Abstract
Abstract 2695
Age and cytogenetics are major determinants of outcome in older patients with AML. Complete response (CR) to induction chemotherapy is observed in less than half of de novo AML patients (excluding APL) older than 60 years. Because unfavorable treatment results are often obtained despite intensive chemotherapy, which carries the potential for significant treatment-induced morbidity and mortality, it would be valuable to be able to predict with high accuracy at the individual patient level who will achieve a CR. Prospective identification of patients unlikely to benefit from anthracycline/cytarabine-based chemotherapy could spare patients from unnecessary treatment-related toxicities and allow consideration for alternative therapeutic strategies. Single Cell Network Profiling (SCNP) is used to measure simultaneously the effects of multiple modulators (including drugs) on signaling cascades at the single cell level. Using SCNP technology we have developed a set of classifiers that predict for likelihood of response to anthracycline/cytarabine-based induction therapy in older patients with AML.
SCNP assays were performed on paired, bone marrow (BM) and peripheral blood (PB), samples from 44 AML patients (de novo, evolved from an antecedent MDS or MPN or treatment related), >60 years old, enrolled on ECOG trial E3999. Based on two previous training studies, 38 combinations of modulators and intra-cellular proteins (signaling nodes along the phosphoinositide 3-kinase (P13K ), the Janus Kinases (Jak) signal transducers and activators of transcription (Stat) and the DNA damage response and apoptosis pathways) were investigated. Basal and modulated protein levels and the effect of modulation on proteins levels in the leukemic blast cells were expressed using a variety of metrics. A total of 64 node/metric combinations (dimensions) were used to build multi-parametric classifiers (ranging from 2 to 10 nodes/metrics) using different modeling methodologies (including random forest, boosting, lasso and a bootstrapped best subsets logistic modeling approach that shrinks regression coefficients (BBLRS)) able to predict the likelihood of response to induction therapy. The performance characteristics of the classifiers built on the BM samples were then evaluated independently on the paired PB samples.
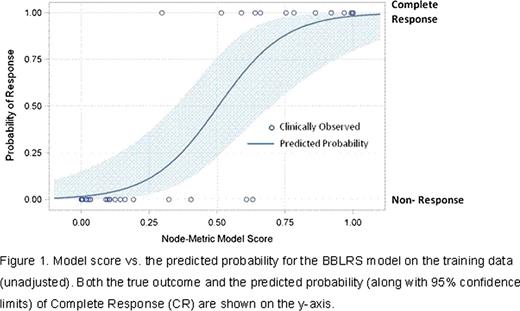
Several promising models with high area under the operator/receiver curve (AUROC) values (indicating strong agreement between actual clinical responses and responses as predicted by the model) were developed based on SCNP proteomic read outs for BM samples. The observed and predicted values from the current best BBLRS model are shown in Figure 1. The unadjusted AUROC of this model is 0.98 and the expected AUROC for the model when applied to an independent (validation) sample is 0.84. Five signaling nodes are represented in this model; they include nodes belonging to growth factor- induced survival pathways (PI3K, RAS/MAPK) as well as DNA damage response and apoptosis pathways. When the predictive accuracy of the lead SCNP classifier was compared to that of a model based on traditional clinical/molecular predictors (i.e. the combination of age, therapy-related AML, and karyotype) the adjusted AUROC of the SCNP classifier far surpassed that of the clinical predictors (adjusted AUROC=0.61 for clinical/molecular predictors vs. adjusted AUROC= 0.84 for the SCNP classifier). Finally, when the nodes in the best BBLRS model developed on data from BM samples were used to model read outs from the paired PB samples, the adjusted AUROC of the resulting BBLRS model was comparable to that of the model fit to BM samples.
This training set data show the value of performing quantitative SCNP under modulated conditions as the basis for developing highly predictive tests for response to induction chemotherapy. Most importantly, the predictions made by the SCNP classifier are independent of established prognostic factors, such as age and cytogenetics The ability of one set of nodes to accurately predict response in paired BM or PB samples from individual patients suggests that the predictive power of the SCNP assay is independent of sample source, further improving the practicality of the test. Independent validation studies are ongoing.
Model score us. the predicted probability for the BBLRS model on the training data (unadjusted). Both the true outcome and the predicted probability (along with 95% confidence limits) of Complete Response (CR) are shown on the y-axis.
Model score us. the predicted probability for the BBLRS model on the training data (unadjusted). Both the true outcome and the predicted probability (along with 95% confidence limits) of Complete Response (CR) are shown on the y-axis.
Cesano:Nodality Inc.: Employment, Equity Ownership. Putta:Nodality Inc.: Employment, Equity Ownership. Gayko:Nodality Inc.: Employment, Equity Ownership. Hackett:Nodality Inc.: Consultancy. Rosen:Nodality Inc.: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal