Abstract
There is increasing evidence that miRNA and transcription factors interact in an instructive fashion in normal and malignant hematopoiesis. We explored the impact of TEL-AML1 (ETV6-RUNX1), the most common fusion protein in childhood leukemia, on miRNA expression and the leukemic phenotype. Using RNA interference, miRNA expression arrays, and quantitative polymerase chain reaction, we identified miRNA-494 and miRNA-320a to be up-regulated upon TEL-AML1 silencing independently of TEL expression. Chromatin immunoprecipitation analysis identified miRNA-494 as a direct miRNA target of the fusion protein TEL-AML1. Using bioinformatic analysis as well as functional luciferase experiments, we demonstrate that survivin is a target of the 2 miRNAs. miRNA-494 and miRNA-320a were introduced to the cells by transfection and survivin expression determined by Western blot analysis. These miRNAs blocked survivin expression and resulted in apoptosis in a similar manner as TEL-AML1 silencing by itself; this silencing was also shown to be Dicer-dependent. miRNAs-494 and -320a are expressed at lower levels in TEL-AML1+ leukemias compared with immunophenotype-matched nonTEL-AML1 acute lymphoblastic leukemia subtypes, and within TEL-AML1+ leukemias their expression is correlated to survivin levels. In summary our data suggest that TEL-AML1 might exert its antiapoptotic action at least in part by suppressing miRNA-494 and miRNA-320a, lowering their expression causing enhanced survivin expression.
Introduction
The t(12;21) is the most frequent chromosomal lesion in childhood B-cell precursor acute lymphoblastic leukemia, occurring with an incidence of 25% overall and generating the TEL-AML1 fusion gene.1 The t(12;21) translocation is present at birth in most children who contract the disease and thus represents presumably an initiating event in leukemogenesis in utero.2-4 The encoded protein contains the 336 amino-terminal region of the TEL Ets protein, fused to the residues 21 to 480 of the tissue-specific transcription factor, AML1. TEL and AML1 are both transcription factors with an important role in hematopoiesis. The resulting fusion protein has the AML1 DNA binding domain and the TEL protein interaction domain and has been shown to maintain transcription factor properties and bind DNA.5-7 TEL-AML1 has been shown to also interfere with apoptosis as its expression affects vigorous antiapoptotic genes, among these survivin.8,9 Together with other chimeric proteins, TEL-AML1 may act as a dominant negative transcription factor that may reduce the expression of tumor suppressor and increase antiapoptotic genes.10 In particular, the frequent inactivation of the AML1 transactivation functions in leukemias, for AML1-ETO and TEL-AML1 chimeric proteins, suggests that AML1 regulates one or more critical genes and miRNAs that when repressed, might promote the development of leukemias.11
miRNAs are single-stranded RNA molecules approximately 21 to 23 nucleotides in length that have the ability to control gene expression; a single miRNA can target multiple genes simultaneously. Different miRNAs have been shown to control fundamental cellular functions such as differentiation and apoptosis. The expression of miRNAs appears to be highly regulated according to the cell's developmental lineage and stage.12-16 Remarkably, aspects of this specificity were found maintained in cancer, and the pattern of miRNA expression varies dramatically across tumor types.14,17,18 In addition, it has been suggested that the expression pattern of a small set of miRNAs was able to define the cancer type better than expression data from thousands of mRNAs.15,19 Leukemia miRNA expression patterns are clearly distinct from solid tumors and, strikingly, they can subgroup leukemias according to their underlying chromosomal lesion/translocations.15,20 Although miRNA expression is believed to play an important role in malignant transformation, little is known about the mechanisms that contribute to abnormal miRNA regulation in cancer. Recent studies have shown that transcription factors bind and regulate miRNA expression. As TEL and AML1 are both transcription factors, we investigated the functional contribution of TEL-AML1 on miRNA expression and identified TEL-AML1 regulated miRNA species that affect a functional gene target, survivin.
Methods
Cell culture
The B-cell precursor leukemic cell line REH (having the TEL-AML1 fusion gene, the second TEL allele deleted and AML1 retained) was grown in 24-well plates (Nunc) at 106 cells/mL. Human embryonic kidney (HEK)293Ts, a cell line that does not express TEL, were cultured in 6-well plates; both were cultured in RPMI with 10% fetal bovine serum.
siRNA-design, miRNA mimics, and transfection of cells
siRNAs targeting the fusion region of TEL-AML1 and controls were designed and synthesized by Dharmacon.8 siRNA duplexes were handled essentially as described.8 The transfection of REH cells with siRNA duplexes at a concentration of 230nM was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The miRNA-494, miRNA-320a mimics and a miRNA mimic with no human targets (control sequence based on cel-miR-67, sequence: UCACAACCUCCUAGAAAGAGUAGA) were synthesized by Dharmacon and used according to the manufacturer's instructions.
Assessment of apoptosis
Apoptotic cell death was evaluated using Annexin V–fluorescein isothiocyanate (FITC)/propidium iodide (PI) (BD Biosciences) and fluorescence-activated cell sorting (FACS) analysis according to the manufacturer's recommendations.
miRNA expression profiling
Total RNA from TEL-AML1 siRNA-treated and control-treated REH cells was isolated using the miRNeasy Mini Kit RNA isolation kit (QIAGEN). Further processing of RNA and hybridization to microarrays was performed by the Gladstone Institute Microarray Facility with methods as described.21 The array used is a printed oligonucleotide array with 5017 probes to 1546 miRNAs of several species (mIRbase v. 10; www.mirbase.org), printed on aminosilane glass. Five micrograms total RNA was labeled with Cy3 (treatment, siRNA to TEL-AML1) and Cy5 (control, siRNA to no known target). A second set of arrays was used with dye swap. Validation of individual miRNA expression in the cell line experiments was performed using Taqman miRNA assays (Applied Biosystems). All miRNA assays were run concurrently with U6 snRNA control which was used as a “housekeeping” gene control, (Applied Biosystems) for normalization. All Taqman miRNA assays were run in triplicate, with comparative threshold cycles calculated, and results are presented with mean and standard error. The comparative threshold cycle calculation for relative quantification was analyzed as described in the Applied Biosystems 7900HT technical manual.22 All microarray data are available on the Gene Expression Omnibus public database under accession numbers GSE23842, GSM587851, and GSM587852.
miRNA, RNA, and protein expression in patient samples
All human sample experiments were performed under University of California, San Francisco Institutional Review Board approved protocols and patients and their families were under full informed consent for biological studies in accordance with the Declaration of Helsinki. Bone marrow samples were obtained from 26 B-cell precursor acute lymphoblastic leukemia (ALL) children at the time of diagnosis and prior to any treatment. The patients were 2-16 years of age, with a median age of 5 years. Eleven children were TEL-AML1 by reverse transcription polymerase chain reaction (RT-PCR). The remaining patients were common ALL immunophenotype (CD19+, CD10+) without TEL-AML1. These other patients were cytogenetically high hyperdiploid (n = 8) and other cytogenetic subtypes including pseudodiploid (n = 7). Mononuclear cells were isolated using Ficoll-Hypaque (Sigma-Aldrich) density gradient centrifugation and RNA was extracted with TRIzol Reagent (Invitrogen). These 26 samples were used to compare miRNA levels in TEL-AML1–positive versus TEL-AML1–negative leukemias.
An additional 28 leukemic bone marrow samples, all TEL-AML1–positive, were analyzed separately to explore the correlation between TEL-AML1, miRNA, and survivin levels. Bone marrow samples were purified over Ficoll to produce a near-pure blast cell population. RNA was isolated using miRNeasy Mini kits (QIAGEN). miRNA was measured in leukemia cells using Taqman assays for individual miRNAs (Applied Biosystems) using U6 snRNA as a calibration control. TEL-AML1 RNA was measured using the Taqman assay as described previously.23 Both miRNA and TEL-AML1 assays were run in duplicate; a mean of the 2 assays was used for statistical analysis. Survivin protein was detected by enzyme-linked immunosorbent assay (ELISA) using the DuoSet IC Human Total Survivin kit (R&D Systems) exactly as described in the kit manual. Cells (2.5 million) were thawed from liquid N2 storage and rinsed twice with phosphate-buffered saline. Proteins were solubilized in lysis buffer (1mM EDTA [ethylenediaminetetraacetic acid], 0.5% Triton X-100, 6M urea, protease inhibitor cocktail [Sigma-Aldrich, 1 mL/100 mL lysis buffer] and phosphate-buffered saline). Before use, the lysate was centrifuged at 2000g for 5 minutes and the supernatant was transferred into a clean tube. Survivin levels were normalized to protein content as measured with the Bradford assay (nanodrop spectrophotometer). ELISA was run in duplicate per sample, and the mean used for analysis. The correlation between TEL-AML1, miRNA levels, and survivin was calculated using Spearman correlation coefficient (a nonparametric rank test).
Western blot analysis
Preparation of cell lysates and Western blot analysis were performed as described previously.8 In brief, protein extracts were separated in 4%-20% and 18% mini gels (Invitrogen) and analyzed by immunoblot analysis using a 1:1000 dilution of primary antibody and a 1:10 000 dilution of a horseradish peroxidase–conjugated secondary antibody or 1:1000 alkaline phosphatase–conjugated secondary antibody from Cell Signaling. Immunocomplexes were visualized with the enhanced chemiluminescence (ECL, Amersham Pharmacia) system or CDP-Star Reagent (New England Biolabs). An antibody for tubulin (Oncogene/EMD Biosciences) was used as a loading control.
Chromatin immunoprecipitation
Cells were harvested, and chromatin was formalin-crosslinked and sheared to approximately 200-1000 bp using a Sonic Dismembrator model 550 at settings 1.5, for 2 to 4 cycles, 50 seconds per cycle, on ice. We followed the microarray application protocol as described in the kit (EZ-ChIP Kit, Millipore) to isolate chromatin DNA complexes. Chromatin immunoprecipitation (ChIP) antibodies to TEL (Santa Cruz Biotechnology) and anti-RNA Polymerase II (Millipore) were used. Chromatin was also purified from crosslinked DNA that had not been immunoprecipitated; this served as an input DNA control for the arrays, which was cohybridized to provide a reference (that is, total input amplicon). To prepare samples for array hybridization, we applied the method developed in Bing Ren's laboratory to carry out ligation-mediated PCR amplification of ChIP DNA.24,25 At least 4 μg of ligation-mediated PCR-amplified DNA was used to carry out the subsequent labeling and hybridization steps. The immunoprecipitation amplicon was labeled with Cy5, and an equal amount of the total input amplicons labeled with Cy3; the 2 are combined to hybridize to the miR-promoter arrays. The promoter array included isothermal probes (Roche Nimblegen) covering only noncoding (nc) RNA at 50-bp spacing: 462 miRNAs, 375 snoRNAs, and 89 piRNAs. Each tiled region spans approximately 15 kb upstream and 10 kb downstream of transcriptional start site for the miRNAs and other ncRNAs. Included also were 12 housekeeping genes and 14 encode regions (HG18) at approximately 100-bp spacing. This array, which we designed, is sold by Roche Nimblegen. The entire experiment was repeated twice, with similar results.
Luciferase assays
Luciferase activity assays were performed using the Bright-GloTM Luciferase Assay System (Promega) using the method described by the manufacturer. Luciferase reporter plasmids were created with survivin 3′-end target sites using oligonucleotides (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) cloned into the multicloning site of the distal end of the luciferase gene reporter vector pMIR-report (Applied Biosystems, using SpeI and HindIII sites). Mismatch versions of the target sites (with 2-bp mismatch) were used as controls. REH cells (2 × 105 cells/well) were plated onto 24-well tissue culture plates containing 0.5 mL serum-free medium. Cotransfection 10pmol of miR-494 or miR-320a and different combinations of plasmid DNA (500 ng luciferase reporter construct mixed with 500 ng β-Gal plasmid pMIR-REPOETTM) was performed using Lipofectamine2000™ transfection reagent (Invitrogen) following standard protocol, with 3 μL of Lipofectamine2000™ per well. Twenty hours after transfection, 1 mL RPMI 1640 medium with 20% fetal bovine serum was added, and cell cultures were maintained at 37°C for 24 hours. Cells were then harvested by centrifugation at 500g and medium aspirated from the cells. The cells were then directly lysed with 200 μL of 1× Glo Lysis Buffer for 5 minutes. Fifty microliters of the cell lysis was mixed with equal amount of Bright-GloTM Luciferase Reagent and Beta-Glo Assay Reagent for 5 minutes, and the FL-800 Microplate Floresence Reader (Bio-Tek Instruments) was used to measure luciferase activity and β-Gal activity in 96-well plates in triplicate. The experiments were repeated 3×. Luciferase activities were normalized against β-Gal activity.
Bioinformatics and data analysis
ChIP-chip data were normalized using the scatter plot smoother lowess based on all probes. For analysis of significant “enrichment,” data were first sorted according to genomic locations, and a window of size of 500 bp moving along the genome was adopted, centering at each probe. For each probe, the median log-ratio of neighbor probes residing in the window was calculated; typically with 10 probes within each window. Significant probes/peaks were selected if they reside in a window within more than 4 probes within the 500-bp window and their signals are higher than the threshold. The threshold was heuristically set at mean + 4 × SD for the 2 TEL-AML1 arrays. We assigned a value to each promoter region based on the highest smoothed values within the tiled region of the promoter.26 The log2 ratios of miRNA promoter pull-downs were graphed against the log2 ratios of the miRNA expression arrays.
Results
siRNA-mediated silencing of TEL-AML1 affects miRNA expression
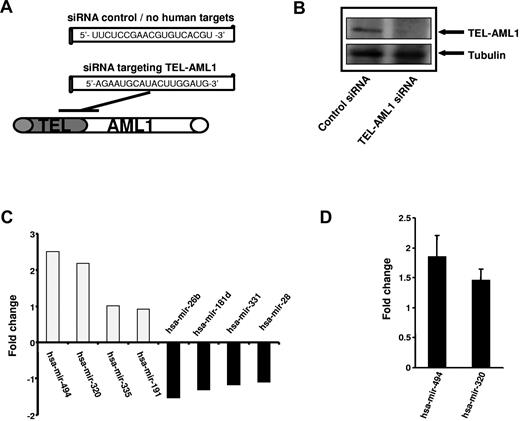
The TEL-AML1 translocation results in the expression of an abnormal hybrid transcription factor which bears elements of TEL and AML1, both important transcription factors involved in hematopoiesis. We here investigated the impact of TEL-AML1 expression upon miRNA expression to identify TEL-AML1 dependent miRNAs. siRNA was used to knock down TEL-AML1 expression and miRNA arrays used to impact upon miRNA expression. siRNA targeting the fusion site of the TEL-AML1 mRNA was used to silence the fusion gene expression, and an siRNA with no human targets was used as control. Efficient silencing of TEL-AML1 in REH cells was performed as described by Diakos et al.8 and protein depletion was confirmed at day 6 of the experiment by Western blot analysis (Figure 1B). Subsequently, total RNA was isolated and miRNA expression was performed using 2-color miRNA arrays. Upon TEL-AML1 silencing miRNA expression was differentially affected; both up-regulation and down-regulation of individual miRNAs were observed. The most prominent alterations observed in this experiment were the increase of miRNA-494 and miRNA-320a (Figure 1C); miRNA quantitative RT-PCR specific for miRNA-494 and miRNA-320a were used to verify these findings (Figure 1D). The data suggest that TEL-AML1 suppresses the expression of these miRNAs and that silencing of the fusion protein is able to reconstitute the expression of miRNA-494 and miRNA-320a.
siRNA-mediated silencing of TEL-AML1 affects miRNA expression. (A) Schematic representation of the siRNA sequence used to target TEL-AML1 and the localization within the TEL-AML1 translocation breakpoint mRNA (position at bp 15-33, accession no. S78496). A siRNA with no human targets was used as control (see Diakos et al.8 ). (B) Depletion of TEL-AML1 using siRNA. Western analysis was performed from REH cell lysates treated with the control siRNA (left lane) or functional siRNA targeting TEL-AML1 (right lane). TEL-AML1 protein was detected using an anti-TEL antibody from ATLAS and an anti-tubulin antibody for loading control. (C) TEL-AML1 affects miRNA expression. The expression profile of REH cell was analyzed after TEL-AML1 depletion by 2 color miRNA arrays. The fold changes of the 4 most up-regulated as well as those of the 4 most down-regulated miRNAs are shown (mean of 2 experiments). (D) miRNA RT-PCR (Applied Biosystems) was performed to verify the changes in miRNA-494 and miRNA-320a expression upon TEL-AML1 silencing. The data shown depicts the average fold change from 3 independent experiments, using U6 snRNA as a calibration control.
siRNA-mediated silencing of TEL-AML1 affects miRNA expression. (A) Schematic representation of the siRNA sequence used to target TEL-AML1 and the localization within the TEL-AML1 translocation breakpoint mRNA (position at bp 15-33, accession no. S78496). A siRNA with no human targets was used as control (see Diakos et al.8 ). (B) Depletion of TEL-AML1 using siRNA. Western analysis was performed from REH cell lysates treated with the control siRNA (left lane) or functional siRNA targeting TEL-AML1 (right lane). TEL-AML1 protein was detected using an anti-TEL antibody from ATLAS and an anti-tubulin antibody for loading control. (C) TEL-AML1 affects miRNA expression. The expression profile of REH cell was analyzed after TEL-AML1 depletion by 2 color miRNA arrays. The fold changes of the 4 most up-regulated as well as those of the 4 most down-regulated miRNAs are shown (mean of 2 experiments). (D) miRNA RT-PCR (Applied Biosystems) was performed to verify the changes in miRNA-494 and miRNA-320a expression upon TEL-AML1 silencing. The data shown depicts the average fold change from 3 independent experiments, using U6 snRNA as a calibration control.
TEL-AML1 directly targets miRNA promoters
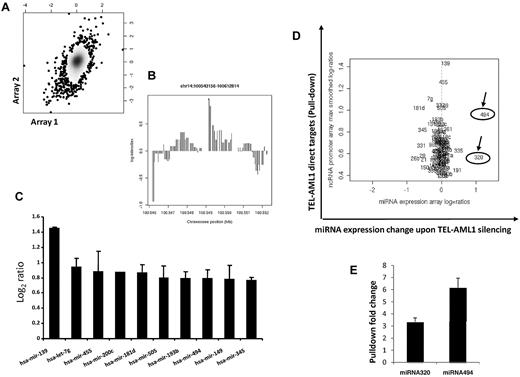
Transcription factors typically bind the promoter regions and thus regulate the expression of their target genes. This is true also for miRNA promoters, as demonstrated by recent data.27,28 Whether the fusion transcription factor TEL-AML1 binds and affects miRNA promoter activity has not been addressed before. We therefore designed a tiling path array covering human miRNA control regions including their host genes, if applicable, to tackle this question. The array includes the human miRNAs registered at the Sanger Data base (Release 10.0, 2007). Using the leukemia cell line (REH) expressing TEL-AML1 but not TEL and a ChIP grade specific antibody against TEL, we performed chromatin ChIP-chip to identify miRNA promoters bound by TEL-AML1. We performed 2 independent experiments and a good correlation between the individual experiments was observed (Figure 2A). The results demonstrate that TEL-AML1 binds miRNA promoters and might also help explain the translocation specific miRNA profiles that have been reported by others in leukemias.15 The miRNA-139 promoter has shown the strongest TEL-AML1 binding (Figure 2B); however TEL-AML1 was able to bind the promoters of a number of miRNAs including miRNA-320a and miRNA-494 (Figure 2C-D).
TEL-AML1 directly targets miRNA promoters. (A) ChIP-chip analysis was performed using the TEL-AML1 expressing cell line REH using an anti-TEL antibody. This cell line has a deletion in the other TEL allele, so only TEL-AML1 targets are pulled-down. Two independent experiments were performed, and anti-TEL-AML1 pull-downs and input DNA controls were cohybridized to the ncRNA tiling arrays. Following normalization and smoothing, the log2-ratios of TEL-AML1 pull-downs versus controls from experiment 1 were graphed against those from experiment 2 on an x/y graph, with density shading. A Pearson correlation coefficient of 0.27 (P < 10−7) between the 2 replicate experiments was observed as shown in this plot, also exemplified by a clustering of data along a slope of x = y (B) Scrutiny of the pull-down peaks revealed TEL-AML1 binding to miRNA regions, the strongest pull-down was detected for the miRNA-139 promoter. Lower but significant signals were found for 99 other miRNAs, shown here is the peak in a 5′ region of miRNA-494. (C) TEL-AML1 binds the promoters of miRNAs. The average log ratio of the binding intensity from 3 experiments was calculated and the 8 miRNAs promoters with the highest affinity to bind TEL-AML1 are shown, among these is miRNA-494. Standard errors are calculated from 3 separate experiments. (D) miRNA-494 and miRNA-320a are direct and functional targets of TEL-AML1. miRNA promoters identified to bind TEL-AML1 (direct targets, y axis; only those targets significantly enriched in ChIP pull downs are shown) were plotted against miRNA expression changes upon TEL-AML1 silencing. Log2 ratios are shown. (E) Using conventional PCR (supplemental Table 1) we analyzed TEL-AML1 pull-down and input from ChIP experiments. We designed primers to test the miRNA-494 and miRNA-320a that were shown to be direct targets of TEL-AML1 in the ChIP experiments. Panel E shows the fold increase of TEL-AML1 binding to the promoters of miRNA-494 and miRNA-320a as it was compared with the input (in triplicate with SE bars), thereby confirming these miRNAs as direct targets of TEL-AML1. The combined data from these experimental approaches distinguishes miRNA-494 and miRNA-320a both as direct and functional targets. miRNA-320a is indicated as miRNA-320 on the figures, as it is termed in the Sanger database.
TEL-AML1 directly targets miRNA promoters. (A) ChIP-chip analysis was performed using the TEL-AML1 expressing cell line REH using an anti-TEL antibody. This cell line has a deletion in the other TEL allele, so only TEL-AML1 targets are pulled-down. Two independent experiments were performed, and anti-TEL-AML1 pull-downs and input DNA controls were cohybridized to the ncRNA tiling arrays. Following normalization and smoothing, the log2-ratios of TEL-AML1 pull-downs versus controls from experiment 1 were graphed against those from experiment 2 on an x/y graph, with density shading. A Pearson correlation coefficient of 0.27 (P < 10−7) between the 2 replicate experiments was observed as shown in this plot, also exemplified by a clustering of data along a slope of x = y (B) Scrutiny of the pull-down peaks revealed TEL-AML1 binding to miRNA regions, the strongest pull-down was detected for the miRNA-139 promoter. Lower but significant signals were found for 99 other miRNAs, shown here is the peak in a 5′ region of miRNA-494. (C) TEL-AML1 binds the promoters of miRNAs. The average log ratio of the binding intensity from 3 experiments was calculated and the 8 miRNAs promoters with the highest affinity to bind TEL-AML1 are shown, among these is miRNA-494. Standard errors are calculated from 3 separate experiments. (D) miRNA-494 and miRNA-320a are direct and functional targets of TEL-AML1. miRNA promoters identified to bind TEL-AML1 (direct targets, y axis; only those targets significantly enriched in ChIP pull downs are shown) were plotted against miRNA expression changes upon TEL-AML1 silencing. Log2 ratios are shown. (E) Using conventional PCR (supplemental Table 1) we analyzed TEL-AML1 pull-down and input from ChIP experiments. We designed primers to test the miRNA-494 and miRNA-320a that were shown to be direct targets of TEL-AML1 in the ChIP experiments. Panel E shows the fold increase of TEL-AML1 binding to the promoters of miRNA-494 and miRNA-320a as it was compared with the input (in triplicate with SE bars), thereby confirming these miRNAs as direct targets of TEL-AML1. The combined data from these experimental approaches distinguishes miRNA-494 and miRNA-320a both as direct and functional targets. miRNA-320a is indicated as miRNA-320 on the figures, as it is termed in the Sanger database.
To uncover which of these target miRNA are functionally affected by TEL-AML1 we combined the expression data from the TEL-AML1 silencing experiment together with the data from ChIP-chip analysis. Although the miR-139 promoter showed the strongest binding for TEL-AML1 its expression changed little upon TEL-AML1 silencing (Figure 2D). miRNA-494 is a member of a miRNA family whose promoter was found among the 10 miRNA promoters with the strongest TEL-AML1 affinity; this binding passed stringent significance criteria (see “Methods” and Figure 2C). In addition, upon TEL-AML1 silencing miRNA-494 showed the most profound change in expression levels as its expression increased more than 2-fold (Figure 2D). The miR-320a promoter was also enriched for TEL-AML1 binding however to a lesser extent than miR-494. In spite of this fact, silencing of TEL-AML1 also resulted in 2-fold increases in miR-320a expression. These 2 experimental approaches in combination suggest that TEL-AML1 directly targets and functionally affects miRNA-494 and miRNA-320a expression. This result does not exclude a role of indirect targeting particularly in the case of miRNA-320a (eg, via the control of other transcription factors with altered expression upon TEL-AML1 silencing). The strong expression changes in miRNA-494 and miRNA-320a were explored further to investigate their role in TEL-AML1-induced leukemogenesis.
miRNA-494 and miRNA-320a target survivin
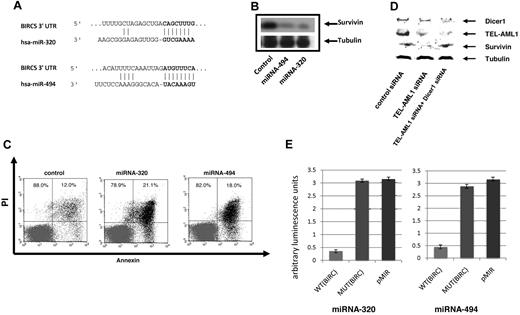
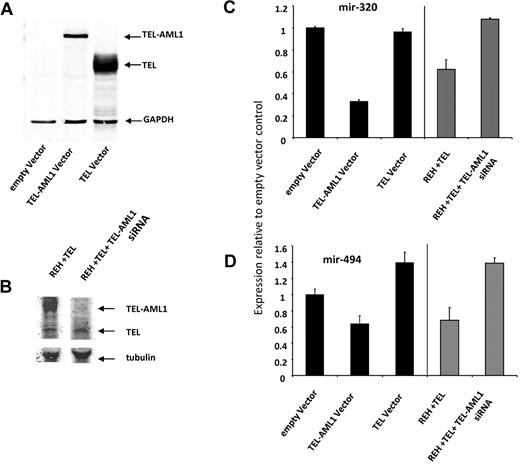
miRNAs regulate gene expression at the posttranscriptional level by blocking the expression of target genes. In an effort to gain further insight in the role of miRNA-494 and miRNA-320a on the biology of TEL-AML1 leukemia we performed a bioinformatics target site analysis for both miRNAs using TargetScan 4.0 predictions.29 Both miRNA-494 and miRNA-320a were interestingly predicted to target survivin, miRNA-494 providing the highest context score of any miRNA predicted to target this protein (Figure 3A). We recently identified survivin as a regulated target of TEL-AML1.8 Survivin is a gene which is highly expressed in leukemia and has been shown to have strong antiapoptotic properties. To determine whether miRNA-494 and miRNA-320a regulate the expression of their predicted target survivin we employed miRNA “mimics” strategy for both miRNA-494 and miRNA-320a. We introduced the mimics into the REH cells and analyzed survivin protein levels by Western blot analysis. Both miRNA-494 and miRNA-320a were able to decrease survivin expression efficiently (Figure 3B) thus implicating survivin as a miRNA-494 and miRNA-320a target. As a control we used a miRNA mimic with no human targets, which did not affect survivin expression (Figure 3B, first lane). In addition we examined the effect of the miRNA-494 and miRNA-320a mimics upon cell survival; survivin depletion has been shown to result in apoptotic cell death, which was associated with TEL-AML1 depletion.8 Both miRNA-494 and miRNA-320a mimics resulted in an efficient increase in early apoptosis as it was measured by annexin V staining and FACS analysis (Figure 3C). DICER1 is a protein component of the miRNA processing machinery and has been shown to be essential for the production of mature and functional miRNA.30,31 If suppression of survivin is mediated by miRNA, then this suppression should be DICER1-dependent. We used siRNA to block DICER1 expression and TEL-AML1 at the same time. Whereas TEL-AML1 silencing decreased survivin expression, the auxiliary targeting of DICER1 prevented this TEL-AML1–dependent effect upon survivin (Figure 3D). This shows that the TEL-AML1 impact upon survivin is DICER1-dependent, and given the established role of DICER1 in miRNA biogenesis, is also miRNA dependent. To provide proof that miRNA-494 and miRNA-320a are targeting survivin, we used luciferase reporter vectors encoding the survivin 3-untranslated region (UTR) indicated to be the targets of these miRNAs; mismatch (mutated) target 3-UTR and empty vectors were used as controls. These experiments demonstrate that miRNA-494 and miRNA-320a both target the 3-UTR of survivin. Taken together these results implicate a role for the control of survivin expression by miRNA-494 and miRNA-320a. We demonstrated further that the TEL gene by itself has no impact on the expression of these 2 miRNAs (Figure 4). HEK293T cells were particularly experimentally amenable to strong enforced expression of TEL and TEL-AML1; only the fusion protein and not TEL alone had an effect on miRNA-494 and miRNA-320a (Figure 4A,C-D). To confirm this result in a leukemia cell line, we introduced the expression of TEL gene in REH cells (which does not contain the native TEL gene), which did not impact expression of the miRNAs (Figure 4B-D).
miRNA-494 and miRNA-320a affect survivin expression and apoptosis. (A) Bioinformatic target prediction analysis identifies survivin as a target of miR-494 and miRNA-320a; the survivin 3′ UTR complementary to the miRNA sequence is shown (using Targetscan). (B) Both miRNA-494 and miRNA-320a can regulate survivin expression. miRNA mimics for miRNA-494 and miRNA-320a were transfected into the REH cells and survivin expression was analyzed by Western blot analysis using a rabbit anti-survivin antibody (Cell Signaling Technology). The miRNA with no human targets were used as control. For loading controls, an equal number of cells from each experimental sample were lysed, and the same volume of the same lysate was applied to a separate gel to probe for tubulin. (C) miRNA-494 and miRNA-320a induce apoptosis. miRNA mimics for miRNA-494 and miRNA-320a were transfected into the REH cells and apoptosis was assessed by FACS analysis of annexin V–FITC/PI staining of control-treated (miRNA with no human targets, see “Methods”), and mimics for miRNA-494 and miRNA-320a. Data are shown from 1 of 3 representative experiments and indicate early apoptosis (annexin V single-positive cells). Percentages of annexin V–positive cells are indicated in the figure (SEs are under 2%). Percentages refer to the cells within the top left and bottom left quadrants, and top right and bottom right, respectively. (D) TEL-AML1 regulation of survivin is Dicer1 dependent. siRNA silencing of Dicer1 restores survivin expression in TEL-AML1 depleted cells indicating the miRNA dependence of this effect. For loading controls, an equal number of cells from each experimental sample were lysed, and the same volume of the same lysate was applied to a separate gel to probe for tubulin. (E) Luciferase vectors containing portions of the survivin (BIRC5) 3-UTR was used to demonstrate that survivin is a direct target of miRNA-494 and miRNA-320a. Both miRNAs were able to block luciferase expression significantly when they were cotransfected into REH cell together with the BIRC5 3-UTR luciferase-expressing vector (WT BIRC5). A mutated sequence and the empty vector were used as controls (MUT BIRC5 and pMir Reporter, respectively), and both had equivalent levels of luciferase activity. The Y-axis displays relative luminescence units, which are normalized per unit of beta-galactosidase activity. SEs from 3 separate experiments are shown. The wild-type target sequence had at minimum average 6-fold lower luminescence compared with the mutant targets, and miRNA targeted to the mutant sequence did not significantly impact luciferase activity compared with the empty vector control. Sequences used to create the plasmids are shown in supplemental Table 1. The mismatch mutant vectors contain 2 mismatch bases from the miRNA seed sequence.
miRNA-494 and miRNA-320a affect survivin expression and apoptosis. (A) Bioinformatic target prediction analysis identifies survivin as a target of miR-494 and miRNA-320a; the survivin 3′ UTR complementary to the miRNA sequence is shown (using Targetscan). (B) Both miRNA-494 and miRNA-320a can regulate survivin expression. miRNA mimics for miRNA-494 and miRNA-320a were transfected into the REH cells and survivin expression was analyzed by Western blot analysis using a rabbit anti-survivin antibody (Cell Signaling Technology). The miRNA with no human targets were used as control. For loading controls, an equal number of cells from each experimental sample were lysed, and the same volume of the same lysate was applied to a separate gel to probe for tubulin. (C) miRNA-494 and miRNA-320a induce apoptosis. miRNA mimics for miRNA-494 and miRNA-320a were transfected into the REH cells and apoptosis was assessed by FACS analysis of annexin V–FITC/PI staining of control-treated (miRNA with no human targets, see “Methods”), and mimics for miRNA-494 and miRNA-320a. Data are shown from 1 of 3 representative experiments and indicate early apoptosis (annexin V single-positive cells). Percentages of annexin V–positive cells are indicated in the figure (SEs are under 2%). Percentages refer to the cells within the top left and bottom left quadrants, and top right and bottom right, respectively. (D) TEL-AML1 regulation of survivin is Dicer1 dependent. siRNA silencing of Dicer1 restores survivin expression in TEL-AML1 depleted cells indicating the miRNA dependence of this effect. For loading controls, an equal number of cells from each experimental sample were lysed, and the same volume of the same lysate was applied to a separate gel to probe for tubulin. (E) Luciferase vectors containing portions of the survivin (BIRC5) 3-UTR was used to demonstrate that survivin is a direct target of miRNA-494 and miRNA-320a. Both miRNAs were able to block luciferase expression significantly when they were cotransfected into REH cell together with the BIRC5 3-UTR luciferase-expressing vector (WT BIRC5). A mutated sequence and the empty vector were used as controls (MUT BIRC5 and pMir Reporter, respectively), and both had equivalent levels of luciferase activity. The Y-axis displays relative luminescence units, which are normalized per unit of beta-galactosidase activity. SEs from 3 separate experiments are shown. The wild-type target sequence had at minimum average 6-fold lower luminescence compared with the mutant targets, and miRNA targeted to the mutant sequence did not significantly impact luciferase activity compared with the empty vector control. Sequences used to create the plasmids are shown in supplemental Table 1. The mismatch mutant vectors contain 2 mismatch bases from the miRNA seed sequence.
The impact of TEL-AML1 silencing upon miRNA-494 and miRNA-320a is independent of TEL expression. (A) HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) with TEL or TEL-AML1 plasmids cloned into pcDNA 3.1 according to manufacturer's protocol exactly as explained in Diakos et al.8 Cells were lysed in radio-immunoprecipitation assay buffer containing proteinase inhibitors (Complete, Roche) with subsequent western analysis. Transient expression of proteins was confirmed by using TEL antibody (AB23465-100 Abcam) antibody and the LI-COR detection system (LI-COR Odyssey). (B) An ETV6 (TEL) expressing vector was introduced into REH cells by lipofection (lane 1) and the TEL-AML1 expression was silenced using siRNA (lane 2). The expression of TEL and TEL-AML1 was analyzed by Western blot analysis using an antibody against TEL. An anti-tubulin antibody was used for loading control on the same gel (using the LI-COR system). (C-D) The expression of miRNA-320 (C) and miRNA-494 (D) were analyzed by miRNA Taqman PCR. The black bars represent the results from lanes 1 to 3 for the HEK293T experiment shown in Figure 5A, and the gray bars represent the result from lanes 1 and 2 for the REH cell experiment in Figure 5B. miRNA levels are displayed relative to the empty vector HEK293a result.
The impact of TEL-AML1 silencing upon miRNA-494 and miRNA-320a is independent of TEL expression. (A) HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) with TEL or TEL-AML1 plasmids cloned into pcDNA 3.1 according to manufacturer's protocol exactly as explained in Diakos et al.8 Cells were lysed in radio-immunoprecipitation assay buffer containing proteinase inhibitors (Complete, Roche) with subsequent western analysis. Transient expression of proteins was confirmed by using TEL antibody (AB23465-100 Abcam) antibody and the LI-COR detection system (LI-COR Odyssey). (B) An ETV6 (TEL) expressing vector was introduced into REH cells by lipofection (lane 1) and the TEL-AML1 expression was silenced using siRNA (lane 2). The expression of TEL and TEL-AML1 was analyzed by Western blot analysis using an antibody against TEL. An anti-tubulin antibody was used for loading control on the same gel (using the LI-COR system). (C-D) The expression of miRNA-320 (C) and miRNA-494 (D) were analyzed by miRNA Taqman PCR. The black bars represent the results from lanes 1 to 3 for the HEK293T experiment shown in Figure 5A, and the gray bars represent the result from lanes 1 and 2 for the REH cell experiment in Figure 5B. miRNA levels are displayed relative to the empty vector HEK293a result.
Expression of miRNA-494 and miRNA-320a in clinical samples
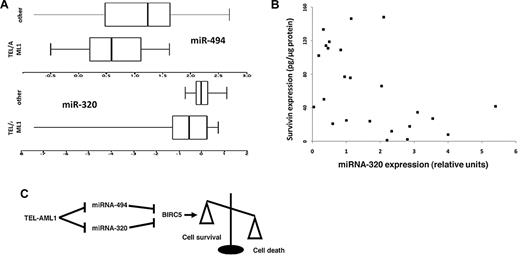
Survivin has been shown to be highly expressed in leukemias. In particular, recent studies suggested a role for survivin in TEL-AML1 leukemia and high levels of survivin expression have been associated with relapse cases.8,32-34 We tested the expression of miRNA-494 and miRNA-320a in TEL-AML1 leukemias and compared them to those of common ALLs of similar immunophenotype and biological characteristics, not carrying this translocation. Despite the small number of patient samples available, we showed that TEL-AML1 leukemias express lower levels of these miRNAs (P = .04 for miRNA-494, and P = .03 for miRNA-320a, Wilcoxon test, Figure 5A). This finding fits well with hypothesis that these miRNAs control survivin expression and might be deregulated at their promoter by the binding and the transcriptional suppression activity of TEL-AML1. We further tested a series of 28 TEL-AML1+ patients to detect whether a relationship existed with TEL-AML1 (RNA levels measured by Taqman) and miRNA-320A and -494 levels (also measured by Taqman), and whether the miRNAs influenced survivin levels (measured by ELISA in the same samples). We were able to detect a negative correlation between TEL-AML1 transcript levels and miRNA-320a levels (Spearman correlation = −0.48, P = .002) but not miRNA-494 (Spearman correlation = −0.02, P = .26). Additionally, a negative correlation existed between both miRNA 320a (Spearman correlation = −0.38, P = .015) and miRNA-494 (Spearman correlation = −0.21, P = .06) and survivin levels. A control miRNA, miRNA-193a, was analyzed in the same samples and was not significantly correlated with either TEL-AML1 or survivin (data not shown).
miR-494 and miR-320a expression in leukemia samples, and a model of action. (A) RNA was isolated from diagnostic leukemia samples from 11 TEL-AML1 positive and 15 TEL-AML1 negative cALL patients, and miRNA expression for miR-494 and miR-320a were analyzed by quantitative PCR. Shown are box-and-whisker plots of log10-transformed data; the box contains 50% of the data separated by the median, and the whiskers contain the remaining 25% at each side. TEL-AML1 leukemias expressed less miRNA-494 (P = .04) and miRNA-320a (P = .03) than other cALLs. (B) Scatterplot of the relationship of miRNA-320a (measured by Taqman) and survivin protein (measured by ELISA) among 28 cryopreserved TEL-AML1 leukemia cell samples. Two samples that were outliers were removed to “center” the remainder of the data. miRNA-320a and survivin were correlated (Spearman corrrelation: −0.38, P = .015). (C) Schematic presentation of the TEL-AML1, miRNA-494, and miRNA-320a impact upon survivin expression and cell survival. TEL-AML1 binds miRNA-494 and miRNA-320a promoters and exerts its transcriptional suppressor activity. These block miRNA-494 and miRNA-320a expression and release miRNA control of survivin expression resulting in increased resistance from apoptosis.
miR-494 and miR-320a expression in leukemia samples, and a model of action. (A) RNA was isolated from diagnostic leukemia samples from 11 TEL-AML1 positive and 15 TEL-AML1 negative cALL patients, and miRNA expression for miR-494 and miR-320a were analyzed by quantitative PCR. Shown are box-and-whisker plots of log10-transformed data; the box contains 50% of the data separated by the median, and the whiskers contain the remaining 25% at each side. TEL-AML1 leukemias expressed less miRNA-494 (P = .04) and miRNA-320a (P = .03) than other cALLs. (B) Scatterplot of the relationship of miRNA-320a (measured by Taqman) and survivin protein (measured by ELISA) among 28 cryopreserved TEL-AML1 leukemia cell samples. Two samples that were outliers were removed to “center” the remainder of the data. miRNA-320a and survivin were correlated (Spearman corrrelation: −0.38, P = .015). (C) Schematic presentation of the TEL-AML1, miRNA-494, and miRNA-320a impact upon survivin expression and cell survival. TEL-AML1 binds miRNA-494 and miRNA-320a promoters and exerts its transcriptional suppressor activity. These block miRNA-494 and miRNA-320a expression and release miRNA control of survivin expression resulting in increased resistance from apoptosis.
Discussion
Although miRNA expression is believed to play an important role in malignant transformation, little is known about the mechanisms that contribute to abnormal miRNA regulation in cancer. In this study we investigated the effect imposed upon miRNA expression by the fusion protein TEL-AML1. Recent studies have shown a close functional relationship between transcription factors and miRNA, and that they form regulatory networks where transcription factors bind and regulate miRNA expression and vice versa.27,28 As TEL and AML1 are both transcription factors we set to clarify whether the TEL-AML1 fusion gene alters the regulation of miRNA expression and by doing so ultimately contributes to malignant transformation. Using a cell line model we were able to identify 2 miRNAs, miRNA-494 and miRNA-320a, to be among the highest differentially expressed miRNA following TEL-AML1 knock-down. These miRNAs were also identified by ChIP-chip analysis as targets of TEL-AML1. In a previous study, we demonstrated a functional relationship between TEL-AML1 and the apoptotic network with a major impact on the expression of the antiapoptotic protein survivin, a potent member of the inhibitor of the apoptosis IAP family.8 In the current study we have linked these miRNAs to the suppression of survivin and also apoptosis in cells expressing these miRNAs. We propose a model in which TEL-AML1 can regulate miRNA-494 and miRNA-320a expression and by doing so can also affect the expression of their mutual target survivin and thus cell vulnerability toward apoptotic cell death (Figure 5C). In an additional experiment we demonstrate that the ability of TEL-AML1 to affect survivin expression is DICER1-dependent and thus provides further evidence that TEL-AML1 employs miRNA to interfere with survivin expression. Interestingly, miRNA-494 is the miRNA with the highest predicted target specificity to survivin (TargetScan) and in our cell line experiments was the most potent inhibitor of survivin (Figure 3B). We observed in clinical samples lower expression levels of miRNA-494 and miRNA-320a among TEL-AML1+ patients compared with cALL of other subtypes, which is compatible with the observations in the cell line experiments. Both miRNA-320a and miRNA-494 were negatively correlated with survivin in a small clinical series of TEL-AML1+ patients, consistent with the cell line experiments; but only miRNA-320 was significant in this regard. Because of the variability of clinical experiments including host differences, leukemia genetic differences, and sample processing/storage/shipping variability, a strong correlation was not expected and this result should be considered exploratory but supportive.
There are limitations to the current study, which should stimulate further research. While we have identified a binding region for the TEL-AML1 protein in miRNA-494 upstream sequence, miRNA-494 is in part of a cluster of 12 miRNA on chromosome 14; while it is the only one in the cluster showing a strong effect of expression related to TEL-AML1 in this study, the remaining miRNAs may also be influenced by TEL-AML1 but were not assessed further.
The TEL-AML1 oncoprotein is generally a transcriptional repressor due to its known ability to recruit chromatin repressors such as histone deacetylases, nuclear receptor corepressors, Sin3a, and repressive activity in in vitro assays (reviewed in35 ). This activity is contributed by the TEL repressor domain which is retained in the chimeric protein. However, AML1 DNA binding and activation domains are also a part of this protein, as nearly all of AML1 is retained. AML1 DNA targets, or in one case, TEL-AML1 protein partners,36 can result in promoter repression. We found, however, that nearly as many miRNAs were enhanced in expression upon removal of TEL-AML1 as were repressed (Figure 1C, and data not shown). Direct miRNA targets were not any more likely to be expressed higher, or lower, upon release of TEL-AML1, and many direct targets were not changed at all, such as the miRNA-139, the strongest TEL-AML1 target (Figure 2D). Our results suggest that a complex picture of miRNA gene regulation by TEL-AML1, and many effects on miRNA are likely not due directly by TEL-AML1 itself but rather by other transcriptional regulators that are altered in expression via TEL-AML1. However another hybrid transcription factor with an AML1 DNA–binding domain (AML1-ETO) was suggested to interfere directly with miRNA. Fazi et al37 showed that this translocation product controls miRNA-223 with an impact upon cell differentiation, thereby providing prior precedence for fusion gene control of miRNA. In other work, PML-RARA was shown to repress miRNA expression within direct targets; this repression was abrogated by retinoic acid.38 Additional experiments in other model systems will be required to consider the totality of miRNA regulation; our current report is largely limited to an explanation for the link between TEL-AML1 and the critical antiapoptotic protein, survivin.
Survivin is involved in control of chromosomal alignment during mitosis, and has a role in preventing apoptosis via caspase inhibition and other activities.39 Survivin is not expressed in most normally differentiated adult tissues, but it appears to be expressed in most cancer types and is a proposed target for anti-cancer vaccines. However, survivin is absolutely essential for various stages of hematopoiesis including erythroid differentiation, and is expressed at low levels in a cell-cycle-specific manner in proliferation and survival of embryonic and hematopoietic stem cells, T cells, neutrophils, and megakaryocytes (reviewed in Fukuda and Pelus40 ). Knockout of survivin in the hematopoietic compartment abrogates bone marrow development.41 Survivin is not a hematopoiesis-specific gene and therefore is likely critical in the kinetic regulation of hematopoiesis rather than functional maintenance and differentiation of specific lineages. The ectopic disruption of survivin regulation by TEL-AML1 as we have shown provides a potential mechanism by which the fusion oncogene mediates some of its known preleukemic effects of enhancement of clonal growth without the complete loss of differentiation or profoundly enhanced proliferation.36,42,43 TEL-AML1 is known to provide a subtle effect on hematopoiesis leading to resistance of TGF-β-mediated growth suppression36 ; however, carrying the fusion gene does not result in disease in animals44 or humans4,45 without secondary events.
Considering the characteristic miRNA profile of the leukemias with different translocations,15 a specific translocation–miRNA regulation might also be an indispensable feature that is shared by other leukemia-associated fusion genes. The fact that miRNA are part of the TEL-AML1 driven network that affects apoptosis opens new opportunities for the development of specific therapeutic intervention at the miRNA level and in our understanding of leukemogenesis by the oncogene.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Hope Street Kids Fund (J.L.W.), Children with Leukemia Fund UK (J.L.W.), Alex's Lemonade Stand Foundation and the Butterfly Foundation (S.Z.), and National Institutes of Health R03-CA137829 (S.Z.) and RO1-CA89032 (J.L.W.). G.K. is supported by a grant from the Jubiläumsfonds Oesterreichischen Nationalbank 13 655. J.L.W. is a scholar of the Leukemia & Lymphoma Society of America. M.S.P.O. is a scholar of the National Council for Scientific and Technological Development #309091/2007, Brazil. G.M.V. is a PhD student supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior #BEX2663/07-4.
National Institutes of Health
Authorship
Contribution: C.D. conceived and performed most of the experiments and wrote the first draft of the manuscript; S.Z. and M.Z. constructed the plasmids and performed the luciferase experiments, and performed the survivin/miRNA/TEL-AML1 transcript comparisons; Y.X. performed bioinformatic analysis and helped write the manuscript; G.M.V. and M.S.P.d.O. collected and characterized the leukemia specimens and isolated and prepared the RNA; G.K. and R.P.-G. assisted with the enforced TEL expression experiment in HEK293 cells; R.-F.Y. assisted with the statistical and bioinformatic analysis; S.Z. helped with the chromatin immunoprecipitation experiments; M.K. maintained cell lines and helped perform quantitative PCR experiments; J.K.W. contributed to experimental design and interpretation; and J.L.W. contributed to experimental design and interpretation, performed some data analysis, assisted in experiments, helped draft and finalize the manuscript, and secured the funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joseph Wiemels, University of California, San Francisco, HD274, 1450 3rd St MC 0520, San Francisco, CA 94158; e-mail: joe.wiemels@ucsf.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal