Abstract
Abstract 2739
Internal tandem duplication mutations of the FMS-like tyrosine kinase 3 receptor (FLT3-ITD) are among the most frequent mutations in de novo acute myeloblastic leukemia (AML) and result in constitutive activation of the receptor tyrosine kinase. Based on data showing that these mutations negatively affect outcome in AML, FLT3 receptor kinase activity is currently being targeted in the clinic. To date, only FLT3 receptor mutant AML patients have been selected for trials involving FLT3 receptor kinase inhibitors, with surprisingly variable clinical responses. Downstream targets of FLT3 receptor activation, whether by receptor mutation or FLT3 ligand binding, involve Stat5, PI3-kinase/Akt and the Ras/Raf/Erk kinase signal transduction pathways which ultimately affect cell survival and proliferation. Functional characterization of those signaling pathways in mutated versus non-mutated FLT3 receptor (FLT3 -WT) myeloblasts has significant potential to reveal heterogeneity among these genetically defined subgroups and to predict for response to kinase inhibition, independent of FLT3 receptor mutation status.
Modulated single cell network profiling (SCNP) was used to evaluate the activation state of intracellular signaling molecules (i.e. nodes), including phosphorylated (p)-Akt, p-Erk, p-S6, p-Stat5 and cleaved-PARP, at baseline and after treatment with specific modulators [including cytokines (such as IL-27) growth factors (such as FLT3 ligand) and drugs (such as cytosine arabinoside)] in 7 healthy bone marrow mononuclear blasts (BMMb) and leukemic myeloblasts, characterized for FLT3 receptor mutation status, from 44 AML patients (38 FLT-WT and 6 FLT3-ITD), aged >60 years (ECOG trial E3999). A total of 64 node-metrics were analyzed.
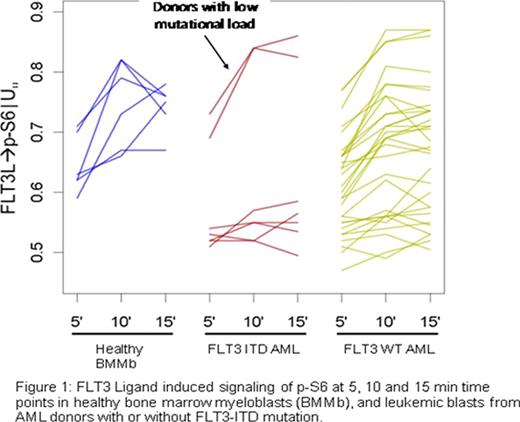
Signaling profiles differed significantly in FLT3-ITD vs. FLT3-WT AML, and in FLT3-WT vs. BMMb (shown in Figure 1 for a representative node, FLT3 ligand induced p-S6). Specifically, compared to BMMb, FLT3-ITD blasts uniformly showed increased basal p-Stat5 levels, decreased FLT3 ligand-induced activation of P13K and Raf/Ras/Erk pathways, minimal IL-27 induced activation of the Jak/Stat pathway, and higher apoptotic responses to DNA-damaging agents. Two AML samples harboring a low FLT3-ITD mutant burden, however, exhibited a signaling pattern similar to FLT3-WT AML samples. By contrast, FLT3-WT samples displayed heterogeneous signaling profiles, overlapping both with those of FLT3-ITD and BMMb samples, suggesting that a fraction of FLT3-WT AML samples exhibit FLT3 receptor pathway deregulation even without FLT3-ITD.
This study showed that SCNP, which provides a detailed view of intracellular signaling networks at the single-cell level, subclassified patients with AML beyond their molecularly determined FLT3 mutation status. In particular, a fraction of FLT3-WT AML samples signaled as if containing a FLT3 receptor length mutation while FLT3-ITD samples with low mutational load signaled like FLT3-WT AML samples. The clinical relevance of this observation, both for disease prognosis and response to kinase inhibitors, will be revealed only if AML patients are accrued to kinase inhibition trials irrespective of FLT3 receptor mutation status. The wide range of signaling responses observed in FLT3-WT AML suggests that disease across FLT3-WT patients is heterogeneous, likely promoted through distinct mutations and alterations, giving rise to distinct signaling profiles in individual patients Our data also provide evidence for the co-existence of differentially signaling blast populations in individual patients. The potential impact of signaling heterogeneity on clinical response needs to be assessed and may require an individualized combination of treatment modalities.
Rosen:Nodality Inc.: Employment, Equity Ownership. Putta:Nodality Inc.: Employment, Equity Ownership. Gayko:Nodality Inc.: Employment, Equity Ownership. Cesano:Nodality Inc.: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal