Abstract
Abstract 670
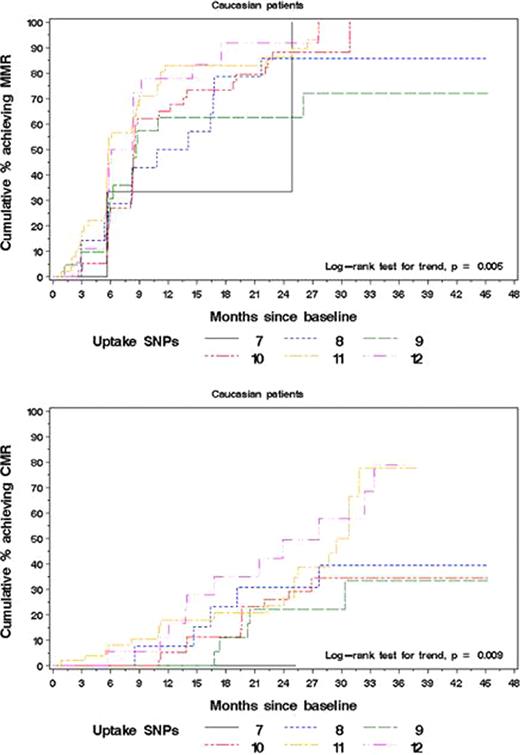
The availability of multiple options for chronic myeloid leukemia (CML) treatment is not paralleled by the availability of biological predictors of outcome allowing to identify patients (pts) who are more likely to benefit from dasatinib or nilotinib rather than imatinib (IM). Pharmacogenetics has proven a potential source of biomarkers given the known influence of polymorphisms in key genes encoding drug transporters and metabolizing enzymes on drug delivery – hence effectiveness. In CML, only two studies had so far explored this field, but both were conducted in heterogeneous populations including pts at different stages of disease, not all receiving IM first-line. We thus aimed to investigate a panel of 20 single nucleotide polymorphisms (SNPs) in ABCB1, ABCG2, SLC22A1, OATP1A2, OCTN1, CYP3A4 and CYP3A5 genes that can be hypothesized to influence IM transport and metabolism in 189 newly diagnosed CML pts enrolled in the TOPS phase III trial (Cortes et al, J Clin Oncol 2010). Pts selection was exclusively based on availability of written informed consent and sufficient amount of archived material. Median age was 46 years; male to female ratio was 103 to 86; 156 (83%) pts were Caucasian and 23 (12%) were Asian; low, intermediate and high Sokal risk pts were 84 (44.4%), 65 (34.4%) and 40 (21.2%), respectively. Baseline demographic/clinical features did not differ significantly from those of the overall population. Treatment outcomes (complete cytogenetic response [CCyR]; major molecular response [MMR] and complete molecular response [CMR]) were compared according to i) each candidate genotype ii) summary measures based on combinations of SNPs in the same gene and iii) summary measures based on combinations of SNPs in functionally related genes (uptake; efflux). CC genotype in OCTN1 had a favorable impact on the achievement of MMR at 12 months (MMR@12m; P = 0.03). With respect to the summary measures, combination of SNPs in the SLC22A1 gene was significantly correlated with MMR@12m (P = 0.03). When considering summary measures of uptake and efflux, the former was found to be associated with both MMR@12m and CMR@12m (P = 0.003 and P = 0.01, respectively). A separate analysis limited to Caucasian pts (n=156) yielded similar results (Table 1). In addition, the analysis in the Caucasian subgroup evidenced a significant association between the CC genotype in ABCB1 rs60023214 and MMR@12m (P = 0.005) (Table 1). Cumulative incidence plots based on the Kaplan-Meier method were also analyzed in the overall population and in Caucasians, with comparable results. Representative plots are shown in Figure 1. There was evidence for difference among MMR cumulative incidence curves for 2 single SNPs and 2 score measures. Presence of the major allele in OCTN1 (CC) and of the minor allele in CYP3A4 rs2740574 (GG) were associated with increased MMR rate (P = 0.028 and P = 0.042, respectively, in the overall population and P = 0.027 and P = 0.038, respectively, in Caucasians). Similarly, an increase in the number of favorable alleles in the SLC22A1 gene was associated with increased MMR rate (P = 0.030 and P = 0.043 in the overall population and in Caucasians, respectively). In addition, the combination of favorable alleles in the genes involved in IM uptake was associated with increased rates of both MMR and CMR (P = 0.004 and P = 0.015, respectively, in the overall population and P = 0.005 and P = 0.009, respectively, in Caucasians). Our results suggest that SNP genotyping might be helpful in selecting pts who are more likely to benefit from first-line use of more potent inhibitors. Further assessment of the SNPs here identified in larger series of pts is warranted.
Summary of significant associations
| . | . | Overall population (n=189) . | Caucasians (n=156) . | ||
|---|---|---|---|---|---|
| Gene, rs . | Outcome . | P . | Hazard ratio[95% CI] . | P . | Hazard ratio[95% CI] . |
| OCTN1 - rs1050152 | MMR@12m | 0.03 | 0.78 (0.63, 0.98) | 0.03 | 0.77 (0.60, 0.97) |
| ABCB1 (MDR1) - rs60023214 | CMR@12m | 0.06 | 0.71 (0.49, 1.02) | 0.005 | 0.56 (0.37, 0.84) |
| SLC22A1 (hOCT1) - SNP combination | MMR@12m | 0.03 | 1.24 (1.02, 1.50) | 0.04 | 1.24 (1.00, 1.54) |
| Uptake - SNP combination | MMR@12m | 0.003 | 1.21 (1.06, 1.37) | 0.005 | 1.24 (1.06, 1.44) |
| Uptake - SNP combination | CMR@12m | 0.01 | 1.30 (1.05, 1.61) | 0.01 | 1.37 (1.08, 1.74) |
| . | . | Overall population (n=189) . | Caucasians (n=156) . | ||
|---|---|---|---|---|---|
| Gene, rs . | Outcome . | P . | Hazard ratio[95% CI] . | P . | Hazard ratio[95% CI] . |
| OCTN1 - rs1050152 | MMR@12m | 0.03 | 0.78 (0.63, 0.98) | 0.03 | 0.77 (0.60, 0.97) |
| ABCB1 (MDR1) - rs60023214 | CMR@12m | 0.06 | 0.71 (0.49, 1.02) | 0.005 | 0.56 (0.37, 0.84) |
| SLC22A1 (hOCT1) - SNP combination | MMR@12m | 0.03 | 1.24 (1.02, 1.50) | 0.04 | 1.24 (1.00, 1.54) |
| Uptake - SNP combination | MMR@12m | 0.003 | 1.21 (1.06, 1.37) | 0.005 | 1.24 (1.06, 1.44) |
| Uptake - SNP combination | CMR@12m | 0.01 | 1.30 (1.05, 1.61) | 0.01 | 1.37 (1.08, 1.74) |
Cumulative incidence of pts achieving MMR and CMR according to the # of major alleles in uptake-related gene (SLC22A1, OATP1A2, OCTN1) SNPs
Cumulative incidence of pts achieving MMR and CMR according to the # of major alleles in uptake-related gene (SLC22A1, OATP1A2, OCTN1) SNPs
Supported by Novartis Oncology, Clinical Development, TOPS Correlative Studies Network
Hughes:Novartis: Honoraria, Research Funding, Speakers Bureau; Bristol-Myers Squibb: Honoraria, Research Funding; Ariad: Honoraria. White:Novartis: Honoraria, Research Funding; Bristol-Myers Squibb: Research Funding. Saglio:Novartis: Consultancy, Honoraria; Bristol-Myers Squibb: Consultancy, Honoraria. Rosti:Novartis: Consultancy, Honoraria; BMS: Consultancy, Honoraria. Hatfield:Novartis: Employment. Martinelli:Novartis: Consultancy, Honoraria; BMS: Honoraria; Pfizer: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal