Abstract
Abstract 1850
Resistance to treatment remains the most important cause of relapse in contemporary acute lymphoblastic leukemia (ALL) therapy which calls for novel drugs to improve treatment outcome. We have shown previously that single agent treatment of BH3-mimetics like ABT-737 and obatoclax (GX15-070) resulted in a dose dependent apoptotic cell death and synergistic with prednisolone. However, little is known about the mechanisms and genomic responses underlying these BH3-mimetics. Since ABT-737 is a Bad-like mimetic while obatoclax is a Bim-like mimetic, we hypothesized that combination of these two different BH3-mimetics will increase the efficacy of cell death and enable reduced doses.
Seven ALL cell lines and a chronic myeloid leukemia cell line (K562) were used in this study. The 8 cells lines were exposed to ABT-737 or obatoclax as well as a combination of both and then subjected to whole genome gene expression analysis using Affymetrix HGU133 Plus 2.0 microarrays. The sets of differentially expressed genes were subsequently used for pathway analyses to identify the associated network functions using Ingenuity software. Cell viability was determined by MTS assay (Promega) and synergism was calculated using CalcuSyn software version 2.1. Western blot was used to detect protein expression level changes of the BCL-2 family members and cleaved PARP. Caspase-3,-8 and -9 activities were measured using Caspase-Glo™ Assay kits (Promega).
Treatment of both ABT-737 and obatoclax resulted in a dose dependent cell death in all 8 cell lines at 24h time point. All 7 ALL cell lines were sensitive to ABT-737 treatment with IC50 ranging between 0.05μM and 1.6μM, while K562 was less sensitive, with an IC50 of 31μM. All the 8 cell lines were sensitive to obatoclax treatment with similar IC50 ranging between 0.6μM and 5.7μM. Simultaneous in vitro exposure of ABT-737 and obatoclax to all cell lines in a 1:10 ratio resulted in synergistic levels of cell death with combination index (CI) values that are distinctly less than one. The levels of cleaved PARP and caspases -3, -8 and -9 activities increased significantly after combination treatment compared to individual treatment of each chemical. Interestingly, we did not observe any change in protein levels of Bcl-2 family members (including Bid, Bim, Bax, PUMA, Bcl-2, Mcl-1 and Bcl-w) after individual or combination treatment, indicating that the mechanism of synergism may be independent of the regulation of Bcl-2 family members.
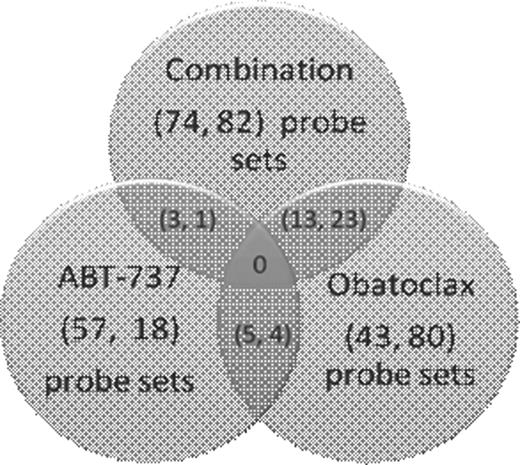
Groups of differentially regulated probe sets (p<0.05) after ABT-737 (75 probe sets), obatoclax (123 probe sets) or combination (156 probe sets) treatment were determined by global gene microarray analysis. Probe sets that were common between individual and combination treatments are represented in the figure 1below, indicating that the mechanisms of ABT-737 and obatoclax-induced cell deaths are different. Relative fold change analysis (genes that changed significantly (p<0.05) after combination treatment relative to single agent treatment) suggests that obatoclax contribute more towards synergism between these two BH3-mimetics. Furthermore, enriched GO terms of differentially expressed genes and the top five network functions associated with the respective treatments also reflect the same trend. The well-known genes that involved in many critical pathways of cell death were evaluated. We found that most genes that were differentially expressed after treatment are associated with the apoptotic pathway.
In this study, we reported that synergism of ABT-737 and obatoclax could be achieved in all 8 leukemia cell lines. Gene microarray analysis suggests that leukemia cells differ in their genomic response to the two drugs, and combination gains over single treatment. Although ABT-737 exhibits higher potency, its sensitivity seems to be cell-type dependent. The administration of BH3-mimetics in the clinic, alone or in combination, may overcome the limitations of single agent treatment and improve the treatment outcome of leukemia.
Probe sets (p<0.05) that were common between the single agent and combination treatments are represented. The values in parentheses indicate number of the probe sets (The left value indicates those were down-regulated and the right, up-regulated). The probe sets were obtained by comparing post-treatment samples to samples without any treatment.
Probe sets (p<0.05) that were common between the single agent and combination treatments are represented. The values in parentheses indicate number of the probe sets (The left value indicates those were down-regulated and the right, up-regulated). The probe sets were obtained by comparing post-treatment samples to samples without any treatment.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal