. Although there was a slight reduction of neutrophil apoptosis at 0 and 4 hours, which may be due to the increased circulating growth factors, there appeared to be no intrinsic defect in neutrophil apoptosis, because at 24 hours neutrophil apoptosis in vitro was nearly complete using neutrophils from the Flipf/d, LysMc/+ mice. Because FLIP protects against death receptor-mediated apoptosis, a reduction of FLIP might be expected to lead to decreased cell survival. However, Flip mRNA was not expressed in circulating wild-type neutrophils (data not shown), further supporting our observations that suggest FLIP does not play a role in neutrophil survival. This interpretation is consistent with known role of the intrinsic mitrochondrial pathway involving Mcl-1, A1, and Bcl-xL in neutrophil apoptosis.26 In contrast to the results observed with FLIP, the deletion of Mcl-1 using LysM-Cre results in the neutropenia with no reduction of macrophages.27,28
Neutrophilia may also occur by increased production or increased release from the bone marrow in response to stress such as infection.29 The Flipf/d, LysMc/+ mice demonstrated extramedullary hematopoiesis associated with an increase of growth factors in the circulation that promote myelopoiesis. However, the data do not support the underlying infection as the cause for the increased neutrophils because 70% of the Flipf/d, LysMc/+ mice were culture negative. Further, although antibiotic treatment reduced the rate of infections, it had no effect on the circulating neutrophils or the phenotype. These observations suggest that the persistent acute inflammation contributed to the stunted growth and premature death in the Flipf/d, LysMc/+ mice.
The marked increase of circulating neutrophils and the multiorgan infiltration with neutrophils was not expected. Prior studies have demonstrated that mice lacking the adhesion molecule CD18 exhibit neutrophilia with increased IL-17 and G-CSF, which contributed to the increased neutrophil counts.30 Further, in these mice, the phagocytosis of apoptotic neutrophils by macrophages and dendritic cells suppressed granulopoiesis, which was mediated by a reduction of IL-23 and subsequently IL-17 and G-CSF.31 However, the Flipf/d, LysMc/+ mice demonstrated a reduction of splenic, lymph node, and peritoneal macrophages, suggesting that increased IL-23 was not responsible. In fact, circulating p40, which is common to both IL-12 and IL-23, was not elevated (data not shown). In contrast, IL-17 and G-CSF, in addition to IL-6, GM-CSF, and M-CSF, were all increased in the Flipf/d, LysMc/+ mice, suggesting that these growth factors may have contributed to the increased granulopoiesis. However, the results of the bone marrow transfer experiments suggest that the primary defect was a cell autonomous defect in myeloid development of FLIP-deficient bone marrow–derived cells, because these cells were capable of transferring the disease in the absence of competitor wild-type cells. The defect in myeloid development resulted in increased growth factors and cytokines, which resulted in the neutrophilia. Further, the mixed chimera experiments demonstrate that Flipf/d, LysMc/+ bone marrow cells were not able to compete with the wild-type cells, suggesting an impaired hematopoietic stem/progenitor cell phenotype. This may be due to excessive cytokines and growth factor production in the Flipf/d, LysMc/+ mice, which may change their ability in homing, repopulating, or potentially exhaust the stem/progenitor cells.32,33 It is unlikely that FLIP deletion in the hematopoietic stem cells was directly responsible, because LysM is weakly expressed and only in a minority of these cells.34
Resident peritoneal macrophages were markedly reduced in the Flipf/d, LysMc/+ mice. When peritoneal macrophages were reduced to < 10%, peritoneal granulocytes increased, suggesting that the reduction of macrophages contributes to the increase of granulocytes. The few Gr1−, F4/80+ macrophages that could be isolated for the peritoneum of the Flipf/d, LysMc/+ mice, or differentiated from bone marrow or spleen, were not deleted for Flip. Therefore, we were unable to examine the nonapoptotic functions of FLIP in macrophages, because any macrophages that could be isolated expressed FLIP. Prior studies demonstrated that under resting conditions neither Gr1+ nor Gr1− monocytes homed to the peritoneum, whereas only Gr1+ monocytes migrated to the peritoneum after thioglycollate.35,36 In the Flipf/d, LysMc/+ mice, resident F4/80mid, Gr1mid cells, consistent with inflammatory monocytes, were increased in the peritoneum. The recruitment of these cells may have been due to the acute inflammation with neutrophils. After the injection of thioglycollate at 20 hours, the inflammatory monocytes and neutrophils were comparable in the Flipf/d, LysMc/+ and control mice. However, in the Flipf/d, LysMc/+ mice, no increase of F4/80+, Gr1− elicited macrophages was observed at 72 hours, consistent with a failure of the inflammatory monocytes to differentiate into macrophages, resulting in a persistent acute inflammatory response. We cannot exclude the possibility that following macrophage differentiation, the cells died. However, even when examined at 48 hours (data not shown), no increase of Gr1− macrophages was observed in the Flipf/d, LysMc/+ mice, suggesting that Flip is essential for macrophage differentiation.
Further documenting the importance of FLIP for macrophage differentiation, infection of c-Kit+ bone marrow cells with a recombinant GFP-Cre retrovirus resulted in a reduction of differentiated CD11b+, F4/80+ macrophages derived from bone marrow of Flipf/d mice. When Flip was deleted, there was also a reduction of CD11b−, F4/80− and CD11b+, F4/80− cells, demonstrating that the defect was not limited to the terminal stage of differentiation. There was no evidence of increased apoptosis to account for the reduction of macrophages. Relevant to this observation, another member of the death receptor pathway, caspase 8, is also necessary for normal macrophage differentiation,37 and the deletion of caspase 8 in myeloid cells prevents macrophage differentiation.38 In contrast, overexpression of FADD, another member of the Fas-mediated death pathway, accelerates the differentiation of macrophages, independent of any death receptor.39 Our observations are consistent with those that identified the role of a complex of FADD, caspase 8, RIP1, and FLIP, which suppressed NF-κB activation and was necessary for M-CSF–mediated macrophage differentiation.39 These observations are also reminiscent of the observation that the germ line deletion of either FLIP, caspase 8, or FADD is embryonic lethal, suggesting that these molecules also cooperate during embryonic development.40
Together, these observations suggest the deletion of FLIP in macrophages resulted in the development of neutrophilia and the inflammatory phenotype. Of interest, a recent study demonstrated that deletion of dendritic cells also resulted in neutrophilia and a multiorgan infiltration with neutrophils.41 Although these mice also exhibited increased Th17 cells, in contrast to the Flipf/d, LysMc/+ mice, they also exhibited an infiltration of CD4+ T cells in intestine and kidney, as well as increased numbers of F4/80+ macrophages.41 Therefore, an intact mononuclear phagocytic system appears necessary for normal granulopoiesis and that deletion of either macrophages or dendritic cells results in neutrophilia. In summary, this study demonstrates that the expression of FLIP in myeloid cells is critical for the differentiation of macrophages, which is a characteristic feature of chronic inflammation, and that the acute inflammatory phenotype is secondary to the reduction of macrophages and an increase of cytokines and growth factors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Guang-Yu Yang, Northwestern University Department of Pathology, for assistance with interpreting the histology of the gastrointestinal tract. We thank Rudina Sobkoviak and Jeff Kraynak for their contributions helping to establish and maintain the Flip conditional knockout mice and Dr Chiang-Ching Huang, Northwestern University Department of Preventive Medicine, for assistance with statistical analysis.
This work was supported in part by National Institutes of Health (NIH) grant nos. AR048269 and AR055240 to R.M.P. and AR050250, AR054796, and AI06759 to H.P., and a Within Our Reach Grant from the American College of Rheumatology to R.M.P. J.D.C. is a Scholar of the Leukemia & Lymphoma Society.
National Institutes of Health
Authorship
Contribution: Q.-Q.H. and R.M.P. conceived the project, designed experiments, performed data analysis, and wrote the manuscript; R.M.P. supervised the project; Q.-Q.H. and R.B. generated and maintained the KO mice and performed the major experiments in genotypic and phenotypic analysis and data acquisition; H.P. and J.D.C. designed experiments, interpreted results, and assisted in manuscript preparation; Z.H. designed and performed CFU and bone marrow infection experiments; L.K. performed immunohistochemistry analysis; S.G. performed the histologic and pathologic identification; H.A. and A.M. assisted with the mixed chimeras and their analysis; and all authors discussed the results and implications and contributed to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard M. Pope, 240 E Huron, Ste M-300, Chicago, IL 60611; e-mail: rmp158@northwestern.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal