Abstract
RapGEF2 is one of many guanine nucleotide exchange factors (GEFs) that specifically activate Rap1. Here, we generated RapGEF2 conditional knockout mice and studied its role in embryogenesis and fetal as well as adult hematopoietic stem cell (HSC) regulation. RapGEF2 deficiency led to embryonic lethality at ∼ E11.5 due to severe yolk sac vascular defects. However, a similar number of Flk1+ cells were present in RapGEF2+/+ and RapGEF2−/− yolk sacs indicating that the bipotential early progenitors were in fact generated in the absence of RapGEF2. Further analysis of yolk sacs and embryos revealed a significant reduction of CD41 expressing cells in RapGEF2−/− genotype, suggesting a defect in the maintenance of definitive hematopoiesis. RapGEF2−/− cells displayed defects in proliferation and migration, and the in vitro colony formation ability of hematopoietic progenitors was also impaired. At the molecular level, Rap1 activation was impaired in RapGEF2−/− cells that in turn lead to defective B-raf/ERK signaling. Scl/Gata transcription factor expression was significantly reduced, indicating that the defects observed in RapGEF2−/− cells could be mediated through Scl/Gata deregulation. Inducible deletion of RapGEF2 during late embryogenesis in RapGEF2cko/ckoERcre mice leads to defective fetal liver erythropoiesis. Conversely, inducible deletion in the adult bone marrow, or specific deletion in B cells, T cells, HSCs, and endothelial cells has no impact on hematopoiesis.
Introduction
G proteins function as molecular switches in signal transduction mechanisms, integrating a variety of stimuli to instruct diverse cellular outcomes. Rap1 (Ras-proximate-1), a Ras-like small GTP-binding protein, functions in signaling pathways that control diverse processes, such as cell adhesion, cell-cell junction formation, cell proliferation, and cell polarity.1-3 Rap1 is expressed as one of the 2 isoforms; Rap1a and Rap1b. Similar to other G proteins, Rap1 cycles between GDP-bound inactive and GTP-bound active forms. These 2 states represent a molecular switch controlled by 2 types of regulators: guanine nucleotide exchange factors (GEFs), the activators, and GTPase-activating proteins (GAPs), the inactivators.
Upon activation by GTP binding, Rap1 triggers several signaling pathways including integrin signaling, B-Raf/MEK/ERK activation, and other effector pathways.1,4,5 A variety of internal as well as external signals, such as cell surface receptors, growth factors, cAMP, and calcium, control the on/off status of Rap1 via the activation or inactivation of GEFs and GAPs.6 Multiple Rap1 GEFs (eg, C3G, EPAC1, EPAC2, PDZGEF2, CalDAG-GEF I, Dock4, AND-34) and GAPs (eg, Rap1GAP1, Rap1GAP2, SPA-1) enable Rap1 to respond to a diverse set of stimuli. Some of the Rap1 GEFs and GAPs are expressed ubiquitously, whereas others are expressed only in certain tissues/cell types or at certain stages of development, thereby permitting Rap1 signaling only in the development and/or differentiation of specific cell types.6
GEFs and GAPs are responsible for maintaining a fine balance in the active and inactive state of Rap1 and any shift in this balance could lead to Rap1 deregulation.7 Recent evidence indicates that deregulation of Rap1 could lead to hematologic malignancies.5 For example, deletion of one of the 2 isoforms of Rap1, Rap1a, resulted in defects in hematopoietic cell adhesion and T-cell polarization in 129Sv background.8 In addition, Rap1b deficiency in mice leads to a marked reduction in marginal zone B cells, severe B cell migration, and homing defects as well as bleeding defects due to defective platelet function, suggesting an essential function for Rap1 in hematopoietic development.9-11 Conversely, Rap1a/1b compound mutant mice die very early during embryogenesis.11 Another study showed that deletion of Rap1a in C57BL/6J background leads to early embryonic lethality at ∼ E7.0,12 indicating that Rap1 signaling is required as early as E7.0 during embryogenesis. Similar to Rap1, deletion of C3G (also known as RapGEF1), a ubiquitously expressed GEF, leads to early embryonic lethality at E7.5, indicating the requirement of C3G-mediated activation of Rap1 in the early stages of embryogenesis.13 In contrast, mice deficient for CalDAG-GEF I showed cell-specific abnormalities and displayed defects in neutrophils and blood platelets.14 On the other hand, in the absence of SPA-1, there is a constitutive activation of Rap1 with concomitant sustained activation of the B-Raf/MEK/ERK pathway that leads to abnormal proliferation of hematopoietic progenitors and the development of myeloid leukemias.15 Thus, deregulation of GEFs or GAPs could lead to hematologic malignancies due to altered Rap1 signaling. These studies demonstrated that the outcome of GEF/GAP deletion could be different from that of Rap1 deletion due to cell-specific expression of GEFs and GAPs and/or a possible functional redundancy within the GEFs or GAPs. Therefore, GEF/GAP knockout mouse models represent a valuable system to investigate the cell specific functions of Rap1.
Recently, RapGEF2 (PDZ-GEF) was identified as a GEF-specific for Rap1.16 We previously showed that deletion of RapGEF2 (Gef26) in Drosophila leads to loss of germ line stem cells from the niche.17 On the basis of RapGEF2's essential role in stem cell regulation in Drosophila and Rap1's important role in hematopoietic development in mice, we hypothesized that RapGEF2/Rap1 signaling might play an important role in hematopoietic stem cell (HSC) regulation in mice. Therefore, we generated RapGEF2 conditional knockout mice and investigated the consequence of loss of RapGEF2 in mouse embryonic development and in fetal as well as adult HSC regulation.
Methods
All the methods can be found in the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the National Cancer Institute at Frederick.
Results
RapGEF2 deletion leads to embryonic lethality due to intra- and extra-embryonic defects
To study the function of RapGEF2 in mice, we generated a RapGEF2 conditional knockout mouse line (RapGEF2cko/cko; Figure 1), which was crossed with β-actin-cre mice to generate heterozygous knockout mice (RapGEF2+/−). No live RapGEF2−/− offspring were obtained from crosses between RapGEF2+/− mice, indicating that they died during embryogenesis. To analyze embryos at different developmental stages, timed matings of RapGEF2+/− mice were initiated. All 3 genotypes were obtained in a Mendelian ratio up to E10.5. At E11.5, fewer RapGEF2−/− embryos were obtained, and the number of resorption sites containing necrotic remnants of RapGEF2−/− embryos increased. At E12.5, no RapGEF2−/− embryos were recovered, and the proportion of resorbed embryos increased to 22%, indicating that the RapGEF2−/− embryos died between E11.5-E12.5 (supplemental Table 1).
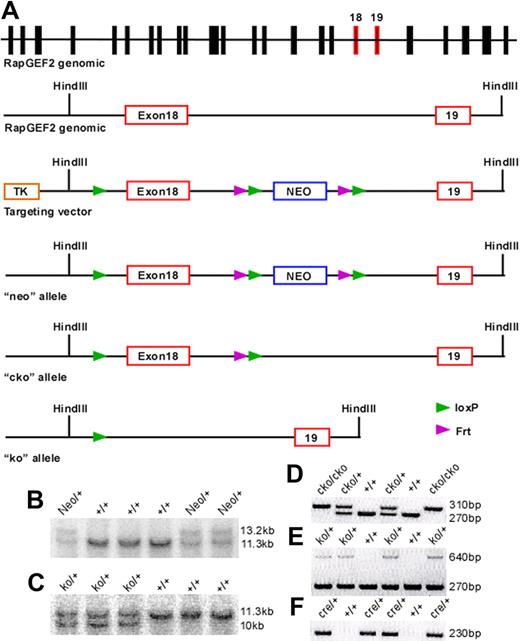
Targeted disruption of the RapGEF2 gene. Strategy for generating the RapGEF2 conditional targeting vector. (A) Schematic representation of the mouse RapGEF2 genomic sequence, targeting vector, neo, cko, and ko alleles. The targeted exon 18 and its adjacent exon 19 are in red. After electroporation of the RapGEF2 targeting vector, embryonic stem (ES) cells and then the neo mice were screened by Southern blotting (B). After crossing the neo mice sequentially with the flp and β-actincre mice, the RapGEF2+/− mice were screened by Southern blotting (C). PCR strategies were used to identify the RapGEF2+/+, RapGEF2cko/+, and RapGEF2cko/cko mice (D), RapGEF2+/+, RapGEF2+/−, and RapGEF2−/− embryos and mice (E), and Cre transgenic mice (F). The RapGEF2+/− mice were fertile and apparently normal, compared with the wild-type (RapGEF2+/+) mice.
Targeted disruption of the RapGEF2 gene. Strategy for generating the RapGEF2 conditional targeting vector. (A) Schematic representation of the mouse RapGEF2 genomic sequence, targeting vector, neo, cko, and ko alleles. The targeted exon 18 and its adjacent exon 19 are in red. After electroporation of the RapGEF2 targeting vector, embryonic stem (ES) cells and then the neo mice were screened by Southern blotting (B). After crossing the neo mice sequentially with the flp and β-actincre mice, the RapGEF2+/− mice were screened by Southern blotting (C). PCR strategies were used to identify the RapGEF2+/+, RapGEF2cko/+, and RapGEF2cko/cko mice (D), RapGEF2+/+, RapGEF2+/−, and RapGEF2−/− embryos and mice (E), and Cre transgenic mice (F). The RapGEF2+/− mice were fertile and apparently normal, compared with the wild-type (RapGEF2+/+) mice.
Analysis of the embryos revealed that until E7.5, there were no visible differences among the RapGEF2+/+, RapGEF2+/−, and RapGEF2−/− embryos. Histologic examination showed normal development of the 3 embryonic germ layers and amnion (not shown). By E9.5-E10.5, however, the RapGEF2−/− embryos were clearly different from the RapGEF2+/+ and RapGEF2+/− embryos; they were smaller and rather translucent, suggesting that development of the embryonic vasculature was impaired. E10.5 RapGEF2−/− embryos were ∼ 50%-60% smaller than the RapGEF2+/+ embryos (Figure 2A-B). In the RapGEF2−/− embryos, organogenesis, including formation of the heart, primitive gut, liver, and brain, was severely affected (supplemental Figure 1A-D).
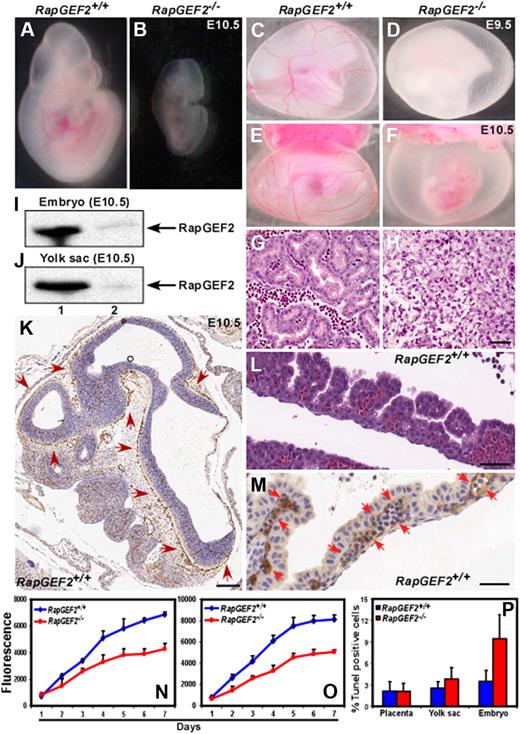
Defective yolk sac vascularization in RapGEF2−/− mice. (A-B) Morphologies of the E10.5 RapGEF2+/+ (A) and RapGEF2−/− (B) embryos. (C-D) Newly formed vitelline vessels of the E9.5 yolk sacs in RapGEF2+/+ (C) and RapGEF2−/− (D) embryos. (E-F) Vitelline vessels of the E10.5 yolk sacs in RapGEF2+/+ (E) and in RapGEF2−/− (F) embryos. The embryos were analyzed under Stemi SV11, Zeiss binocular microscope and photographed by Sony DSC-S85 digital camera. (G-H) H&E-stained transverse section of an E10.5 RapGEF2+/+ yolk sac showing blood vessels filled with red blood cells (G), and through a RapGEF2−/− yolk sac, which lacked blood vessels (H). Scale bar, 100 μm. (I-J) Northern blots showing the expression level of RapGEF2 in E10.5 RapGEF2+/+ (lane 1) and RapGEF2−/− (lane 2), embryos (I) and yolk sacs (J). (K) In situ hybridization with a RapGEF2-specific probe showing the expression pattern of RapGEF2 RNA in an E10.5 RapGEF2+/+ embryo sagittal section. Weak expression of RapGEF2 was detected throughout the embryo, but much stronger expression (indicated by red arrows) was observed in the major blood vessels. Scale bar, 500 μm. (L) H&E-stained sagittal section of an E10.5 RapGEF2+/+ yolk sac showing the yolk sac blood vessels filled with erythroblasts. Scale bar, 50 μm. (M) In situ hybridization with a RapGEF2-specific probe showing the expression pattern of RapGEF2 RNA in an E10.5 RapGEF2+/+ yolk sac. RapGEF2 was predominantly detected in the blood vessels (indicated by red arrows) of the yolk sac. Scale bar, 50 μm. (N-O) Line graph displaying the proliferation rate of RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac cells (N) and embryonic cells (O) measured by AlamarBlue proliferation assay. (P) Histogram showing the percentage of Tunel-positive apoptotic cells in E10.5 RapGEF2+/+ and RapGEF2−/− placenta, yolk sac, and embryos.
Defective yolk sac vascularization in RapGEF2−/− mice. (A-B) Morphologies of the E10.5 RapGEF2+/+ (A) and RapGEF2−/− (B) embryos. (C-D) Newly formed vitelline vessels of the E9.5 yolk sacs in RapGEF2+/+ (C) and RapGEF2−/− (D) embryos. (E-F) Vitelline vessels of the E10.5 yolk sacs in RapGEF2+/+ (E) and in RapGEF2−/− (F) embryos. The embryos were analyzed under Stemi SV11, Zeiss binocular microscope and photographed by Sony DSC-S85 digital camera. (G-H) H&E-stained transverse section of an E10.5 RapGEF2+/+ yolk sac showing blood vessels filled with red blood cells (G), and through a RapGEF2−/− yolk sac, which lacked blood vessels (H). Scale bar, 100 μm. (I-J) Northern blots showing the expression level of RapGEF2 in E10.5 RapGEF2+/+ (lane 1) and RapGEF2−/− (lane 2), embryos (I) and yolk sacs (J). (K) In situ hybridization with a RapGEF2-specific probe showing the expression pattern of RapGEF2 RNA in an E10.5 RapGEF2+/+ embryo sagittal section. Weak expression of RapGEF2 was detected throughout the embryo, but much stronger expression (indicated by red arrows) was observed in the major blood vessels. Scale bar, 500 μm. (L) H&E-stained sagittal section of an E10.5 RapGEF2+/+ yolk sac showing the yolk sac blood vessels filled with erythroblasts. Scale bar, 50 μm. (M) In situ hybridization with a RapGEF2-specific probe showing the expression pattern of RapGEF2 RNA in an E10.5 RapGEF2+/+ yolk sac. RapGEF2 was predominantly detected in the blood vessels (indicated by red arrows) of the yolk sac. Scale bar, 50 μm. (N-O) Line graph displaying the proliferation rate of RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac cells (N) and embryonic cells (O) measured by AlamarBlue proliferation assay. (P) Histogram showing the percentage of Tunel-positive apoptotic cells in E10.5 RapGEF2+/+ and RapGEF2−/− placenta, yolk sac, and embryos.
Whole-mount immunohistochemical staining using the endothelial marker platelet endothelial cell adhesion molecule-1 (PECAM1) revealed that the RapGEF2−/− embryos lacked major blood vessels, suggesting a severe defect in embryonic vascularization. This was in sharp contrast with the well-developed normal vascularization of the RapGEF2+/+ embryos (supplemental Figure 1E-F). Analysis of the extra-embryonic tissues of E9.5 and E10.5 embryos showed that the RapGEF2−/− yolk sacs lacked the distinctive networks of branching vitelline vessels (Figure 2D,F), which were clearly visible in the RapGEF2+/+ yolk sacs (Figure 2C,E), and appeared anemic. Hematoxylin and eosin (H&E)-stained transverse sections of yolk sacs revealed large vitelline collecting vessels and well-differentiated branching capillary vessels filled with erythroid cells in the RapGEF2+/+ yolk sacs, but these structures were almost undetectable in the RapGEF2−/− yolk sacs (Figure 2G-H). These findings indicated that RapGEF2 is essential for blood vessel formation during embryogenesis.
We next examined the expression level and pattern of RapGEF2 by Northern blotting and in situ hybridization of E10.5 yolk sacs and embryos. Northern blots revealed abundant RapGEF2 expression in the E10.5 RapGEF2+/+ embryos and yolk sacs, but none in the RapGEF2−/− embryos, as expected (Figure 2I-J). In situ hybridization showed that RapGEF2 was widely expressed in the RapGEF2+/+ embryos, with strong expression at the vascular endothelial cells of major vessels (Figure 2K). In the yolk sac, its expression was largely restricted to the vitelline vessels (Figure 2L-M) supporting the evidence that RapGEF2 is required for yolk sac vascularization.
RapGEF2 deficiency leads to impaired cell proliferation and migration
Rap1 signaling affects important cellular functions including cell proliferation, adhesion, and migration.1,2,4,5 Because RapGEF2 is an upstream activator of Rap1, loss of RapGEF2 could potentially alter Rap1 signaling. Therefore, we determined the impact of RapGEF2 deletion on cell proliferation, adhesion, migration, and apoptosis. AlamarBlue cell proliferation assays revealed that the RapGEF2−/− yolk sac and embryonic cells proliferated at a much slower rate. On an average, a ∼ 40%-50% reduction in the overall proliferation rate was observed compared with RapGEF2+/+ cells (Figure 2N-O). Apoptosis, measured by Tunel staining on yolk sac sections, showed no difference in the percentage of apoptotic cells between RapGEF2+/+ and RapGEF2−/− yolk sacs. Conversely, a higher percentage (∼ 10%) of Tunel-positive cells were detected in RapGEF2−/− embryos compared with RapGEF2+/+ (∼ 3%) embryos (Figure 2P).
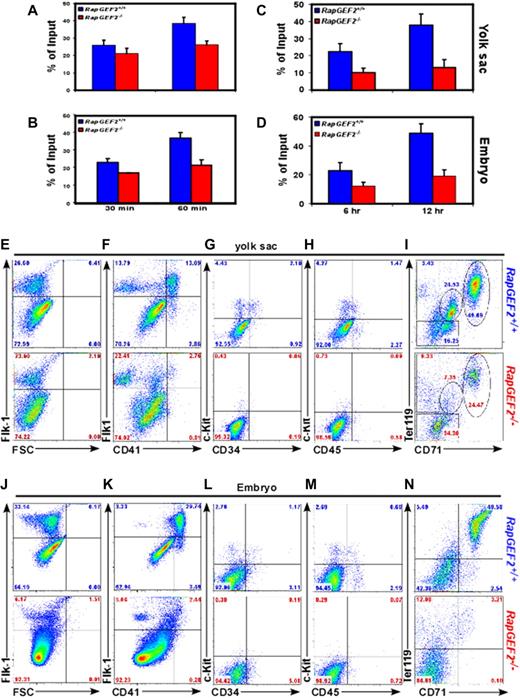
To determine whether loss of RapGEF2 could also lead to adhesion and migration defects, RapGEF2−/− cells were subjected to in vitro adhesion and migration assays. Adhesion assays revealed that the adhesion levels of RapGEF2−/− yolk sac and embryonic cells was reduced by ∼ 20% and ∼ 30% after 30 and 60 minutes of seeding in fibronectin coated plates (Figure 3A-B), indicating that the adhesion property of the cells is slightly affected in the absence of RapGEF2. However, cell migration assays disclosed a more profound defect in the migratory properties of RapGEF2−/− cells. The migration of RapGEF2−/− yolk sac and embryonic cells was ∼ 50% decreased compared with RapGEF2+/+ cells (Figure 3C-D). These results suggest that RapGEF2 could play an essential role in the proliferation and migration of cells during embryogenesis.
CD41+ population is reduced in response to RapGEF2 deletion. (A-B) Bar graph showing the percentage of adherent RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac (A) and embryonic cells (B) after 30 and 60 minutes of seeding in the fibronectin (FN)-coated plate. (C-D) Histogram displaying the in vitro migration levels of RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac (C) and embryonic cells (D) after 6 and 12 hours. (E-I) Representative FACS images showing the distribution of Flk1+ (E), Flk1+CD41+ (F), c-Kit+CD34+ (G), c-Kit+CD45+ (H), and CD71+Ter119+ (I) fractions in the E10.5 RapGEF2+/+ and RapGEF2−/− yolk sacs. (J-N) Representative FACS images showing the distribution of Flk1+ (J), Flk1+CD41+ (K), c-Kit+CD34+ (L), c-Kit+CD45+ (M), and CD71+Ter119+ (N) in the E10.5 RapGEF2+/+ and RapGEF2−/− embryos.
CD41+ population is reduced in response to RapGEF2 deletion. (A-B) Bar graph showing the percentage of adherent RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac (A) and embryonic cells (B) after 30 and 60 minutes of seeding in the fibronectin (FN)-coated plate. (C-D) Histogram displaying the in vitro migration levels of RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac (C) and embryonic cells (D) after 6 and 12 hours. (E-I) Representative FACS images showing the distribution of Flk1+ (E), Flk1+CD41+ (F), c-Kit+CD34+ (G), c-Kit+CD45+ (H), and CD71+Ter119+ (I) fractions in the E10.5 RapGEF2+/+ and RapGEF2−/− yolk sacs. (J-N) Representative FACS images showing the distribution of Flk1+ (J), Flk1+CD41+ (K), c-Kit+CD34+ (L), c-Kit+CD45+ (M), and CD71+Ter119+ (N) in the E10.5 RapGEF2+/+ and RapGEF2−/− embryos.
RapGEF2 is required for embryonic hematopoiesis
Embryonic lethality of RapGEF2−/− mice, due to yolk sac vascular defects and lack of erythroid cells, could be a sign of severe impairment of embryonic hematopoiesis, since yolk sac is one of the primary sites of hematopoiesis during early embryogenesis.18 Moreover, an efficient and controlled proliferation, differentiation, and migration of cells are required to establish a functional vascular and hematopoietic system during embryogenesis, and RapGEF2−/− cells displayed defects in both proliferation and migration. To determine whether endothelial/hematopoietic development was affected in the RapGEF2-deficient mice, E10.5 yolk sacs were analyzed for molecular markers expressed on endothelial as well as hematopoietic progenitors. Similar levels of Flk1+ cells were observed in the RapGEF2+/+ and RapGEF2−/− E10.5 yolk sacs, indicating that the early progenitors that give rise to endothelial and hematopoietic lineages were in fact generated in RapGEF2−/− yolk sacs19,20 (Figure 3E). However, a dual staining revealed that the cell populations expressing Flk1+ and CD41+ (markers of the early hematopoietic lineage) were significantly reduced in the RapGEF2−/− yolk sacs compared with the RapGEF2+/+ yolk sacs (Figure 3F). Previous studies demonstrated that the expression of CD41 marks the initiation of definitive hematopoiesis during mouse embryogenesis.21,22 Therefore, reduced levels of CD41 expressing cells in the RapGEF2-deficient yolk sacs suggest that the maintenance of definitive hematopoiesis was impaired. Further analyses using hematopoietic progenitor cell markers, c-Kit/CD34 and c-Kit/CD45, revealed a significant reduction in the percentage of the c-Kit+, c-Kit+CD34+, and c-Kit+CD45+ cell populations in RapGEF2−/− yolk sacs compared with RapGEF2+/+ yolk sacs (Figure 3G-H). Soon after the initiation of blood island vasculogenesis, a subset of CD41+ cells begin to express Ter119 leading to the development of erythroid lineage.22 To determine whether a reduction in the CD41 expressing cells in the RapGEF2−/− yolk sacs in turn affected erythropoiesis, we stained for CD71/Ter119, markers of erythroid progenitors, and found a significant reduction in the Ter119+ progenitors in the RapGEF2−/− yolk sacs compared with RapGEF2+/+ yolk sacs (Figure 3I). These data together suggest that the deletion of RapGEF2 leads to a defect in the development of the hematopoietic lineage.
We tried to analyze the effect of RapGEF2 loss on hematopoietic development in the aorta-gonad-mesonephros (AGM) region of the embryos, but were unable to dissect this region from the RapGEF2−/− embryos due to their severe underdevelopment. Therefore, we prepared single-cell suspensions from E10.5 embryos and stained and analyzed their expression of hematopoietic markers. The percentage of Flk1+ progenitors (∼ 6%) was significantly lower in the RapGEF2−/− embryos than in the RapGEF2+/+ embryos (∼ 33%), although there were similar number of Flk1+ cells in the RapGEF2−/− yolk sacs (Figure 3J). This indicates that the defect in the earlier expansion of the progenitors could be much more profound in the intra-embryonic sites compared with yolk sac. Furthermore, a higher percentage of the Flk1+ cells were also positive for CD41+ (∼ 30%), and only a small fraction had the Flk1+CD41− molecular phenotype (∼ 3.5%) in the RapGEF2+/+ embryos, but almost all of the Flk1+ cells were Flk1+CD41− in the RapGEF2−/− embryos (Figure 3K). The c-Kit+CD34+ and c-Kit+CD45+ hematopoietic progenitor cells were also dramatically reduced in the RapGEF2−/− embryos compared with the RapGEF2+/+ embryos (Figure 3L-M). Similarly, as in yolk sacs, there were significantly fewer CD71+Ter119+ (Figure 3N) and Gr1+Mac1+ (not shown) progenitors in RapGEF2−/− embryos compared with RapGEF2+/+ embryos. These data suggest that similar to yolk sac hematopoiesis, intra-embryonic hematopoietic development was also defective in the absence of RapGEF2 shown by a significant reduction in the CD41 expressing cells.
In vitro differentiation of RapGEF2−/− hematopoietic progenitors is impaired
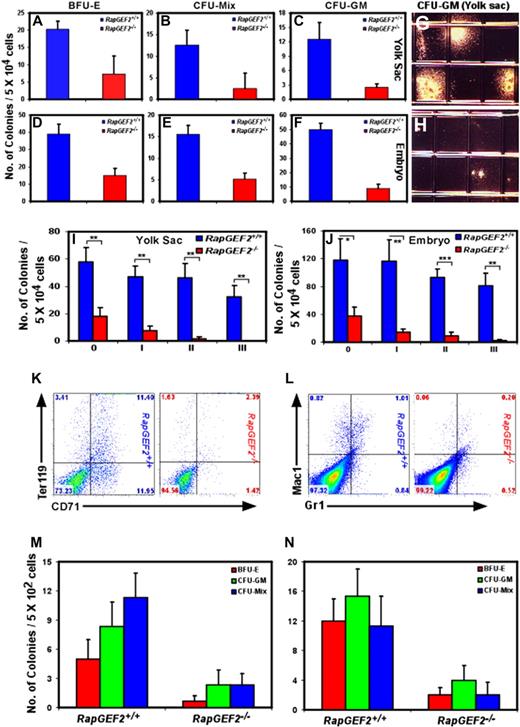
To confirm that RapGEF2 affects hematopoietic development, we determined if hematopoietic progenitor cells (HPCs) were present in RapGEF2−/− yolk sacs, placentas, and embryos by evaluating the growth differentiation potential in vitro. We tested cells from the RapGEF2−/− and RapGEF2+/+ E10.5 yolk sac, placenta, and embryo for their ability to form in vitro erythroid (erythroid burst-forming units; BFU-E), mixed (mixed colony-forming units; CFU-Mix), and granulocyte/macrophage (granulocyte-macrophage colony-forming units; CFU-GM) colonies. RapGEF2−/− yolk sac cells formed many fewer BFU-E, CFU-Mix, and CFU-GM colonies (4- to 5-fold less) than RapGEF2+/+ yolk sac cells (Figure 4A-C). Cells harvested from the RapGEF2−/− embryos also displayed a similar limited potential to form colonies, also with a 4- to 5-fold reduction in number (Figure 4D-F). The RapGEF2−/− colonies were also significantly smaller than the RapGEF2+/+ colonies (Figure 4G-H). These assays revealed that the RapGEF2−/− cells could form hematopoietic colonies, but their number and size were greatly reduced compared with those formed by the RapGEF2+/+ cells. Next, we evaluated the proliferation and self-renewal capacity of the RapGEF2−/− HPCs by colony replating assays. The number of hematopoietic colonies was reduced (∼ 4-fold less) after initial plating of RapGEF2−/− yolk sac and embryonic cells compared with RapGEF2+/+ cells. By the third replating, RapGEF2−/− cells failed to form any colonies, whereas, RapGEF2+/+ cells continue to produce higher number of colonies (Figure 4I-J). Thus, the self-renewal capacity of the RapGEF2−/− HPCs is severely impaired.
Impaired self-renewal and differentiation of RapGEF2−/− hematopoietic progenitors in vitro. The number of BFU-E, CFU-Mix, and CFU-GM colonies formed from RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac cells (A-C) or embryonic cells (D-F). The average and standard deviation were derived from 3 independent colony-formation assays. (G-H) Representative photographs showing the size of the CFU-GM colonies derived from RapGEF2+/+ (G) and RapGEF2−/− (H) E10.5 yolk sac cells. (I-J) Histograms showing the number of colonies formed in the initial plating 0′ and the subsequent replating i to iii of E10.5 yolk sac (I) or embryonic (J) cells. The average, standard deviation, and statistical significance were derived from 3 independent assays. *P < .05, **P < .005, ***P < .0005. (K-L) Flow cytometry images showing the distribution of CD71+Ter119+ (K) and Gr1+/Mac1+ (L) cell populations. (M-N) Bar graphs displaying the number of BFU-E, CFU-GM, and CFU-Mix colonies formed after plating FACS-sorted c-Kit+/CD34+ double-positive HPCs from E10.5 yolk sac (M) or embryonic cells (N). Compared with RapGEF2+/+, 5 times more number of RapGEF2−/− yolk sacs and embryos were pooled to obtain sufficient number of c-Kit+/CD34+ HPCs.
Impaired self-renewal and differentiation of RapGEF2−/− hematopoietic progenitors in vitro. The number of BFU-E, CFU-Mix, and CFU-GM colonies formed from RapGEF2+/+ and RapGEF2−/− E10.5 yolk sac cells (A-C) or embryonic cells (D-F). The average and standard deviation were derived from 3 independent colony-formation assays. (G-H) Representative photographs showing the size of the CFU-GM colonies derived from RapGEF2+/+ (G) and RapGEF2−/− (H) E10.5 yolk sac cells. (I-J) Histograms showing the number of colonies formed in the initial plating 0′ and the subsequent replating i to iii of E10.5 yolk sac (I) or embryonic (J) cells. The average, standard deviation, and statistical significance were derived from 3 independent assays. *P < .05, **P < .005, ***P < .0005. (K-L) Flow cytometry images showing the distribution of CD71+Ter119+ (K) and Gr1+/Mac1+ (L) cell populations. (M-N) Bar graphs displaying the number of BFU-E, CFU-GM, and CFU-Mix colonies formed after plating FACS-sorted c-Kit+/CD34+ double-positive HPCs from E10.5 yolk sac (M) or embryonic cells (N). Compared with RapGEF2+/+, 5 times more number of RapGEF2−/− yolk sacs and embryos were pooled to obtain sufficient number of c-Kit+/CD34+ HPCs.
We also assayed the hematopoietic potential of E10.5 RapGEF2+/+ and RapGEF2−/− yolk sac cells grown on an OP9 stroma, which supports myeloerythroid differentiation. We assayed the myeloerythroid differentiation of the progenitors obtained from this culture on methylcellulose. The RapGEF2−/− cells had a limited ability to generate BFU-E, CFU-GM, and CFU-Mix colonies compared with RapGEF2+/+ cells, and the number and size of the colonies were greatly reduced (not shown). To examine the different lineages derived from the yolk sac progenitors, colonies were extracted from the culture, stained with myeloerythroid cell-surface markers, and analyzed by flow cytometry. The c-Kit+, Sca1+, and c-Kit+Sca1+ fractions (hematopoietic progenitors) contained fewer cells in the RapGEF2−/− colonies compared with the RapGEF2+/+ colonies (not shown). Likewise, the number of CD71+Ter119+ progenitors (Figure 4K) and Gr1+Mac1+ cells (granulocyte/macrophage; Figure 4L) in the RapGEF2−/− colonies was small compared with those in the RapGEF2+/+ colonies. Furthermore, obvious CD41+ and CD45+ populations were observed in the RapGEF2+/+ colonies, but almost none of these cells were found in the RapGEF2−/− colonies (not shown). These in vitro assays indicated that RapGEF2 is essential for the expansion and differentiation of hematopoietic progenitors into hematopoietic lineages.
However, our fluorescence-activated cell sorting (FACS) analysis (Figure 3F-H,K-M) shows that the HPC number itself is reduced in RapGEF2−/− yolk sacs and embryos. Therefore, reduced number of colonies observed in the above in vitro assays is due to the presence of lower number of HPCs in the RapGEF2−/− yolk sacs and embryos. To directly determine whether the RapGEF2−/− HPCs have impaired in vitro differentiation and a lower CFU number is not due to reduced number of HPCs, we FACS-sorted c-Kit+/CD34+ double-positive early hematopoietic progenitors from RapGEF2+/+ and RapGEF2−/− yolk sacs and embryos. An equal number of c-Kit+/CD34+ (5 × 102 cells/dish) HPCs were plated into methylcellulose to evaluate their colony-forming potential. A significantly reduced number (∼5-fold less) of BFU-E, CFU-GM, and CFU-Mix colonies were detected in RapGEF2−/− yolk sac or embryonic HPC-seeded dishes (Figure 4M-N). These results together suggest that the number of HPCs is significantly reduced in RapGEF2−/− yolk sacs and embryos and that majority of the RapGEF2−/− HPCs were dysfunctional and failed to proliferate and differentiate.
Inducible RapGEF2 deletion leads to defective fetal liver erythropoiesis
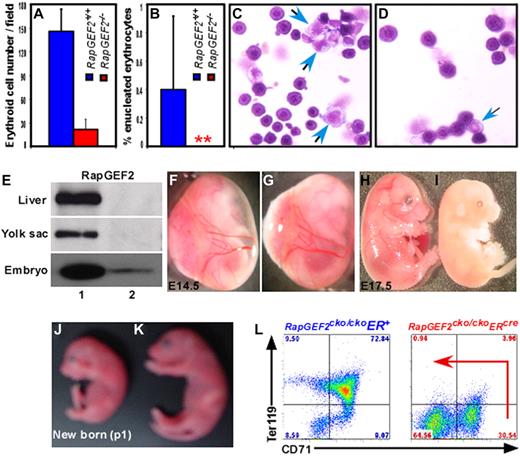
To further investigate the impact of RapGEF2 deletion on erythroid development, embryonic peripheral blood was analyzed. During the course of embryogenesis, the nucleated erythroid cells mature into enucleated erythrocytes.23 Due to embryonic lethality of RapGEF2−/− genotype at ∼ E11.5, we were able to analyze peripheral blood only up to E10.5. The E10.5 embryonic blood contains predominantly nucleated erythroid cells in RapGEF2+/+ and RapGEF2−/− embryos (Figure 5A-D). Nevertheless, the number of nucleated erythroid cells was significantly reduced in RapGEF2−/− embryos (Figure 5A,C-D). No enucleated erythro-cytes were detected in the RapGEF2−/− embryonic blood. However, a relatively very small percentage (∼ 0.5%) of enucleated erythrocytes was detected in RapGEF2+/+ peripheral blood (Figure 5B). This observation is in line with previous reports that enucleation and maturation of erythroblasts into erythrocytes occur mainly at ∼ E12.5-E13.5 of embryogenesis.23
Defective fetal liver hematopoiesis in RapGEF2−/− mice. (A-B) Histogram showing the average number of nucleated (A) and enucleated (B) erythroid cells in the peripheral blood of E10.5 RapGEF2+/+ and RapGEF2−/− embryos. (C) Representative photographs displaying the cytospined and stained peripheral blood of E10.5 RapGEF2+/+ (C) and RapGEF2−/− (D) embryos. Blue arrows indicate myeloid progenitors whereas remaining cells are nucleated erythroid cells. (E) Northern blot showing the expression level of RapGEF2 in the E16.5 liver, yolk sac, and embryo of RapGEF2cko/ckoER+ (lane 1) and RapGEF2cko/ckoERcre (lane 2) mice. Females were injected with tamoxifen on day 11.5 and 13.5 of pregnancy. (F-G) Photographs showing the yolk sac blood vessels of E14.5 RapGEF2cko/ckoER+ (F) and RapGEF2cko/ckoERcre (G) embryos. (H-I) Photographs of E17.5 RapGEF2cko/ckoER+ (H) and RapGEF2cko/ckoERcre (I) embryos. The embryos were analyzed under Stemi SV11, Zeiss binocular microscope and photographed by Sony DSC-S85 digital camera. The E17.5 RapGEF2cko/ckoERcre embryos were pale, soft, fragile, and devoid of blood vessels. Timed matings were set up between the RapGEF2cko/+ERcre heterozygotes, and the pregnant females were given tamoxifen by injection to induce RapGEF2 deletion. First dose of tamoxifen was injected at E11.5, when yolk sac vascularization is mostly complete. Second dose was give at E13.5. If the induced deletion of RapGEF2 at this stage leads to embryonic lethality, we could conclude that the lethality in RapGEF2cko/ckoERcre mice was not due to yolk sac vascular defects. (J-K) Day-1 RapGEF2cko/ckoERcre (J) and RapGEF2cko/ckoER+ (K) neonates. Pregnant females were given tamoxifen by injection on days 17.5 and 19.5 of pregnancy. (L) Representative FACS images showing the distribution of the CD71+Ter119+ population in the E16.5 fetal liver cells of RapGEF2cko/ckoER+ and RapGEF2cko/ckoERcre embryos.
Defective fetal liver hematopoiesis in RapGEF2−/− mice. (A-B) Histogram showing the average number of nucleated (A) and enucleated (B) erythroid cells in the peripheral blood of E10.5 RapGEF2+/+ and RapGEF2−/− embryos. (C) Representative photographs displaying the cytospined and stained peripheral blood of E10.5 RapGEF2+/+ (C) and RapGEF2−/− (D) embryos. Blue arrows indicate myeloid progenitors whereas remaining cells are nucleated erythroid cells. (E) Northern blot showing the expression level of RapGEF2 in the E16.5 liver, yolk sac, and embryo of RapGEF2cko/ckoER+ (lane 1) and RapGEF2cko/ckoERcre (lane 2) mice. Females were injected with tamoxifen on day 11.5 and 13.5 of pregnancy. (F-G) Photographs showing the yolk sac blood vessels of E14.5 RapGEF2cko/ckoER+ (F) and RapGEF2cko/ckoERcre (G) embryos. (H-I) Photographs of E17.5 RapGEF2cko/ckoER+ (H) and RapGEF2cko/ckoERcre (I) embryos. The embryos were analyzed under Stemi SV11, Zeiss binocular microscope and photographed by Sony DSC-S85 digital camera. The E17.5 RapGEF2cko/ckoERcre embryos were pale, soft, fragile, and devoid of blood vessels. Timed matings were set up between the RapGEF2cko/+ERcre heterozygotes, and the pregnant females were given tamoxifen by injection to induce RapGEF2 deletion. First dose of tamoxifen was injected at E11.5, when yolk sac vascularization is mostly complete. Second dose was give at E13.5. If the induced deletion of RapGEF2 at this stage leads to embryonic lethality, we could conclude that the lethality in RapGEF2cko/ckoERcre mice was not due to yolk sac vascular defects. (J-K) Day-1 RapGEF2cko/ckoERcre (J) and RapGEF2cko/ckoER+ (K) neonates. Pregnant females were given tamoxifen by injection on days 17.5 and 19.5 of pregnancy. (L) Representative FACS images showing the distribution of the CD71+Ter119+ population in the E16.5 fetal liver cells of RapGEF2cko/ckoER+ and RapGEF2cko/ckoERcre embryos.
To overcome embryonic lethality and to determine the role RapGEF2 in mid- to late embryogenesis or in the later stages of hematopoiesis, we crossed RapGEF2cko/cko mice with tamoxifen-inducible β-actin-ERcre mice to produce RapGEF2cko/ckoERcre mice, which can be used to delete RapGEF2 at any stage during development. We induced RapGEF2 deletion by giving injections of tamoxifen at E11.5 and E13.5 and analyzed embryos at E14.5-E20.5 as well as neonates. Northern blot analysis of the RNA extracted from the E16.5 yolk sacs, liver, and embryos showed the efficient deletion of RapGEF2 in the RapGEF2cko/ckoERcre embryos (Figure 5E). No embryonic lethality was observed from E14.5-E16.5, and the vitelline vessels of the yolk sacs were not disrupted in response to RapGEF2 deletion (Figure 5F-G). However, the majority (∼ 73%) of the RapGEF2cko/ckoERcre mice died between E17.5 and E18.5. The remaining RapGEF2cko/ckoERcre mice (∼ 27%) died between E19.5-E20.5, and no neonates were obtained. The E17.5 RapGEF2cko/ckoERcre embryos were pale, fragile, and appeared to lack all the major embryonic blood vessels (Figure 5H-I). However, ∼ 15% of the RapGEF2cko/ckoER+ control mice also died between E18.5 to E20.5, suggesting tamoxifen toxicity could have made a minor contribution to the RapGEF2cko/ckoERcre embryonic death. In contrast, when pregnant RapGEF2cko/+ERcre heter-ozygous mice were given tamoxifen at E17.5 and E19.5, the RapGEF2cko/ckoERcre mice were born and survived like the RapGEF2cko/ckoER+ control mice, although the RapGEF2cko/ckoERcre neonates were slightly smaller than their RapGEF2cko/ckoER+ littermates (Figure 5J-K). These results suggest that RapGEF2 is not only essential at the early stages of embryonic hematopoiesis, but may also contribute to the differentiation of other cell types and the development of other tissues.
To investigate whether the RapGEF2 deletion had any impact on later stages of hematopoietic development, such as fetal liver hematopoiesis, we analyzed the E16.5 livers of RapGEF2cko/ckoERcre and control RapGEF2cko/ckoER+ embryos. For these experiments, RapGEF2 was deleted by tamoxifen injections at 11.5 and 13.5 days of pregnancy. The RapGEF2cko/ckoERcre E16.5 livers were pale. Flow cytometry analysis of cells harvested from them revealed a significant reduction of the c-Kit+Sca1+ cell population, in clear contrast to this cell population in RapGEF2cko/ckoER+ livers. The RapGEF2cko/ckoERcre fetal livers also had significantly fewer Gr1+Mac1+ cells (not shown). Most strikingly, CD71/Ter119 double staining revealed only a CD71+ population in the RapGEF2cko/ckoERcre livers; the CD71+Ter119+ double-positive population was completely missing. In contrast, the livers of RapGEF2cko/ckoER+ mice showed a high percentage (∼72%) of cells in the CD71+Ter119+ fraction. Thus, the deletion of RapGEF2 inhibited the differentiation of erythroid progenitors into mature Ter119+ erythrocytes (Figure 5L).
Transcriptional deregulation of SCL/Gata in response to RapGEF2 deletion
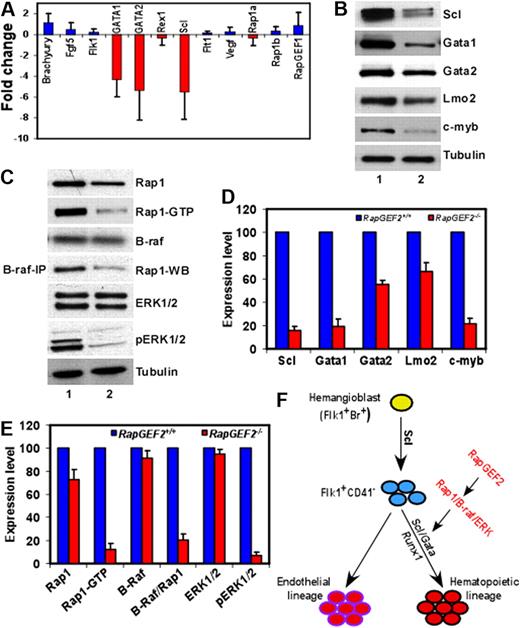
Our results show that the deletion of RapGEF2 leads to severe defects in the early stages of embryonic hematopoiesis. To investigate how the RapGEF2 deletion contributed to the observed phenotypes, we performed quantitative polymerase chain reaction (Q-PCR) on RNAs isolated from the E10.5 yolk sac cells of RapGEF2+/+ and RapGEF2−/− embryos and examined changes in the gene expression of Brachyury, Fgf5, Flk1, Gata1, Gata2, Rex1, Scl, Flt1, Vegf, which were previously shown to be essential for hematopoietic development.24-28 The Q-PCR assays revealed a significant down-regulation (5- to 6-fold) of the transcription factors, Scl, Gata-1, and Gata-2 in the RapGEF2−/− yolk sac cells compared with those of RapGEF2+/+ (Figure 6A). In contrast, the expression level of Brachyury, essential for the differentiation of primitive mesoderm, was not altered between RapGEF2+/+ and RapGEF2−/− cells (Figure 6A). Similarly, the levels of primitive hematopoietic and endothelial markers such as Flk1, Flt1, Fgf5, and Rex1 were not significantly different between the RapGEF2+/+ and RapGEF2−/− yolk sac cells. Moreover, the RNA level of Vegf, which is essential for both embryonic hematopoiesis and angiogenesis, was unchanged between the RapGEF2+/+ and RapGEF2−/− cells (Figure 6A). Thus, the expression of Scl and Gata genes are significantly reduced in the absence of RapGEF2.
Deregulation of SCL/Gata transcription factors in RapGEF2−/− cells. (A) Differences in the expression level of hematopoietic marker, Brachyury, Fgf5, Flk1, Gata1, Gata2, Rex1, Scl, Flt1, and Vegf, and of Rap1a, Rap1b, and RapGEF1 in RapGEF2−/− E10.5 yolk sac cells compared with RapGEF2+/+ yolk sac cells, by Q-PCR. (B) Western blots showing the expression of the transcription factors SCL, Gata1, Gata2, and Lmo2, and the proto-oncogene c-myb in RapGEF2+/+ (lane 1) and RapGEF2−/− (lane 2) E10.5 yolk sac cells. Tubulin was used as a loading control. (C) Representative Western blots showing the level of Rap1, Rap1GTP, and B-raf, and the results of a B-raf/Rap1 pull-down assay showing the amount of B-raf-bound Rap1 (fourth panel from top), and the expression level of ERK1/2 and phosphorylated pERK1/2 in RapGEF2+/+ (lane 1) and RapGEF2−/− (lane 2) E10.5 yolk sac cells. Tubulin was used as a loading control. (D-E) The expression level of proteins (intensity of protein bands) was quantified by Imagequant software and presented as percent expression in RapGEF2−/− cells compared with RapGEF2+/+ cells, where the expression level was set as 100%. Then, the statistical analysis was shown as histograms. (F) Drawing displaying the possible role of RapGEF2 in the development of hematopoietic lineage. In this model, SCL/Gata transcription factors are required for the development of hematopoietic lineage, and RapGEF2 might regulate SCL through the Rap1/B-raf/ERK pathway.
Deregulation of SCL/Gata transcription factors in RapGEF2−/− cells. (A) Differences in the expression level of hematopoietic marker, Brachyury, Fgf5, Flk1, Gata1, Gata2, Rex1, Scl, Flt1, and Vegf, and of Rap1a, Rap1b, and RapGEF1 in RapGEF2−/− E10.5 yolk sac cells compared with RapGEF2+/+ yolk sac cells, by Q-PCR. (B) Western blots showing the expression of the transcription factors SCL, Gata1, Gata2, and Lmo2, and the proto-oncogene c-myb in RapGEF2+/+ (lane 1) and RapGEF2−/− (lane 2) E10.5 yolk sac cells. Tubulin was used as a loading control. (C) Representative Western blots showing the level of Rap1, Rap1GTP, and B-raf, and the results of a B-raf/Rap1 pull-down assay showing the amount of B-raf-bound Rap1 (fourth panel from top), and the expression level of ERK1/2 and phosphorylated pERK1/2 in RapGEF2+/+ (lane 1) and RapGEF2−/− (lane 2) E10.5 yolk sac cells. Tubulin was used as a loading control. (D-E) The expression level of proteins (intensity of protein bands) was quantified by Imagequant software and presented as percent expression in RapGEF2−/− cells compared with RapGEF2+/+ cells, where the expression level was set as 100%. Then, the statistical analysis was shown as histograms. (F) Drawing displaying the possible role of RapGEF2 in the development of hematopoietic lineage. In this model, SCL/Gata transcription factors are required for the development of hematopoietic lineage, and RapGEF2 might regulate SCL through the Rap1/B-raf/ERK pathway.
Therefore, we further analyzed the expression of the Scl and Gata transcription factors at the protein level. SCL was significantly down-regulated (∼ 80% less) in the RapGEF2−/− yolk sac cells compared with the RapGEF2+/+ cells (Figure 6B,D). As a result, SCL's downstream target Gata129 was also significantly down-regulated (∼ 75% less), whereas, Gata2 expression was reduced by ∼50% (Figure 6B,D). In contrast, Lmo2, SCL's partner in the transcription complex,30 was only slightly reduced (∼35% less) in the absence of RapGEF2. Furthermore, the proto-oncogene c-myb, which is essential for hematopoietic development, was also significantly down-regulated (∼ 75% less) in the RapGEF2−/− cells (Figure 6B,D). Thus, the levels of SCL, Gata1, and c-myb were significantly reduced, whereas, Gata2 expression is declined by half in the absence of RapGEF2 and could potentially contribute to the observed hematopoietic defects in RapGEF2−/− mice, because these transcription factors are required for early embryonic hematopoietic development.24,26,29,31
RapGEF2 is an activator of the Ras-family small GTPase Rap1, which is involved in multiple effector pathways, including the integrin signaling and B-Raf/MEK/ERK activation pathways, among others.4,5,32 To investigate which pathway might mediate RapGEF2's regulation of hematopoietic development, we first analyzed the effect of RapGEF2 loss on the activation status of Rap1. As predicted, in the absence of RapGEF2, Rap1 activation was severely repressed. That is, the level of its active form, Rap1GTP, was significantly reduced in the E10.5 RapGEF2−/− yolk sac cells compared with RapGEF2+/+ yolk sac cells, whereas the level of total Rap1 was only slightly reduced (Figure 6C,E).
In B-raf–expressing cells, Rap1GTP can activate B-raf by direct binding; the Rap1GTP/B-raf complex mediates the phosphorylation and activation of ERK1/2 through MEK1.33-37 We compared B-raf expression in the RapGEF2+/+ and RapGEF2−/− yolk sac cells (Figure 6C,E) and detected it in both genotypes, suggesting that Rap1GTP/B-raf-mediated signaling might not be prevented in the cells of either genotype. Next, we analyzed the effect of the impaired Rap1 activation (reduced Rap1GTP level) in RapGEF2−/− cells on the activation of the B-raf/ERK pathway. B-raf/Rap1 coimmunoprecipitation assays revealed a direct interaction between Rap1 and B-raf in RapGEF2+/+ yolk sac cells. Significantly less Rap1 (∼ 80% less) was detected in the B-raf/Rap1 coimmunoprecipitation assay of RapGEF2−/− yolk sac lysates (Figure 6C,E), indicating that the reduced Rap1 activation resulted in a concomitant defect in Rap1's interaction with B-raf. Furthermore, although the downstream targets of B-raf/Rap1GTP, ERK1/2, were expressed in both RapGEF2+/+ and RapGEF2−/− yolk sac cells, the active forms of ERK1/2, pERK1/2, were almost completely undetectable in the RapGEF2−/− cells (Figure 6C,E). Thus, in the absence of RapGEF2, ERK1/2 phosphorylation and activation were severely impaired. Therefore, the disrupted Rap1/B-raf/ERK signaling in RapGEF2−/− cells could have contributed to the deregulation of Scl/Gata transcription factors that in turn lead to defective embryonic hematopoiesis (Figure 6F).
Additional results can be found in the supplemental materials.
Discussion
RapGEF2 is one of many GEFs involved in the activation of Rap1. In the current study, by deleting RapGEF2 we discovered a novel function for Rap1; RapGEF2-mediated Rap1 signaling is required for yolk sac vasculogenesis and the maintenance of definitive hematopoiesis. The RapGEF2-deficient mice died at ∼ E11.5 with severe defects in embryonic and yolk sac vascularization. In contrast to Rap1 or C3G (also known as RapGEF1) deletion where embryonic lethality occurs at ∼ E7.0,12,13 RapGEF2-deficient embryos did not show any significant visible defects until E7.5 but died at ∼ E11.5. This suggests that the requirement for C3G and RapGEF2 could be different, and that RapGEF2-mediated Rap1 activation might occur in a different cell type at a later stage during embryogenesis. RapGEF2−/− yolk sac and embryonic cells displayed reduced levels of proliferation and migration and slightly elevated levels of apoptosis in the embryos, pointing to potential contribution to embryonic lethality. However, RapGEF2 is predominantly expressed in the yolk sac and embryonic blood vessels. The RapGEF2-deficient embryos and yolk sacs lacked the normal honeycomb-like meshwork of blood vessels (Figure 2).38 This suggests a specific requirement for RapGEF2-mediated Rap1 activation during embryonic endothelial/hematopoietic development.
Surprisingly, none of the RapGEF2 embryonic phenotypes were observed in any of the known Rap1-specific GEF/GAP knockout mice. Most interestingly, the defective yolk sac vasculogenesis phenotype and the timing of embryonic lethality observed in RapGEF2-deficient mice is similar to that of a set of transcription factor-deficient (Scl, Gata, Runx1, Lmo2) mice. Deletion of Scl, Gata, Runx1, or Lmo2 in mice leads to embryonic lethality due to severe yolk sac vascular defects.24,39,40 Later studies proved that these transcription factors are extremely essential for the onset and development of primitive and definitive hematopoiesis during embryogenesis.27,39,40 Moreover, SCL is also reported to be essential for the angiogenic remodeling of the yolk sac capillary network into the complex vitelline vessels.41 The striking similarities between RapGEF2 and Scl, Gata knockout yolk sac phenotypes prompted us to investigate whether RapGEF2/Rap1 signaling is also required for the development of embryonic hematopoiesis.
To this end, we analyzed RapGEF2−/− yolk sacs and embryos using specific markers of hematopoiesis, to identify the possible defects in hematopoietic development. Flk1+/Brachyury+ cells (hemangioblasts) represent the first population derived from embryonic mesoderm that give rise to both endothelial and hematopoietic lineages.42,43 Later, the cells lose Brachyury but continue to express Flk1 and give rise to endothelial lineage. A subset of Flk1+ cells start to express hematopoietic markers such as CD41, which give rise to the hematopoietic lineage.21 Flk1 deficiency leads to a failure in the maturation of both endothelial and hematopoietic lineages.19,20 We detected an almost similar number of Flk1+ cells in RapGEF2+/+ and RapGEF2−/− yolk sacs, indicating that the Flk1+ early progenitors that give rise to hematopoietic and endothelial lineages are in fact present normally in the absence of RapGEF2. It appears, however, that the endothelial cells were unable to coalesce to form primitive yolk sac vasculature. A dual staining by Flk1/CD41 disclosed a significant reduction of CD41 expressing cells in RapGEF2-deficient yolk sacs and embryos. In addition, a dramatic reduction in the CD34+/c-Kit+, CD45+/c-Kit+, and Ter119+ hematopoietic populations were detected in RapGEF2-deficient yolk sacs and embryos. Previous studies demonstrate that CD41 expression marks the onset of definitive hematopoiesis during mouse embryogenesis.21,22 Our study revealed a dramatic reduction of the CD41 expressing population in the RapGEF2−/− mice indicating impairment in the maintenance of definitive hematopoiesis. Interestingly, a similar lack of CD41 expression is also identified in Scl-ES cells and knockout yolk sacs.21 Further studies showed that SCL is required for the generation of all hematopoietic lineages, and loss of Scl leads to complete block in hematopoietic development and in vitro colony formation.44,45 Interestingly, the expression level of SCL is significantly reduced in RapGEF2-deficient yolk sac cells. Although SCL is down-regulated in RapGEF2−/− yolk sacs, we still detected some hematopoietic activity in the RapGEF2−/− yolk sacs, such as reduced numbers of CD41+, CD34+/c-Kit+, CD45+/c-Kit+, and Ter119+ hematopoietic cells. Moreover, in vitro proliferation, colony formation, and colony replating assays revealed that RapGEF2-deficient yolk sac and embryonic cells were able to proliferate, albeit at a much reduced rate, and also produced fewer numbers of hematopoietic colonies. Similarly, FACS-sorted RapGEF2−/− HPCs (c-Kit+/CD34+) produced significantly reduced number of hematopoietic colonies compared with RapGEF2+/+ HPCs. This could be because SCL expression is significantly reduced but not completely lost in the RapGEF2−/− cells. Moreover, the expression level of Lmo2, SCL's interacting partner, is only slightly reduced in RapGEF2−/− cells, pointing to the possibility of some activity for this transcriptional complex. This reduced activity could be sufficient to initiate the development of hematopoietic lineage but not enough to drive CD41 expression and go beyond definitive hematopoiesis.
To identify a possible connection between RapGEF2/Rap1 signaling and Scl regulation, expression levels of SCL transcription factor complex was analyzed in RapGEF2-deficient yolk sac cells. Expression levels of Scl and Gata1 are significantly down-regulated at the RNA and protein level, and the Gata2 protein level is reduced by approximately 50% in RapGEF2−/− cells. It appears that the RapGEF2/Rap1 signaling mediates its effects through Scl/Gata transcription factors. Previous studies have shown that the B-raf/ERK signaling is one of the immediate downstream targets of Rap1.5,35 Activated ERK induces the transcriptional activation of a wide range of genes that are involved in cellular differentiation and cell division. Another study demonstrates that activated ERK can phosphorylate and activate the transcription factor SCL. SCL in turn transcriptionally regulates Gata1 and Gata2.46,47 To identify how the B-raf/ERK signaling is operated in the absence of RapGEF2 and whether a disrupted B-raf/ERK signaling is one of the possible reasons for SCL deregulation, we analyzed the activation status of this pathway in RapGEF2−/− yolk sac cells. We detected impaired activation of Rap1, defective interaction between Rap1 and B-Raf, which in turn resulted in defective ERK activation. Therefore, Rap1/B-raf/ERK signaling is impaired in RapGEF2-deficient cells. These data together suggest that RapGEF2 controls the hematopoietic development by possibly regulating SCL at 2 levels; first, by regulating Scl gene expression through an ERK-independent pathway and second, by regulating SCL protein activation through an ERK-dependent pathway. The hypothesis that B-raf/ERK signaling regulates SCL is further strengthened by the observation that ERK2 is essential for mesoderm differentiation and B-raf deficiency leads to defective extra-embryonic vascularization and a block in the development of endothelial lineage due to defective ERK activation.48-50 These functional similarities between ERK signaling and SCL transcriptional network in the specification of mesoderm indirectly pointing to a possible connection between these pathways. Nevertheless, many more studies are required to understand multiple steps involved in the regulation of the SCL transcription complex by Rap1 signaling through the MAPK/ERK pathway during hematopoietic commitment.
To determine the role of RapGEF2 in mid-to-late embryogenesis or in the later stages of hematopoiesis, RapGEF2 was deleted in late-stage embryos by using inducible β-actin-ERcre. This leads to impaired fetal liver hematopoiesis and embryonic lethality. There are 2 possible explanations for this phenotype: (1) in contrast to the yolk sac cells where RapGEF2 is required intrinsically, in the fetal liver RapGEF2 may have an extrinsic function during hematopoietic development; and (2) alternatively, RapGEF2 might be necessary for fetal liver development and function, with the loss of RapGEF2 resulting in organ failure, which in turn leads to defective fetal liver hematopoiesis. Although RapGEF2cko/ckoβ-actin-ERcre embryos displayed defects in fetal liver hematopoiesis, other defects may be responsible, at least in part, for the embryonic lethality of the RapGEF2cko/ckoERcre mice. For example, when RapGEF2cko/cko mice were crossed with NestinCre mice to delete RapGEF2 in neuronal cells, the RapGEF2cko/ckoNestincre mice died soon after birth, indicating that RapGEF2 plays a role in brain development (A.S. and S.X.H., unpublished results, July 2009). SCL also has an important role in neurogenesis, and whether RapGEF2 is an upstream regulator of SCL in this cell type needs to be explored.
In contrast to severe defects in embryonic hematopoiesis in response to global deletion of RapGEF2 by β-actincre, specific deletion of RapGEF2 in several hematopoietic or endothelial lineages surprisingly did not lead to any significant defects. Specific deletion of RapGEF2 in endothelial cells or in fetal liver, and adult hematopoietic stem and progenitor cells (HSPCs), by crossing RapGEF2cko/cko mice to VE-cadcre or Vavcre mice, respectively, did not result in any unusual phenotype. This suggests that RapGEF2 function is required in the commitment of primitive progenitors into endothelial and hematopoietic progenitors but not required in the latter stages, such as development and expansion of the already committed endothelial and hematopoietic progenitors. Similarly, inducible deletion of RapGEF2 in adult bone marrow cells by crossing RapGEF2cko/cko mice with Mx1cre or specific deletion from T cells and B cells by crossing to CD4cre and CD19cre mice, respectively, resulted in no detectable defects in T-cell or B-cell development or adult hematopoiesis, although RapGEF2 expression was detected in all of these cell types. Interestingly, deletion of RapGEF2 did not affect the activation status of Rap1 in these cell types. It appears that RapGEF2 is either not required, or its function is fully compensated by other GEFs in these cell types. Nevertheless, further studies using double and triple deletion of GEFs and GAPs are required to fully understand the specific functions and the compensatory mechanisms among GEFs and GAPs in different cell types.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Holly Morris, Rob Koogle, and Debbie Swing for animal care, Eileen Southon for assistance with blastocyst injections, Helen Shin for mice genotyping, Donna Butcher, Roberta Smith, and the Pathology/Histotechnology Laboratory, NCI-Frederick, for tissue sectioning and staining.
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), NCI, Center for Cancer Research.
National Institutes of Health
Authorship
Contribution: A.S. and K.O.G. designed and performed the experiments and wrote the manuscript; X.C. generated the RapGEF2-deficient mouse; V.C. and L.T. contributed to the generation of the mouse and critically reviewed the manuscript; and J.R.K. and S.X.H. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for V.C. is Department of Molecular Viralogy, Immunology and Medical Genetics, The Ohio State University, Columbus, OH.
Correspondence: Steven X. Hou, Mouse Cancer Genetics Program, Center for Cancer Research, NCI-Frederick, Bldg 560/12-70, 1050 Boyles St, Frederick, MD 21702-1201; e-mail: hous@mail.nih.gov; or Jonathan R. Keller, Basic Research Program, SAIC-Frederick, Center for Cancer Research, National Cancer Institute-Frederick, Bldg 560/12-03, 1050 Boyles St, Frederick, MD 21702-1201; e-mail: kellerjo@mail.nih.gov.
REFERENCES
Author notes
A.S., K.O.G., and X.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal