Abstract
ADAMTS13 modulates von Willebrand factor (VWF) platelet-tethering function by proteolysis of the Tyr1605-Met1606 bond in the VWF A2 domain. To examine the role of the metalloprotease domain of ADAMTS13 in scissile bond specificity, we identified 3 variable regions (VR1, -2, and -3) in the ADAMTS family metalloprotease domain that flank the active site, which might be important for specificity. Eight composite sequence swaps (to residues in ADAMTS1 or ADAMTS2) and 18 single-point mutants were generated in these VRs and expressed. Swapping VR1 (E184-R193) of ADAMTS13 with that of ADAMTS1 or ADAMTS2 abolished/severely impaired ADAMTS13 function. Kinetic analysis of VR1 point mutants using VWF115 as a short substrate revealed reduced proteolytic function (kcat/Km reduced by 2- to 10-fold) as a result of D187A, R190A, and R193A substitutions. Analysis of VR2 (F216-V220) revealed a minor importance of this region. Mutants of VR3 (G236-A261) proteolysed wild-type VWF115 normally. However, using either short or full-length VWF substrates containing the P1′ M1606A mutation, we identified residues within VR3 (D252-P256) that influence P1′ amino acid specificity, we hypothesize, by shaping the S1′ pocket. It is concluded that 2 subsites, D187-R193 and D252-P256, in the metalloprotease domain play an important role in cleavage efficiency and site specificity.
Introduction
Von Willebrand factor (VWF) is a large multimeric plasma glycoprotein with 2 important haemostatic roles. Its primary function is mediating the recruitment of platelets following damage to the vascular endothelium.1 It also acts as a carrier molecule for FVIII, thereby protecting it from degradation and extending its otherwise short plasma half-life.2 VWF circulates in plasma as multimers of varying sizes, ranging from 2-40 covalently linked VWF subunits. VWF normally circulates in a globular form, which conceals its platelet-binding sites. However, when vascular damage causes the exposure of subendothelial collagen, VWF is specifically and rapidly recruited to these sites through a collagen-binding site in the A3 domain. When bound to collagen, VWF is exposed to increased shear forces exerted by the flowing blood, which causes unfolding of the molecule and in turn exposure of platelet-binding site in the A1 domain.3 Once unraveled, VWF can then bind to the GPIb-IX-V receptor complex present on the surface of circulating platelets.4 This mechanism serves to capture circulating platelets, which once recruited can become activated, and in turn, lead to the presentation of GPIIbIIIa, which binds more stably to VWF through its C domains.
VWF multimeric size is a primary determinant of its platelet-tethering function because larger multimers not only have more collagen- and platelet-binding sites but also unfold more readily to reveal their platelet-binding sites.5 VWF multimeric size (and therefore also its ability to tether platelets) is regulated by the plasma metalloprotease, ADAMTS13.6 ADAMTS13 hydrolyses the Tyr1605-Met1606 peptide bond within the VWF A2 domain, converting VWF into smaller multimeric forms. The importance of ADAMTS13 function is highlighted by the thrombotic microangiopathy that arises in individuals with ADAMTS13 deficiency.6 These individuals develop microvascular platelet-rich thrombosis attributable to the comparatively high concentrations of ultra-large VWF species in plasma that persist in the absence of ADAMTS13 function.
ADAMTS13 is expressed predominantly in the liver by hepatic stellate cells.7 It has also been found in platelets,8 cultured endothelial cells,9 and glomerular podocytes.10 ADAMTS13 is secreted into the blood as an active enzyme and circulates at a plasma concentration of ∼ 5nM.11,12 ADAMTS13 is a multidomain metalloprotease. From its N-terminus, it consists of a metalloprotease domain (MP), disintegrin-like domain (Dis), a thrombospondin type 1 (TSP1) repeat, cysteine-rich domain, spacer domain, and 7 additional TSP1 repeats. Uniquely among ADAMTS family members, ADAMTS13 also has 2 C-terminal CUB domains.13,14
The MP domain is homologous to those of the metzincin family of proteases, containing a highly conserved active site sequence motif HEXXHXXGXXH and an adjacent Met-turn that provide a Zn2+-binding pocket. The bound Zn2+ in conjunction with the active site Glu225 residue coordinate a water molecule that drives hydrolysis of the scissile bond.15 In ADAMTS13, a high-affinity Ca2+-binding site close to the active site is also essential for efficient proteolysis of VWF to occur.16
VWF is the only known substrate of ADAMTS13, which it cleaves at just a single site (Tyr1605-Met1606) in the A2 domain. Unlike many plasma proteases, ADAMTS13 is constitutively active (ie, it is neither specifically released or specifically activated from a latent form upon demand) and also has a very long plasma half-life (estimated 2-3 days).17 For these reasons, it is essential that ADAMTS13 is highly specific to ensure that it does not proteolyze other targets nonspecifically. ADAMTS13 can bind to globular VWF, but proteolysis does not occur.18,19 Proteolysis of VWF takes place once it has been unfolded by rheological shear, because the scissile bond normally remains buried while VWF is in its circulating globular conformation. Once VWF has been unfolded, extensive interactions between VWF and ADAMTS13 domains are required for efficient cleavage. These interactions appear, at least in part, to confer high substrate specificity.18,20-25
ADAMTS13 binds to unfolded VWF with high affinity (KD ∼ 10-20nM).26,27 A critical step in both VWF binding and proteolysis is the docking of the spacer domain (involving residues Arg568, Arg660, Tyr661, and Tyr665)20,24 onto the C-terminal end of the VWF A2 domain (residues 1659-1668).21,28-31 It seems that this interaction mediates much of the tight binding between VWF and ADAMTS13. However, although this approximates the 2 molecules, these interactions take place some distance (∼ 110 Å) from the scissile bond and active site, respectively. The Dis domain also plays an essential role by enabling specific proteolysis22,23,28 through an interaction with residues close (∼ 26 Å) to the cleavage site involving ionic interactions between Asp1614 in VWF and Arg349 that helps place the Tyr1605-Met1606 bond into the active site cleft of the MP domain.23 Without the presence of these ancillary domains, the MP domain binds VWF with very low affinity and also loses some cleavage site specificity.26 However, despite the importance of these binding sites for proteolysis, the MP domain must also necessarily interact with the substrate in the vicinity of the cleavage site and harbor sites that contribute to scissile bond specificity. Indeed, low-affinity interactions between the MP domain and VWF residues spanning the scissile bond appear to be critical for VWF proteolysis because deletions in VWF close to the cleavage site abolish proteolysis22 and mutagenesis of the Tyr1605 (P1) and Met1606 (P1′) residues to alanine also greatly reduces the proteolytic efficiency.32 These findings suggest that the MP domain contains subsites that accommodate residues close to the cleavage site that are critical for cleavage of the VWF Tyr1605-Met1606 bond. Because the location and precise role of these ADAMTS13 subsites are unknown, we have investigated 3 candidate sites in this manuscript.
Methods
Molecular modeling of ADAMTS13 MP-Dis
Expression of recombinant ADAMTS13 MP domain variants
The expression vector for wild-type ADAMTS13 has been described elsewhere.36 Generation of MP domain mutations (E184A, L185A, D187A, R190A, Q191A, V192N, R193A, T196A, Q197A, D217N, L218A, V220A, T221A, A250G, S251A, D252N, R257A, and L260A) in the full-length ADAMTS13 expression vector was performed using the Quikchange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions.
Eight ADAMTS13 variants were generated in the variable regions (VR) within the MP domain that were not conserved among ADAMTS family members, and parts of the VR were swapped for the corresponding regions in either ADAMTS1 or ADAMTS2. Regions E184-R193 (VR1), F216-T221 (VR2), A237-A261 (VR3), A237-248 (VR3A), D252-A261 (VR3B), D252-P256 [VR3B(I)], and R257-A261 [VR3B(II)] were swapped with the corresponding residues in ADAMTS1. Region E184-R190 (VR1) was also swapped with the corresponding residues in ADAMTS2. These VR swap variants were generated by inverse polymerase chain reaction using primers that included the ADAMTS1 or ADAMTS2 sequence to be cloned. The resulting polymerase chain reaction products were purified by agarose gel electrophoresis, phosphorylated with T4 Kinase (Invitrogen), and blunt-end ligated using T4 Ligase (Invitrogen), according to manufacturers' instructions. Before further use, all expression vectors were verified by DNA sequencing.
Wild-type ADAMTS13 and all ADAMTS13 mutants were transiently expressed in HEK293T cells using linear polyethylenimine for transfection (Polysciences Inc), as previously described.16,37 Expression and secretion of ADAMTS13 was confirmed by Western blotting using a polyclonal rabbit anti-ADAMTS13 antibody.11 After 3-4 days, conditioned medium was harvested, cleared, and concentrated using 50-kDa molecular weight cutoff spin columns (Amicon). ADAMTS13 concentration in these samples was determined using a specific in-house ADAMTS13 enzyme-linked immunosorbent assay. A polyclonal anti-ADAMTS13 [affinity depleted of anti-ADAMTS13 TSP1(2-4) antibodies] was used as the capture antibody, and an affinity purified biotinylated rabbit polyclonal anti-ADAMTS13 TSP1(2-4) was used as for detection, as previously described.11,12
Expression and purification of VWF115 and VWF115 mutants
The VWF A2 domain fragment substrates, VWF115 (spanning VWF residues 1554-1668), and also cleavage site variants VWF115(M1606A) and VWF115(Y1605A/M1606A) generated by site-directed mutagenesis were expressed in Rosetta DE-3 Escherichia coli (Novagen), purified, and quantified as previously described.38
Expression of recombinant full-length VWF and the VWF(M1606A) mutant
An expression vector for full-length human VWF has been previously described.27 Generation of the M1606A mutation in this vector was performed using the Quikchange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. Recombinant full-length wild-type VWF and VWF(M1606A) were expressed transiently in HEK293T cells using linear polyethylenimine (Polysciences Inc), as previously described.16,37 The concentration of VWF was determined by a specific VWF enzyme-linked immunosorbent assay,39 and VWF multimer analysis was assessed using agarose gel electrophoresis and Western blotting for VWF, as previously described.27,39
ADAMTS13 activity assays
Qualitative analysis of the activity of wild-type ADAMTS13 and the different full-length ADAMTS13 MP domain mutants was carried out using 1nM ADAMTS13 in 20mM Tris-HCl (pH 7.8), 150mM NaCl, 5mM CaCl2, which was pre-incubated at 37°C for 1 hour without substrate. To start the reaction, 6μM VWF115 was added, and subsamples were removed and stopped with (ethylenedinitrilo)tetraacetic acid (EDTA) (40mM final concentration) at different time points (0-5 hours). Thereafter, proteolysis of VWF115 was visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. For more sensitive detection of the 7-kDa VWF115 cleavage product (to detect trace amounts of proteolysis), Western blotting was performed using a monoclonal antibody that recognizes the C-terminal sequence of the VWF A2 domain (R&D Systems). To confirm findings, all assays were performed at least 3 times using different batches of expressed ADAMTS13 or ADAMTS13 variants.
Quantitative analysis of VWF115 proteolysis was performed by reactions that were set up exactly as in the preceding paragraph, except 1μM VWF115 was used. To quantify proteolysis samples were analyzed by high-pressure liquid chromatography (HPLC), as previously described.38 From these experiments, the catalytic efficiencies (kcat/Km) were derived, as previously described.38
Analysis of the proteolysis of multimeric VWF was performed under denaturing conditions. For the analysis of ADAMTS13 VR1 mutants, purified plasma-derived VWF was used. Briefly, VWF was incubated with 1.5 M guanidine-HCl at 37°C for 30 minutes before starting the reaction. The denatured VWF was then diluted 10-fold to a final concentration of 10 μg/mL into 20mM Tris-HCl, 0.5% bovine serum albumin, 5mM CaCl2 buffer, in which 2.1-2.8nM ADAMTS13 had been pre-incubated for 60 minutes. Digestion of VWF was performed at 37°C for 90 minutes. For the analysis of the ADAMTS13 VR3B-AD1 mutant, recombinant full-length VWF(M1606A) mutant (containing the P1′ ADAMTS13 cleavage site substitution) was used. ADAMTS13 (0.5nM) was preincubated in 20mM Tris-HCl, 0.5% bovine serum albumin, 5mM CaCl2 buffer. In this reaction, 0.4 μg/mL full-length VWF(M1606A) was incubated at 37°C in the presence of 1.5 M urea. Subsamples were removed at various time points (0-100 minutes), and the reaction was stopped with EDTA (40mM). In all cases, changes in VWF multimeric distribution caused by ADAMTS13 proteolysis were analyzed by agarose gel electrophoresis and Western blotting for VWF, as previously described27,39
Results
Design and expression of ADAMTS13 MP domain mutants
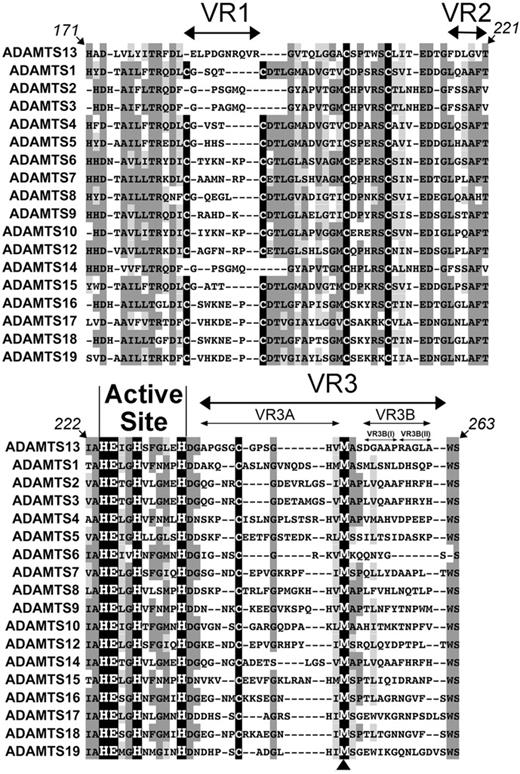
To aid in the identification of potentially unique MP domain subsites, the amino acid sequences of ADAMTS13 and all other human ADAMTS family members were aligned (Figure 1A). This alignment shows highly conserved regions, including the active site containing 3 Zn2+-binding histidines (His224, His228, and His234) and the catalytic glutamic acid (Glu225). The methionine residue involved in the characteristic Met-turn (Met249) is also perfectly conserved (see arrowhead beneath Figure 1). As each family member recognizes and cleaves different target proteins, the homologous regions in this alignment most likely represent conserved structural elements required for the MP folding rather than determinants of enzyme specificity. However, diverse amino acid sequences flank these conserved regions and form 3 “variable regions” (VR1, -2, and -3) that we hypothesized might be involved in conferring specificity to the different ADAMTS family MP domains for their respective substrates.
Identification of variable regions (VR) within the ADAMTS13 MP domain. Amino acid sequence alignment of ADAMTS13 MP domain (residues 171-263) with other ADAMTS family MP domains. The active site HEXHXXXXXH sequence is labeled, and the perfectly conserved Met-turn methionine is indicated (arrowhead below alignment). Conserved (dark gray) and homologous (light gray) regions are highlighted; Cys residues are shown in black. Variable/nonconserved regions VR1, -2, -3, -3A, -3B, -3B(I), and -3B(II) are depicted by arrows.
Identification of variable regions (VR) within the ADAMTS13 MP domain. Amino acid sequence alignment of ADAMTS13 MP domain (residues 171-263) with other ADAMTS family MP domains. The active site HEXHXXXXXH sequence is labeled, and the perfectly conserved Met-turn methionine is indicated (arrowhead below alignment). Conserved (dark gray) and homologous (light gray) regions are highlighted; Cys residues are shown in black. Variable/nonconserved regions VR1, -2, -3, -3A, -3B, -3B(I), and -3B(II) are depicted by arrows.
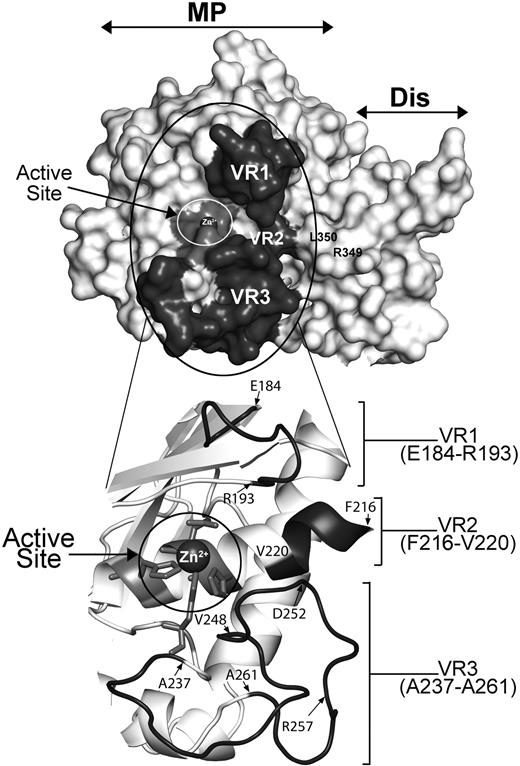
To first assess the viability of this hypothesis, we modeled the structure of the ADAMTS13 MP domain upon the available crystal structures of ADAMTS1, -4, and -534,35 (Figure 2). Our model suggests that VR1, -2, and -3 are optimally positioned to interact with residues surrounding the scissile bond and therefore potentially dictate the specificity/efficiency of ADAMTS13 proteolysis of the Tyr1605-Met1606 cleavage site in the VWF A2 domain.
Molecular model of ADAMTS13 MP-Dis based on the crystal structures of ADAMTS1, -4, and -5. The active site containing a Zn2+ ion is circled in white and labeled. R349 and L350 in the Dis domain known to be important for VWF proteolysis are labeled. VR1, -2, and -3 (shown in dark gray) flank the active site cleft. The region circled in black is enlarged and shown in cartoon below to better depict the position of VR1, -2, and -3. Amino acids at the start and end of each VR region are identified.
Molecular model of ADAMTS13 MP-Dis based on the crystal structures of ADAMTS1, -4, and -5. The active site containing a Zn2+ ion is circled in white and labeled. R349 and L350 in the Dis domain known to be important for VWF proteolysis are labeled. VR1, -2, and -3 (shown in dark gray) flank the active site cleft. The region circled in black is enlarged and shown in cartoon below to better depict the position of VR1, -2, and -3. Amino acids at the start and end of each VR region are identified.
Based on these observations, we targeted VR1, -2, and -3 for mutagenesis. Eight ADAMTS13 variants were generated in which VR (or parts of the VR) were swapped for the corresponding regions in either ADAMTS1 or ADAMTS2. In addition, 18 single-point mutations of amino acids within these VR were also generated (Figure 3).
ADAMTS13 MP domain variants. The wild-type ADAMTS13 MP domain sequence for VR1, -2, and -3 and flanking amino acids are shown. Single residues targeted for mutagenesis are shown in large bold type (point mutants). The different VR swaps are shown with the residues in the replaced sequence in bold/underlined.
ADAMTS13 MP domain variants. The wild-type ADAMTS13 MP domain sequence for VR1, -2, and -3 and flanking amino acids are shown. Single residues targeted for mutagenesis are shown in large bold type (point mutants). The different VR swaps are shown with the residues in the replaced sequence in bold/underlined.
Analysis of ADAMTS13 MP domain VR1 mutants (E184-R193)
All ADAMTS13 VR1 variants were transiently expressed in HEK293T cells. The ADAMTS13 VR1 swaps containing the corresponding regions of either ADAMTS1 (VR1-AD1) or ADAMTS2 (VR1-AD2) were both expressed to similar levels as wild-type ADAMTS13, as determined by Western blot analysis of conditioned medium (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This was perhaps somewhat surprising given that VR1-AD1 contains an additional pair of Cys residues that might form a disulphide bond, a pair that is not conserved in ADAMTS2, -3, -13, or -14. The VR1-AD2 variant was also expressed, despite the shortening of the VR1 loop with this swap. With the exception of V192N, the other 8 single-point mutants in VR1 were expressed and secreted normally (supplemental Figure 1B).
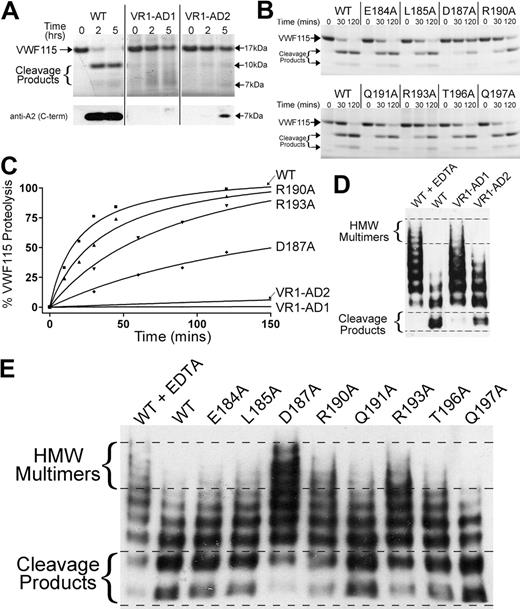
To examine the functional importance of VR1, the activity of ADAMTS13 variants VR1-AD1 and VR1-AD2 was assessed using VWF115 as a substrate. Reactions were analyzed by SDS-PAGE to visualize the 10-kDa and 7-kDa VWF115 cleavage products (Figure 4A). Substitution of E184-R193 with the corresponding 9 amino acids in ADAMTS1 (VR1-AD1) abolished all detectable activity. When VR1 (E184-R190) was swapped to the corresponding 5 residues in ADAMTS2 (VR1-AD2), proteolytic function was also severely affected. However, trace cleavage product bands were evident, which was confirmed by Western blotting using a monoclonal antibody that detects the C-terminal part of the VWF A2 domain. This identified the 7-kDa VWF115 cleavage product, which was evident after incubation for 5 hours with the VR1-AD2 mutant (see Figure 4A bottom panel). This suggests that the cleavage activity of this variant was not entirely ablated. The loss/marked reduction of ADAMTS13 activity upon substitution of VR1 demonstrated the functional importance of this loop that flanks the active site.
Functional characterization of ADAMTS13 VR1 (E184-R193) MP domain mutants. (A-B) Proteolysis of VWF115 over time by ADAMTS13/VR1 swap variants (A) or VR1 point mutants (B) was visualized by SDS-PAGE and Coomassie staining. For more sensitive detection of the 7-kDa VWF115 cleavage product (to detect trace amounts of proteolysis), a Western blot is included below Figure 4A. (C) Proteolysis of VWF115 was quantified over time by HPLC for those variants with reduced proteolytic function. See Table 1 for the derived kcat/Km values. (D-E) Multimeric, plasma-derived full-length VWF was proteolyzed by 2.8nM VR1 swap variants (D) or 2.1nM point mutants (E) and analyzed by agarose gel electrophoresis and Western blotting for VWF. HMW, high molecular weight; WT, wild type.
Functional characterization of ADAMTS13 VR1 (E184-R193) MP domain mutants. (A-B) Proteolysis of VWF115 over time by ADAMTS13/VR1 swap variants (A) or VR1 point mutants (B) was visualized by SDS-PAGE and Coomassie staining. For more sensitive detection of the 7-kDa VWF115 cleavage product (to detect trace amounts of proteolysis), a Western blot is included below Figure 4A. (C) Proteolysis of VWF115 was quantified over time by HPLC for those variants with reduced proteolytic function. See Table 1 for the derived kcat/Km values. (D-E) Multimeric, plasma-derived full-length VWF was proteolyzed by 2.8nM VR1 swap variants (D) or 2.1nM point mutants (E) and analyzed by agarose gel electrophoresis and Western blotting for VWF. HMW, high molecular weight; WT, wild type.
We next examined the influence of the 9 single-point mutants in VR1 (with residue changes E184A, L185A, D187A, R190A, Q191A, V192N, R193A, T196A, and Q197A) upon enzyme function. The activities of ADAMTS13 VR1 single-point mutants were screened using SDS-PAGE analysis of VWF115 proteolysis. By comparison to wild-type ADAMTS13, all single-point mutants exhibited normal activity with the exception of D187A, R190A, and R193A (Figure 4B). D187A and R193A mutants proteolyzed VWF115 appreciably more slowly than wild-type ADAMTS13, as evidenced by the reduced appearance of the 10-kDa and 7-kDa bands and the increased amounts of uncleaved VWF115 after 120 minutes. The reduction in the proteolytic function of the ADAMTS13(R190A) mutant was modest. To more accurately quantify the influence of these VR1 single-point mutations upon VWF115 proteolysis, time-course experiments were set up and cleavage was quantified by HPLC (Figure 4C). Comparison of the catalytic efficiencies (kcat/Km) of VWF115 proteolysis revealed that D187A, R190A, and R193A exhibited ∼ 10-fold, ∼ 2-fold, and ∼ 4-fold reduced catalytic efficiencies (n = 3), respectively (Table 1), consistent with the data from the SDS-PAGE analysis (Figure 4A-B). The VR1-AD1 and VR1-AD2 variants had such greatly reduced activities that it was not possible to assign values for kcat/Km.
Catalytic efficiencies (kcat/Km) of VWF115 proteolysis by wild-type ADAMTS13 and ADAMTS13 VR1 variants derived from time-course reactions
| . | Time course, kcat/Km, ×105 M−1s−1 . | Fold reduction . | P . |
|---|---|---|---|
| WT | 12.5 ± 3.92 | ||
| D187A | 1.27 ± 0.38 | 10 | 0.004 |
| R190A | 5.70 ± 0.42 | 2 | 0.051 |
| R193A | 3.03 ± 0.40 | 4 | 0.007 |
| VR1-AD1 | 0 | ∞ | |
| VR1-AD2 | n.d. | >25 |
| . | Time course, kcat/Km, ×105 M−1s−1 . | Fold reduction . | P . |
|---|---|---|---|
| WT | 12.5 ± 3.92 | ||
| D187A | 1.27 ± 0.38 | 10 | 0.004 |
| R190A | 5.70 ± 0.42 | 2 | 0.051 |
| R193A | 3.03 ± 0.40 | 4 | 0.007 |
| VR1-AD1 | 0 | ∞ | |
| VR1-AD2 | n.d. | >25 |
The fold reduction for each variant and the P value derived from an unpaired, 1-tailed t test is given (n = 3, except R190A for which n = 2).
WT indicates wild type; and n.d., not determined.
To generalize the results obtained using VWF115, we assessed the function of all ADAMTS13 VR1 variants using plasma-derived multimeric VWF as a substrate under denaturing conditions. As in the VWF115 assay, the VR1-AD1 variant exhibited no detectable VWF cleaving activity (Figure 4D). The ADAMTS13 variant in which E184-R190 was swapped with ADAMTS2 (VR1-AD2) and that cleaved VWF115 very slowly, proteolyzed full-length VWF, albeit appreciably less effectively than wild-type ADAMTS13 (Figure 4D).
In the multimeric VWF assays in which the VR1 point mutants were compared, wild-type ADAMTS13 proteolysis caused the higher-molecular-weight VWF multimers to disappear and lower-molecular-weight cleavage products became evident after 90 minutes (Figure 4E). The ADAMTS13 mutants, D187A, R190A, and R193A, all exhibited reduced proteolytic function.
Analysis of ADAMTS13 MP domain VR2 mutants (F216-T221)
Western blot analysis of conditioned media demonstrated that the ADAMTS13 VR2-AD2 swap variant in which residues F216-T221 were swapped for the corresponding 5 amino acids in ADAMTS1 was not secreted. The ADAMTS13 VR2 sequence is predicted to form an α-helix that runs along the base of the active site cleft. Some of the residues in this helix therefore face toward the inside of the MP domain, and it is likely that substitution of these are responsible for disruption of domain structure and intracellular retention of this variant. The ADAMTS13 VR2 single-point mutant, L218A, was also not secreted. The other VR2 single-point mutants (D217N, V220A, and T221A) were expressed normally (supplemental Figure 1C), suggesting that the mutations did not by themselves induce any gross structural changes that influenced enzyme secretion. When these 3 VR2 single-point mutants were analyzed functionally, all exhibited very modest reductions in the rate of VWF115 proteolysis (< 2-fold reduced catalytic efficiency; supplemental Figure1E), suggesting that these residues were not major determinants of ADAMTS13 function.
Functional analysis of ADAMTS13 MP domain VR3 (G236-A261)
To examine the importance of the larger variable sequence (VR3) that spans amino acids G236-A261 (Figure 1), 5 variants were made in which the whole or parts of this region were swapped for the corresponding residues in ADAMTS1: these were VR3-AD1, VR3A-AD1, VR3B-AD1, VR3B(I)-AD1, and VR3B(II)-AD1 (Figures 1,Figure 2–3). After expression of these variants in HEK293T cells, neither VR3-AD1 nor VR3A-AD1 swap variants were detected in the conditioned media. VR3B-AD1, VR3B(I)-AD1, and VR3B(II)-AD1 variants, however, were all expressed and secreted normally (supplemental Figure 1D). Of the 5 single-point mutants generated in VR3, A250G and S251A were both secreted poorly. These 2 amino acids are very well conserved among ADAMTS family members, suggesting their importance in the correct folding of these domains. Variants D252N, R257A, and T260A were all secreted similarly to wild-type ADAMTS13 (data not shown).
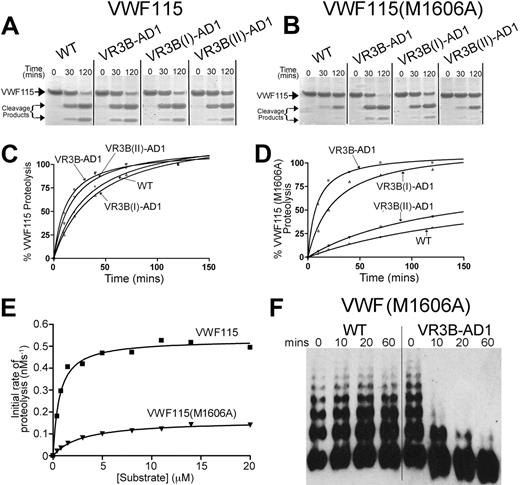
Successfully expressed ADAMTS13 variants VR3B-AD1, VR3B(I)-AD1, and VR3B(II)-AD1 were assessed functionally using VWF115 as a substrate. Rather surprisingly, given the extent of the amino acids changes, we detected essentially normal proteolytic function for all 3 of these VR3 swaps (Figure 5A). Consistent with this was the finding that the 3 point mutants generated in this region also exhibited normal enzymatic activity (data not shown).
Functional characterization of ADAMTS13 VR3 (G236-A261) MP domain mutants. (A-B) Proteolysis of VWF115 (A) or VWF115(M1606A) (B) over time by wild-type (WT) ADAMTS13 or VR3B swap variants was visualized by SDS-PAGE and Coomassie staining. (C-D) Proteolysis of VWF115 (C) or VWF115(M1606A) (D) by ADAMTS13 and VR3B swap variants was quantified over time by HPLC and represented graphically. See Table 2 for the derived kcat/Km values. (E) To establish the individual kinetic constants Km and kcat for VWF115(M1606A) proteolysis, the initial rate of proteolysis (< 15% cleavage) was analyzed by HPLC as a function of substrate concentration. The derived individual parameters are presented in Table 3. (F) 0.5nM wild-type ADAMTS13 or VR3B swap variant was used to proteolyze multimeric, recombinant VWF(M1606A) under denaturing conditions. From 0 to 60 minutes, reactions were stopped with EDTA, and changes in VWF multimeric composition were monitored by SDS agarose gel electrophoresis and Western blotting for VWF.
Functional characterization of ADAMTS13 VR3 (G236-A261) MP domain mutants. (A-B) Proteolysis of VWF115 (A) or VWF115(M1606A) (B) over time by wild-type (WT) ADAMTS13 or VR3B swap variants was visualized by SDS-PAGE and Coomassie staining. (C-D) Proteolysis of VWF115 (C) or VWF115(M1606A) (D) by ADAMTS13 and VR3B swap variants was quantified over time by HPLC and represented graphically. See Table 2 for the derived kcat/Km values. (E) To establish the individual kinetic constants Km and kcat for VWF115(M1606A) proteolysis, the initial rate of proteolysis (< 15% cleavage) was analyzed by HPLC as a function of substrate concentration. The derived individual parameters are presented in Table 3. (F) 0.5nM wild-type ADAMTS13 or VR3B swap variant was used to proteolyze multimeric, recombinant VWF(M1606A) under denaturing conditions. From 0 to 60 minutes, reactions were stopped with EDTA, and changes in VWF multimeric composition were monitored by SDS agarose gel electrophoresis and Western blotting for VWF.
From our molecular modeling, the VR3B region is predicted to form part of a pocket adjacent to the active site (see “Discussion”) and is potentially able to interact with the P1′ of the substrate. We therefore explored whether VR3B plays a direct role in ADAMTS13 cleavage site specificity.
Because the ADAMTS13 VR3B-AD1 variant contains the corresponding sequence found in ADAMTS1, we hypothesized that this might have altered the scissile bond specificity of this variant, directing it toward the preferred substrate of ADAMTS1. ADAMTS1 preferentially proteolyzes the Glu441-Ala442 bond of versican40 (and also Gln-Phe, Glu-Gly, and Glu-Leu bonds in aggrecan41 and a Glu-Leu bond in thrombospondin-2 precursor42 ). The P1′ specificity of ADAMTS1 is therefore broader than that of ADAMTS13, for which only one peptide bond (ie, Tyr1605-Met1606 in VWF) has been reported to be cleaved. Therefore, to test whether the ADAMTS13 VR3B swap variants also exhibit broader P1′ specificity, proteolysis was assessed of VWF115 in which the P1′ residue was mutated to alanine [VWF115(M1606A)].
The VWF115(M1606A) substitution resulted in reduced rate of proteolysis by wild-type ADAMTS13 (Figure 5, compare panel B with panel A). This is in accordance with a previous report that showed that VWF115(M1606L) and VWF115(Y1605A/M1606A) were not efficiently proteolyzed by ADAMTS13.32 Interestingly, the ADAMTS13 VR3B-AD1 variant proteolyzed VWF115(M1606A) much more efficiently than wild-type ADAMTS13 (Figure 5B)—very similar to the proteolysis of wild-type VWF115 (Figure 5A). This suggests that introduction of the ADAMTS1 VR3B amino acid sequence (D252-P261) does indeed broaden the P1′ residue specificity. We obtained a similar result using the ADAMTS13 VR3B(I)-AD1 variant, but not using the VR3B(II)-AD1 variant (Figure 5B), suggesting that, more specifically, residues D252-P256 [VR3B(I)-AD1] interact with the P1′ rather than residues R257-A261 [VR3B(II)-AD1].
The molecular weights of the cleavage products of VWF115(M1606A) generated by ADAMTS13 VR3B-AD1 appear unaltered (ie, 10 kDa and 7 kDa) by SDS-PAGE analysis. This suggests that proteolysis occurred at the Y1605-A1606 bond, as expected. That the VR3B-AD1 swap variant does not proteolyze a different site was further corroborated by the finding that a VWF115(Y1605A/M1606A) mutant with both cleavage site residues substituted was proteolyzed much more slowly than VWF115(M1606A), suggesting that the substitution of the P1 residues still influenced proteolysis by ADAMTS13 VR3B-AD1 (supplemental Figure 1F). To further confirm the identity of the site cleaved by ADAMTS13 VR3B-AD1, the VWF115(M1606A) cleavage fragments generated by ADAMTS13 VR3B-AD1 proteolysis were isolated by HPLC, and thereafter, tryptic peptides were analyzed by mass spectrometry (MS; Biopolymer Mass Spectrometry Lab, Imperial College London). All tryptic peptides were detected as [M+H]+ molecular ions by matrix-assisted laser desorption ionization time-of-flight analyses. Most importantly, the VWF115 fragment 1584YLSDHSFLVSQGDREQAPNLVY1605 ([M+H]+ (m/z): 2539.2), corresponding to the peptide N-terminal to the cleavage site, and the fragment 1606AVTGNPASDEIKR1618 ([M+H]+ (m/z): 1358.7), representing the C-terminal peptide, were both identified, which together, confirmed proteolysis of the Y1606-A1606 bond. Cleavage fragments were also sequenced by MS/MS using matrix-assisted laser desorption ionization time-of-flight /time-of-flight equipment. The results, again, demonstrated the Y1605-A1606 peptide bond as the site of proteolysis (data not shown).
To confirm and quantify the effect of the swaps made in the VR3B region, proteolysis of VWF115 and VWF115(M1606A) was analyzed kinetically by HPLC (Figure 5C-D). As observed with the SDS-PAGE analyses, the ADAMTS13 VR3B-AD1 variant proteolyzed VWF115 at a similar rate as wild-type ADAMTS13. The kcat/Km of VWF115(M1606A) proteolysis by wild-type ADAMTS13 was 18-fold reduced compared with cleavage of wild-type VWF115 (Table 2, n = 3). The ADAMTS13 VR3B(II)-AD1 variant cleaved VWF115(M1606A) 11-fold more slowly compared with VWF115. The VR3B-AD1 and VR3B(I)-AD1 swap variants, however, proteolyzed VWF115 and VWF115(M1606A) at a similar rate.
Catalytic efficiencies (kcat/Km) of VWF115 and VWF115(1606A) proteolysis by wild-type ADAMTS13 and ADAMTS13 VR3B variants derived from time course reactions (n=3)
| . | Time course, kcat/Km, ×105 M−1s−1) . | Fold reduction . | |
|---|---|---|---|
| VWF115 . | VWF115 (M1606A) . | ||
| WT | 7.2 ± 2.9 | 0.4 ± 0.1 | 18 |
| VR3B-AD1 | 11.9 ± 4.9 | 11.5 ± 4.2 | 0 |
| VR3B(I)-AD1 | 4.3 ± 0.7 | 4.0 ± 1.2 | 0 |
| VR3B(II)-AD1 | 7.8 ± 3.4 | 0.7 ± 0.3 | 11 |
| . | Time course, kcat/Km, ×105 M−1s−1) . | Fold reduction . | |
|---|---|---|---|
| VWF115 . | VWF115 (M1606A) . | ||
| WT | 7.2 ± 2.9 | 0.4 ± 0.1 | 18 |
| VR3B-AD1 | 11.9 ± 4.9 | 11.5 ± 4.2 | 0 |
| VR3B(I)-AD1 | 4.3 ± 0.7 | 4.0 ± 1.2 | 0 |
| VR3B(II)-AD1 | 7.8 ± 3.4 | 0.7 ± 0.3 | 11 |
WT indicates wild type.
To confirm these changes in catalytic efficiencies independently and also to establish the individual kinetic constants Km and kcat for VWF115(M1606A) proteolysis, the initial rate of proteolysis (< 15% cleavage) was analyzed as a function of substrate concentration (Figure 5E and Table 3). These data showed a 15-fold reduction in kcat/Km, consistent with the time-course data. Furthermore, these results revealed that the reduced kcat/Km of VWF115(M1606A) arose as a consequence of both increased Km and reduced kcat.
Michaelis-Menten constants kcat and Km and the catalytic efficiencies (kcat/Km) of VWF115 and VWF115(1606A) proteolysis by wild-type ADAMTS13 and ADAMTS13 VR3B variants derived from kinetic analysis from the initial rates of substrate proteolysis
| . | Michaelis-Menten constants (WT ADAMTS13) . | Fold difference . | |
|---|---|---|---|
| VWF115 . | VWF115 (M1606A) . | ||
| Km, μM | 0.65 | 2.9 | 4.6 |
| kcat, s−1 | 0.52 | 0.16 | 3.3 |
| kcat/Km, ×105 M−1s−1) | 8.21 | 0.55 | 15 |
| . | Michaelis-Menten constants (WT ADAMTS13) . | Fold difference . | |
|---|---|---|---|
| VWF115 . | VWF115 (M1606A) . | ||
| Km, μM | 0.65 | 2.9 | 4.6 |
| kcat, s−1 | 0.52 | 0.16 | 3.3 |
| kcat/Km, ×105 M−1s−1) | 8.21 | 0.55 | 15 |
We investigated next whether these changes in scissile bond specificity were also observed using full-length VWF as a substrate. Recombinant full-length VWF(M1606A) was expressed normally and exhibited the same multimeric distribution as recombinant wild-type VWF. Reactions were set up with 0.5nM wild-type ADAMTS13 and ADAMTS13 VR3B-AD1 variant with recombinant VWF(M1606A) as a substrate under denaturing conditions. In these assays, no cleavage of VWF(M1606A) by wild-type ADAMTS13 was detected (Figure 5F). In contrast, the ADAMTS13 VR3B-AD1 variant proteolyzed VWF(M1606A) rapidly, as visualized by disappearance of high-molecular-weight VWF multimers (Figure 5F).
Discussion
In this study, we examined the role of ADAMTS13 MP domain subsites in the proteolysis of VWF. Often, functionally important residues in proteins can be derived from the study of missense mutations that cause deficiencies and result in human disease. The vast majority of ADAMTS13 missense mutations that cause thrombotic thrombocytopenic purpura (TTP), however, are type 1 mutations that result in intracellular retention and are therefore characterized by low plasma levels of ADAMTS13 rather than by impaired enzyme function.43 We therefore used amino acid sequence alignments with other ADAMTS family members and structural homology modeling to identify potentially important enzyme subsites.
Amino acid sequence alignments revealed 3 MP domain regions that are poorly conserved among ADAMTS family members (Figure 1) and that might therefore form functional sites that impart specificity to the different ADAMTS enzymes. Molecular modeling of the ADAMTS13 MP domain based on the crystal structures of ADAMTS1, -4, and -534,35 revealed that the 3 nonconserved VR are predicted to flank the ADAMTS13 active site (Figure 2). These 3 VR in the MP domain are therefore potentially placed to interact with VWF in proximity to the cleavage site.
We assessed each VR by targeted mutagenesis and functional characterization. Substitution of the region E184-R193 (VR1) with the corresponding 9 amino acids in ADAMTS1 abolished proteolysis of VWF, and substitution with the sequence found in ADAMTS2 greatly reduced enzyme activity, demonstrating the importance of this region for ADAMTS13 function (Figure 4A). Of the 8 single-point mutants in this region, 3 (D187A, R190A, and R193A) exhibited reduced catalytic efficiencies (kcat/Km) for proteolysis of VWF115 (∼ 10-fold, ∼ 2-fold, and ∼ 4-fold reduced, respectively; Figure 4B-C; Table 1). We previously reported the involvement of Asp187 in a functionally important high-affinity Ca2+-binding site,16 which likely modulates the shape of this functional loop. Using full-length multimeric VWF as a substrate, we confirmed that the effects of these substitutions also apply to the physiologic target of ADAMTS13. The VR1 swap with ADAMTS2 sequence clearly reduced proteolysis of VWF multimers, but less so than its proteolysis of VWF115. It is difficult to explain this discrepancy between the 2 substrates, but it may suggest that the mild denaturing conditions used in the full-length multimeric VWF assay in some way alleviates the detrimental effects of the VR1 swap with ADAMTS2 sequence. It is important to note, however, that reduced proteolysis by the single-point mutants D187A, R190A, and R193A was observed with both VWF115 and full-length VWF. Interestingly, the R193W and T196I substitutions have been reported as causative hereditary TTP mutations.6,44 For the R193W mutation, both reduced secretion and reduced activity of ADAMTS13 were reported.44 Although the hydrophobic nature of tryptophan may cause misfolding of the MP domain in this TTP mutant, this substitution may also impair a functionally important subsite in the VR1 region close to the active site of ADAMTS13. We found that the point mutant R193A was secreted efficiently but that activity was reduced (Figure 4B,E), suggesting that Arg193 may contribute to efficient proteolysis by direct interactions with VWF. This contention seems plausible given the location of R193 immediately adjacent to the active site (Figure 2).
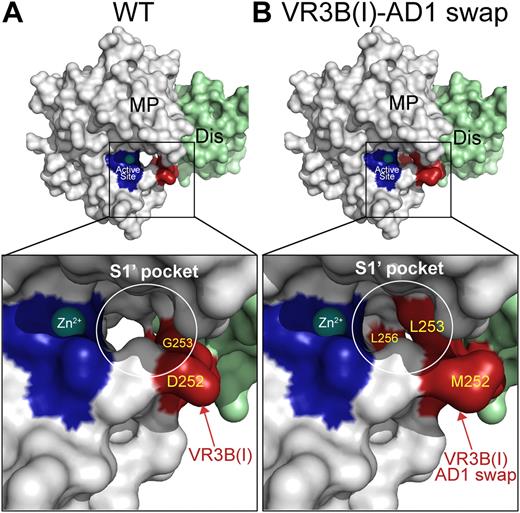
With regard to the VR3 (G236-A261) in the MP domain, our first intriguing finding was that the VR3B (D252-A261) variant proteolyzed VWF115 efficiently despite the substitution of 10 amino acids in close proximity to the active site (Figure 3B,D), as did 3 single-point mutants (D252N, R257A, and L260A) in this VR3B region (data not shown). From our model of the ADAMTS13 MP domain, part of the VR3B region is predicted to form a pocket adjacent to the active site (Figure 6A). This region is potentially favorably located for interacting with the P1′ residue (Met1606) of the VWF substrate. The protease pocket or subsite that interacts with the P1′ is commonly referred to as S1′. For another subclass of metalloproteases, the matrix metalloproteases, the S1′ pockets and their role in substrate and cleavage site specificity have been extensively studied in pursuit of specific inhibitors. The characterization of matrix metalloprotease S1′ pockets has involved X-ray crystallography in the presence/absence of inhibitors/ligands, complemented by functional studies. For the ADAMTS family members, crystal structures have only been resolved for the MP domains of ADAMTS1, -4, and -5.34,35 These crystal structures revealed that although their S1′ pockets are quite different in shape, they all appear to be flexible (ie, S1′ is only resolved upon inhibitor binding). Our structural homology model, together with our VR3B swap variants, therefore provided excellent tools to investigate the possible location and role of the ADAMTS13 S1′ pocket.
Molecular models of the ADAMTS13 S1′ subsite. (A) Model of the ADAMTS13 MP-Dis domains based on the crystal structures of the corresponding domains from ADAMTS1, -4, and -5. The active site histidines are shown in dark blue and the bound Zn2+ is shown as a sphere. Enlarged boxed area shows the active site in more detail. The S1′pocket that accommodates the P1′ residues in VWF (Met1606) is circled in white. The VR3B(I) region is shown in red with visible residues labeled. (B) Model as in panel A in the same orientation except that the VR3B(I) region (D252-P256) in ADAMTS13 MP domain has been substituted for the corresponding 5 residues in ADAMTS1. Changing these amino acids appreciably alters the shape/depth of the S1′ pocket.
Molecular models of the ADAMTS13 S1′ subsite. (A) Model of the ADAMTS13 MP-Dis domains based on the crystal structures of the corresponding domains from ADAMTS1, -4, and -5. The active site histidines are shown in dark blue and the bound Zn2+ is shown as a sphere. Enlarged boxed area shows the active site in more detail. The S1′pocket that accommodates the P1′ residues in VWF (Met1606) is circled in white. The VR3B(I) region is shown in red with visible residues labeled. (B) Model as in panel A in the same orientation except that the VR3B(I) region (D252-P256) in ADAMTS13 MP domain has been substituted for the corresponding 5 residues in ADAMTS1. Changing these amino acids appreciably alters the shape/depth of the S1′ pocket.
The VR3B-AD1 swap variant contains ADAMTS1 residues at a site that potentially forms the S1 pocket. We therefore examined whether the swap had altered and redirected P1′ specificity toward that of ADAMTS1. ADAMTS1 preferentially cleaves before Ala (but also before Phe, Gly, and Leu).40,45 Consequently, we examined the proteolysis of VWF115(M1606A) by ADAMTS13 and the different VR3B swap mutants. We first showed that wild-type ADAMTS13 proteolyzed VWF115(M1606A) inefficiently (15- to 18-fold reduced kcat/Km), consistent with the P1′ residue being an important determinant of substrate specificity (Table 2). Others have previously shown that VWF115(M1606L) is also inefficiently proteolyzed by ADAMTS13.32 When we analyzed the ability of the VR3B-AD1 swap variant to proteolyze VWF115(M1606A), we found that, in contrast to wild-type ADAMTS13, this ADAMTS13 variant cleaved VWF115(M1606A) as efficiently as it did wild-type VWF115, despite the substitution of the P1′ residue (Tables 2–3). The location of cleavage was further confirmed by MS/MS analysis. When we endeavored to more closely delineate those residues responsible for efficient cleavage of VWF115(M1606A), we found that a similar proteolytic efficiency was seen for the ADAMTS13 VR3B(I)-AD1 swap (D252-P256) but not for the ADAMTS13 VR3B(II)-AD1 (R257-A261) variant. When recombinant multimeric full-length VWF containing the M1606A P1′ mutation was used as a substrate, the VR3B swap variant again proteolyzed the substrate efficiently, whereas wild-type ADAMTS13 did not. These results strongly suggest that the VR3B region, and more specifically residues D252-P256 in VR3B(I), help form the S1′ pocket and influence P1′ specificity. From Figure 6, it appears that the shape of the S1 pocket is likely to change considerably in response to this substitution.
We also found that proteolysis of the VWF115 mutant (Y1605A/M1606A), in which both the P1 and P1′ residues were substituted for alanine, was even less efficient than VWF115(M1606A), highlighting the important contribution of the P1 residue (Tyr1605) in VWF in proteolysis (supplemental Figure 1F). The ADAMTS13 VR3B-AD1 swap variant proteolyzed the VWF115 mutant (Y1605A/M1606A) more slowly than it did VWF115(M1606A), but in both cases proteolysis was faster than in wild-type ADAMTS13. Interestingly, disruption of the S1′ was recently postulated to be a contributing factor in development of TTP in carriers of the ADAMTS13 S119F mutation. Although this residue is most likely not directly involved in the S1′ pocket, because it is predicted to face toward the inside of the MP domain, the S119F substitution may alter the folding of residues that contribute to the shape of the S1′ loop.46
Could S1′ specificity be important physiologically? Two recent studies have suggested that oxidation of Met1606 to methionine sulfoxide inhibits proteolysis by ADAMTS13.47,48 Reactive oxygen species such as HOCl capable of oxidizing Met1606 are released by neutrophils during inflammatory responses. This process may be responsible for the elevated plasma VWF and low ADAMTS13 activity that have been reported during certain inflammatory states, including sepsis and acute respiratory distress syndrome.48,49 This could be attributable to the loss of VWF proteolysis resulting from the high degree of specificity conferred by the S1′ pocket in the ADAMTS13 MP domain.
It is becoming increasingly clear that VWF proteolysis by ADAMTS13 is preceded by multiple binding interactions between the 2 molecules. An early interaction appears to be the binding of C-terminal ADAMTS13 domains to globular VWF.18,19 Proteolysis does not occur on binding because the scissile bond remains hidden in the core of the folded VWF monomer. Upon the unfolding of VWF, the ADAMTS13 spacer domain can bind,20,21,30,31 possibly in 2 steps,50 followed by Dis domain binding,22,23,28 which aids the positioning of the scissile bond into the active site cleft. In a final step before proteolysis occurs, MP domain subsites, including S1′, play an essential role by interacting with VWF residues adjacent to the cleavage site. Our data strongly suggest that the specificity of the ADAMTS13 S1′ subsite is determined by MP domain region D252-P256.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kevin Canis (Imperial College London) for help with the mass spectrometry and also Dr T. McKinnon (Imperial College London) for providing plasma purified VWF.
This work was supported by British Heart Foundation grants FS/06/002, PG/09/038, RG/06/007 and by an unrestricted grant from Amgen funding a studentship.
Authorship
Contribution: R.d.G. designed the research, performed experiments, analyzed results, and wrote the paper; D.A.L. designed the research, analyzed results, and wrote the paper; and J.T.B.C. designed the research, performed experiments, analyzed results, wrote the paper, and made the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rens de Groot, Department of Haematology, Imperial College London, 5th Fl, Commonwealth Bldg, Hammersmith Hospital Campus, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: r.degroot06@imperial.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal