Abstract
Ldb1 and erythroid partners SCL, GATA-1, and LMO2 form a complex that is required to establish spatial proximity between the β-globin locus control region and gene and for transcription activation during erythroid differentiation. Here we show that Ldb1 controls gene expression at multiple levels. Ldb1 stabilizes its erythroid complex partners on β-globin chromatin, even though it is not one of the DNA-binding components. In addition, Ldb1 is necessary for enrichment of key transcriptional components in the locus, including P-TEFb, which phosphorylates Ser2 of the RNA polymerase C-terminal domain for efficient elongation. Furthermore, reduction of Ldb1 results in the inability of the locus to migrate away from the nuclear periphery, which is necessary to achieve robust transcription of β-globin in nuclear transcription factories. Ldb1 contributes these critical functions at both embryonic and adult stages of globin gene expression. These results implicate Ldb1 as a factor that facilitates nuclear relocation for transcription activation.
Introduction
The β-globin locus control region (LCR) activates the adult globin genes (major and minor) over a distance of approximately 20 kb. Activation is accompanied by the establishment of proximity between these elements1,2 through any of several mechanisms, including looping, linking, or tethering to a common nuclear substructure.3 Intrachromosomal associations between genes and enhancers have also been observed in the α-globin locus in erythroid cells,4 at the TH2, interferon-γ, and major histocompatibility complex loci in T cells, the IgH and Igκ loci in B cells, and the growth hormone locus in pituitary cells, among others.5,6 Furthermore, close interactions can exist between chromosomes to regulate TH1 and TH2 cytokine and odorant receptor choice during differentiation.7,8 How these interactions between regulatory elements and genes form and how they function to increase transcription remain unclear.
The erythroid activators EKLF, GATA-1, and FOG-1, as well as the widely expressed nuclear factor lim domain binding protein 1 (Ldb1) are required to establish spatial proximity between the LCR and β-globin gene.9-11 Ldb1, in a complex with LMO2 and DNA-binding partners GATA-1 and SCL, occupies the LCR and β-globin promoter in induced murine erythroleukemia (MEL) cells and fetal liver erythroid cells of mice, and shRNA-mediated reduction of Ldb1 prevents interaction between these elements. Ldb1 is the murine homolog of Drosophila melanogaster Chip and is proposed to be a general “facilitator” of long-range chromatin interactions.12 Proximity between distant elements that are Ldb1-binding sites may depend on Ldb1 self-interaction, although this has not been firmly established.13,14 It is not known whether all functions of Ldb1 involve long-range associations or whether all Ldb1 functions in erythroid cells are carried out through its interaction with DNA-binding partner GATA-1. However, there is a high correlation between positive GATA-1 regulatory activity and co-occupancy of GATA sites by SCL and Ldb1.15
During the transcription cycle, the c-terminal domain (CTD) of RNA pol II (pol II) is phosphorylated on alternative serine residues with different consequences.16 The Ser5P form of pol II is competent for initiation and promoter escape, and the LCR is important for Ser5P pol II localization at the β-globin gene.17 The Ser2P form of pol II is required for productive elongation and is the form that predominates in promoter distal regions of actively transcribed genes. Positive elongation factor b (P-TEFb), a complex containing cyclin T1 (CycT1) and cyclin-dependent kinase 9 (Cdk9), is recruited to promoters by transcription factors and functions to enhance elongation by phosphorylation of the pol II CTD Ser2 residue. P-TEFb advances along transcribed genes together with hyperphosphorylated pol II and the facilitates chromatin transcription (FACT) complex, which is involved in nucleosome disassembly during transcription.18
In mammalian nuclei, induced genes and genes that are stimulated to higher transcription levels often undergo migration away from the nuclear periphery to a more central position where they become associated with transcription factories (TFs) that are repositories of hyperphosphorylated pol II.19 For example, the CFTR gene is located in the nuclear periphery when inactive and relocates to a more interior position when active.20 Similarly, the IgH locus resides in the nuclear periphery in lymphoid progenitors before activation but migrates to the nuclear interior during B-cell development when the locus is rearranged and becomes transcriptionally active.21 The LCR is required for migration of the β-globin locus to a more central nuclear position in differentiating mouse fetal liver erythroid cells, which corresponds with the transition from limited to very robust β-globin transcription.22 The nuclear factors involved in this migration are unknown, although sumoylation of GATA-1 appears to be important for the process.23 Nor is it clear how nuclear migration relates to the establishment of proximity between the LCR and β-globin gene.
To gain insight into the transcriptional regulatory mechanisms influenced by Ldb1, we reduced its expression by shRNA in MEL cells and observed that Ldb1 is required to stabilize the Ldb1/GATA-1/SCL/LMO2 complex on β-globin chromatin. Ldb1 is also required for enrichment of P-TEFb in the locus and for RNA pol II phosphorylation to the fully elongation competent Ser2P form at the β-globin gene after dimethyl sulfoxide (DMSO)-induced differentiation. Furthermore, Ldb1 reduction results in failure of the globin locus to migrate to an interior nuclear position as is normally the case during differentiation. In contrast to Ldb1 reduction, in erythroid cells of mice homozygous for a deletion of the LCR, enrichment of the Ldb1 complex and pTEF-b at the β-globin promoter is not compromised and thus does not require the LCR. However, residence of the Ldb1 complex at the β-globin promoter in the absence of the LCR is insufficient for high-level transcription. Thus, both the LCR and Ldb1 are required for nuclear relocalization and robust globin gene transcription. These results point collectively to a shared function of these components, possibly the establishment of LCR/β-globin proximity, as a prerequisite for nuclear migration and full transcription activation of the β-globin gene.
Methods
Cell culture
Stable clones for control or Ldb1-knockdown MEL cells were successfully established using one of the lentiviral shRNAs available from Open Biosystems. Cells were cultured and differentiated as described previously.11
LCR deleted mice
Wild-type controls and mice homozygous for a deletion of the β-globin LCR were used in this study,24 and definitive erythroid cells were collected from E14.5 fetal liver as described previously.24,25 All experiments were carried out under protocols approved by the National Institutes of Health Animal Welfare Committee.
EB differentiation
Wild-type control embryonic stem (ES) cells and Ldb1-null mutant (Ldb1−/−) cells were kindly provided by Dr Paul Love (National Institute of Child Health and Human Development).26 Our studies were carried out using one clone of many established by these investigators with similar phenotypic behavior. ES cells were maintained and differentiated as described previously with some modification.27 Briefly, undifferentiated ES cells were maintained on mouse embryonic fibroblast feeder layers (Open Biosystems) in Dulbecco modified Eagle medium supplemented with 15% fetal bovine serum (FBS; Hyclone), 0.1% β-mercaptoethanol (Sigma-Aldrich), and 1 ng/mL leukemia inhibitory factor (Chemicon). To generate embryoid bodies (EBs), cultures at a concentration of 1 × 105 cells per milliliter were incubated in EB medium (Iscove modified Dulbecco medium supplemented with 15% FBS, 50 ng/mL L-ascorbic acid, Sigma-Aldrich; 300 μg/mL transferrin, Sigma-Aldrich; 5% PFHM II, Invitrogen; 4.5 × 10−4M monothiolglycerol, Sigma-Aldrich) for 4 to 5 days without feeder cells. The EBs were then harvested and dissociated with 0.25% trypsin (Invitrogen) A total of 1 × 106 cells per milliliter of single cells were replated in C4 medium (Iscove modified Dulbecco medium supplemented with 10% AdvanceSTEM Serum Replacement, Hyclone; 5% FBS, 10% PFHM II, 1.5 × 10−4M monothiolglycerol) plus 20 ng/mL of recombinant mouse erythropoietin (R&D Systems). The C4 medium was replaced every 2 days, and cells were harvested at the indicated times.
RT-PCR and Western blot analysis
ChIP and quantitative real-time PCR
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described,25,28 except that 50 U/mL of micrococcal nuclease was used to digest chromatin to nucleosome-sized fragments. Real-time polymerase chain reaction (PCR) using either TaqMan (Applied Biosystems) or SYBR Green (Bio-Rad) chemistry was performed to determine enriched DNA or cDNA using the ABI Prism 7900HT (Applied Biosystems).29 The enrichment of target DNA over input was calculated by the ΔΔCt method with results presented as mean plus or minus SEM. Primer and probe sequences are available on request.
Antibodies
Ldb1 (SC-11198), GATA-1 (SC-1233), Pol-II (SC-899), Cdk9 (SC-484), and CycT1 (SC-10750) were obtained from Santa Cruz Biotechnology. Anti-acetylhistone H3 (06-599) and anti-trimethylhistone H3 (Lys4) (07-473) were from Upstate Biotechnology, and RNA polymerase II CTD repeat YSPTSPS (phospho S2) antibody was from Abcam. LMO2 antibodies were from Cell Signaling Technology, and SCL antibodies were kindly provided by Dr Catherine Porcher.
Three-dimensional immuno-FISH
Three-dimensional immuno–fluorescence in situ hybridization (FISH) was performed essentially as described.30 The murine β-globin locus was detected with a nick-translated BAC probe (RP22-370E12); approximately 100 ng of probe (DIG-labeled) and 5 μg of mouse Cot-1 DNA were used per hybridization. FISH spots were visualized with a fluorescein isothiocyanate–conjugated monoclonal antibody to DIG (Sigma-Aldrich). The nuclear lamina was detected using a goat anti–Lamin B1 antibody (Santa Cruz Biotechnology) and a donkey Cy3 or Alexa-594 conjugated anti–goat antibody (Jackson ImmunoResearch Laboratories, Invitrogen)
Image acquisition and analysis
Image stacks of MEL cell samples (Figure 5) mounted in Tris buffered 90% glycerol (pH 8.8, DABCO antifade) were captured on an Olympus IX71 microscope (100×/1.40, PL Apo oil objective) equipped with a cooled Photometrics Coolsnap HQ scientific grade CCD camera using Deltavision SoftWorx software V3.5.1 (Applied Precision). Image convolution and 3-dimensional colocalization analysis of the globin locus with the lamina was also performed using SoftWorx V3.5.1. FISH spots were judged to contact the lamina if at least several pixels in one plane overlapped the Lamin B1 stain. More than 100 alleles were scored for each condition.
Results
Recruitment of P-TEFb and Ser2 phosphorylation of RNA pol II in the β-globin locus
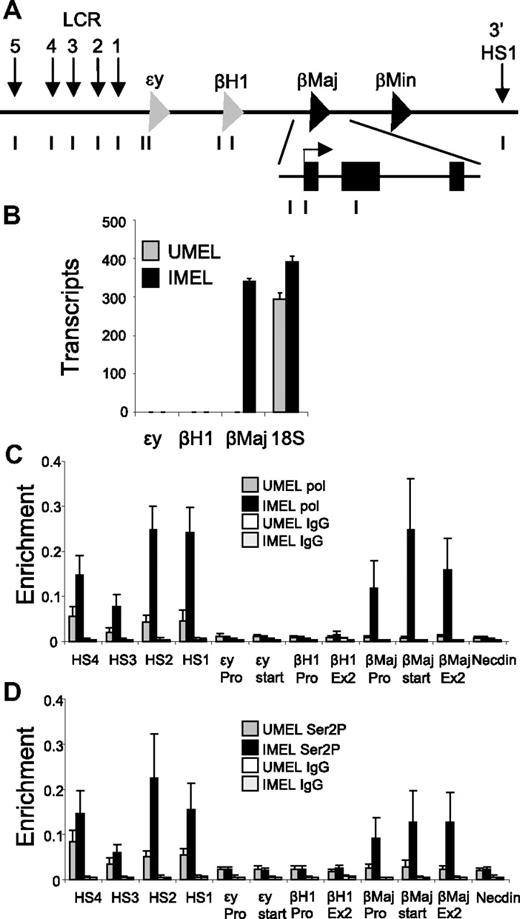
To begin to investigate the relationship between localization of Ldb1, and thus the establishment of LCR-β-globin proximity and productive transcription elongation, we asked at what point during MEL cell differentiation the fully elongation competent form of pol II, which is phosphorylated at Ser2P of the CTD is detected in the β-globin locus (Figure 1A).
Recruitment of elongation competent Ser2P RNA pol II to the β-globin locus during mouse MEL cell differentiation. (A) The mouse β-globin locus is diagrammed, and the positions of TaqMan probes used for real-time PCR are indicated below and named on the graphs. (B) MEL cells were treated with 2% DMSO for 4 days (IMEL) or without DMSO (UMEL), and globin mRNA was analyzed by quantitative real-time RT-PCR. Data are mean ± SEM, and 18S ribosomal RNA transcripts served as a control. (C-D) ChIP was performed MEL cell chromatin and antibodies to (C) RNA pol II or (D) Ser2P pol II. Three different chromatin preparations were analyzed by quantitative PCR. Error bars represent the SEM. Necdin, not transcribed in these cells, served as a negative control.
Recruitment of elongation competent Ser2P RNA pol II to the β-globin locus during mouse MEL cell differentiation. (A) The mouse β-globin locus is diagrammed, and the positions of TaqMan probes used for real-time PCR are indicated below and named on the graphs. (B) MEL cells were treated with 2% DMSO for 4 days (IMEL) or without DMSO (UMEL), and globin mRNA was analyzed by quantitative real-time RT-PCR. Data are mean ± SEM, and 18S ribosomal RNA transcripts served as a control. (C-D) ChIP was performed MEL cell chromatin and antibodies to (C) RNA pol II or (D) Ser2P pol II. Three different chromatin preparations were analyzed by quantitative PCR. Error bars represent the SEM. Necdin, not transcribed in these cells, served as a negative control.
Transcription of β-globin goes from undetectable to robust on DMSO-induced differentiation of MEL cells, whereas embryonic ϵγ and βH1 are not activated (Figure 1B). High-resolution ChIP and quantitative PCR were carried out with mononucleosome-sized fragments (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and an antibody recognizing both modified and unmodified forms of pol II. The enzyme can be detected at low levels in LCR before induction (Figure 1C).17,31 However, high levels of pol II are only apparent at the LCR and β-globin gene after differentiation, similar to the kinetics of Ldb1 binding and the establishment of proximity, or chromatin looping, between the LCR and β-globin gene.11 Surprisingly, ChIP with an antibody that recognizes primarily Ser2P pol II revealed that low levels of elongation competent pol II are detectable at the LCR DNase I hypersensitive sites before MEL cell induction (Figure 1D). Similar to bulk pol II, Ser2P pol II is only detected at the β-globin gene after induction, and its binding is distributed throughout the gene, as expected for the elongation competent form of pol II.
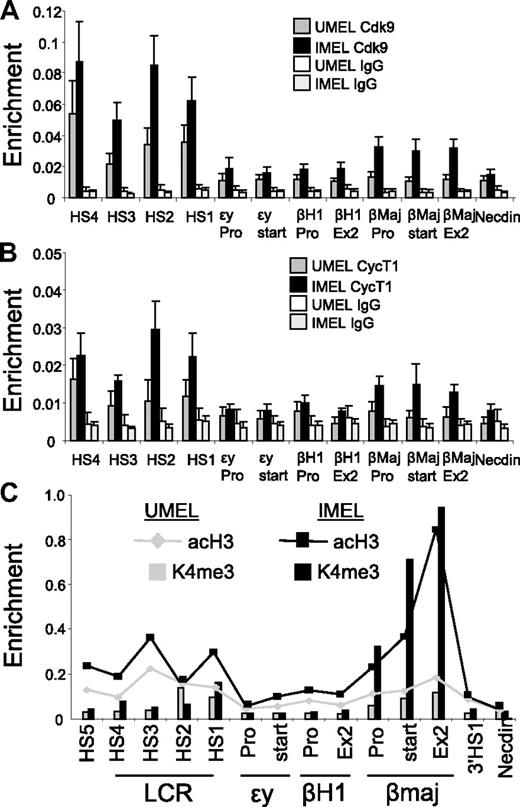
To confirm enrichment of Ser2P pol II within the LCR, we performed ChIP with antibodies to Cdk9 and CycT1, subunits of the P-TEFb complex that catalyzes this phosphorylation event required for productive elongation. Both these proteins were detected at the LCR in undifferentiated MEL cells (Figure 2A-B). Thus, transcription elongation-related components occupy the LCR at low levels before differentiation and might reflect a poised state of the locus before activation of the β-globin gene. Similar to the Ldb1 complex and Ser2P pol II, the P-TEFb complex is detected at the β-globin gene only after induction of transcription by DMSO. P-TEFb is distributed across the gene body in a pattern similar to Ser2P pol II and to that observed for P-TEFb in the active α-globin gene.4 The association of Ser2P RNA pol II and P-TEFb with the β-globin gene coincides with enrichment of acetylated histone H3 and H3K4 trimethylation, consistent with active transcription (Figure 2C).32,33
Enrichment of P-TEFb kinase at the β-globin LCR and promoter region in differentiating MEL cells and alterations in H3ac and H3K4me3. (A-C) Chromatin was prepared from MEL cells before and after 4 days of 2% DMSO treatment. ChIP was performed with antibodies to (A) Cdk9 or (B) CycT1. Necdin served as a negative control. Data are mean ± SEM. (C) Antibodies to acH3 or H3K4me3 were used in ChIP and graphed together by setting the highest value obtained with each antibody equal to 1. Lines represent AcH3; and bars, H3K4me3.
Enrichment of P-TEFb kinase at the β-globin LCR and promoter region in differentiating MEL cells and alterations in H3ac and H3K4me3. (A-C) Chromatin was prepared from MEL cells before and after 4 days of 2% DMSO treatment. ChIP was performed with antibodies to (A) Cdk9 or (B) CycT1. Necdin served as a negative control. Data are mean ± SEM. (C) Antibodies to acH3 or H3K4me3 were used in ChIP and graphed together by setting the highest value obtained with each antibody equal to 1. Lines represent AcH3; and bars, H3K4me3.
Ldb1 is important for stability of the GATA-1/SCL/LMO2 complex and is required for Ser2 pol II phosphorylation at the β-globin locus.
Our observations thus far raise the question of the involvement of Ldb1 in P-TEFb recruitment. Ldb1 might have a direct role in formation of an elongation competent Ser2P pol II complex at the β-globin promoter. Alternatively, the GATA-1/SCL/LMO2 components of the Ldb1 complex might individually or collectively suffice to recruit P-TEF-b, whereas Ldb1 might be required solely for establishment of LCR-gene proximity. To investigate this issue, we studied MEL cell clones in which Ldb1 had been knocked down by shRNA.
Ldb1 was reduced substantially in 2 shRNA expressing MEL cell clones as judged by analysis of protein by Western blot (Figure 3A). Reduction of Ldb1 had no obvious effect on the levels of GATA-1 protein, a DNA-binding component of the Ldb1 complex. However, under these conditions, β-globin transcription after DMSO induction was very strongly reduced (5% of the level in control shRNA-treated cells; Figure 3B) as was bulk pol II recruitment to the gene (supplemental Figure 2). Transcription of α-globin was also reduced after Ldb1 reduction, in accord with a role for Ldb1 in enhancer function in this locus.4 However, numerous other genes normally induced by DMSO in MEL cells were unaffected by the knockdown, indicating that Ldb1 loss did not result in a general block to differentiation (supplemental Figure 3).34 Consistent with the change in transcription, enrichment of acetylated histone H3 and H3K4 trimethylation across the β-globin gene was reversed to a pattern that resembled undifferentiated MEL cells (compare Figure 3C with Figure 2C) on reduction of Ldb1,
Ldb1 knockdown inhibits enrichment of the Ldb1 complex and Ser2P RNA pol II at the β-globin promoter and LCR in MEL cells. (A) MEL cells were stably transduced with Ldb1 shRNA or with an empty virus (Ctrl) and individual clones were isolated. Reduced Ldb1 expression in 2 stable Ldb1 knockdown clones (KD1 and KD2) was confirmed by Western blot analysis before (−) and after 4 days of 2% DMSO treatment (+). GATA-1 and α-tubulin served as positive and internal controls, respectively. (B) Stable clones were treated with 2% DMSO. Total RNA was isolated at day 4, and β-globin expression was analyzed by quantitative real-time RT-PCR. Each value was normalized with 18S ribosomal RNA. Three RNA preparations were analyzed. Error bars represent the SEM. (C) ChIP was carried out using KD1 cells after 4 days of 2% DMSO induction using antibodies to acH3 and H3K4me3, and the data were graphed together by setting the highest level for each antibody equal to 1. (D) The ChIP assay was performed with KD1 cells after 4 days of 2% DMSO induction using antibodies to Ldb1, GATA-1, LMO2, and SCL. Error bars represent the SEM for multiple independent chromatin preparations. (E-F) ChIP was carried out as for panel D using antibodies to Ser2P pol II (E) or Cdk9 (F). Necdin served as a negative control. Error bars represent the SEM for multiple independent chromatin preparations. ChIP assays with KD2 cells gave similar results (not shown) to those in panels C through F.
Ldb1 knockdown inhibits enrichment of the Ldb1 complex and Ser2P RNA pol II at the β-globin promoter and LCR in MEL cells. (A) MEL cells were stably transduced with Ldb1 shRNA or with an empty virus (Ctrl) and individual clones were isolated. Reduced Ldb1 expression in 2 stable Ldb1 knockdown clones (KD1 and KD2) was confirmed by Western blot analysis before (−) and after 4 days of 2% DMSO treatment (+). GATA-1 and α-tubulin served as positive and internal controls, respectively. (B) Stable clones were treated with 2% DMSO. Total RNA was isolated at day 4, and β-globin expression was analyzed by quantitative real-time RT-PCR. Each value was normalized with 18S ribosomal RNA. Three RNA preparations were analyzed. Error bars represent the SEM. (C) ChIP was carried out using KD1 cells after 4 days of 2% DMSO induction using antibodies to acH3 and H3K4me3, and the data were graphed together by setting the highest level for each antibody equal to 1. (D) The ChIP assay was performed with KD1 cells after 4 days of 2% DMSO induction using antibodies to Ldb1, GATA-1, LMO2, and SCL. Error bars represent the SEM for multiple independent chromatin preparations. (E-F) ChIP was carried out as for panel D using antibodies to Ser2P pol II (E) or Cdk9 (F). Necdin served as a negative control. Error bars represent the SEM for multiple independent chromatin preparations. ChIP assays with KD2 cells gave similar results (not shown) to those in panels C through F.
ChIP analysis revealed that in uninduced MEL cells the normally very low levels of Ldb1 at the LCR were reduced to undetectable levels (data not shown). After DMSO induction, Ldb1 was diminished by 2- to 3-fold at the LCR and β-globin promoter in Ldb1 knockdown cells, although the reduction was 7-fold at HS3 (Figure 3D). Interestingly, occupancy of the Ldb1 complex members LMO2, SCL, and GATA-1 was also reduced at these sites between 2- and 7-fold on knockdown of Ldb1, indicating that Ldb1 plays a role in stabilizing the entire complex on chromatin, even though it is not one of the DNA-binding components. However, at HS2 the reduction of GATA-1 and SCL was weaker (1.3- to 1.7-fold). This result suggests that chromatin binding by the latter factors might be sufficiently strong, at least at some locations, to be only modestly sensitive to reduction of Ldb1and is consistent with the idea that GATA-1 occupancy is an early event in β-globin activation.35,36
Knockdown of Ldb1 and failure to enrich the Ldb1 complex resulted in a striking reduction in recruitment of P-TEFb to the β-globin locus and in reduced Ser2 phosphorylation of pol II (Figure 3E-F). These transcription elongation components were reduced both at the LCR and within the β-globin gene. Together, these data suggest that Ldb1 is required to recruit and/or stabilize the components of the Ldb1 complex on globin locus chromatin and that Ldb1 is necessary for P-TEFb enrichment and thus for Ser2 phosphorylation of pol II both at the LCR and at the β-globin promoter.
The Ldb1 complex occupies the β-globin promoter independently of the LCR in vivo
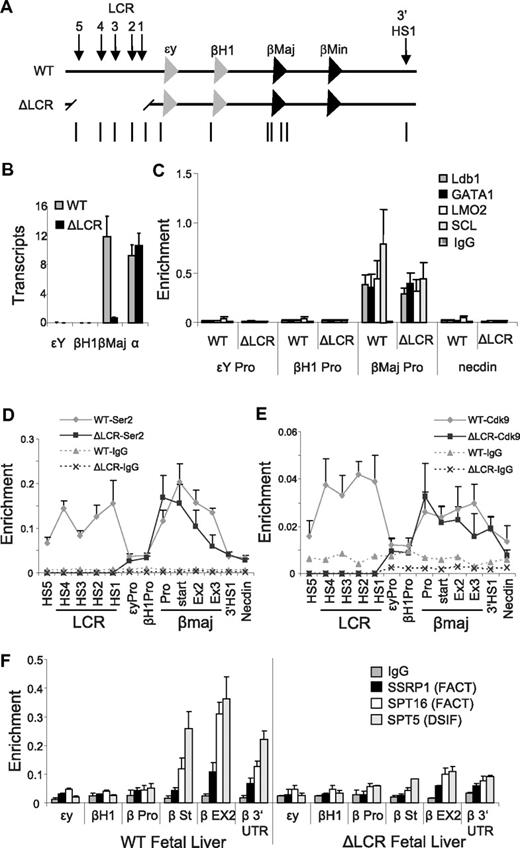
Because Ldb1 occupies the LCR but not the β-globin promoter in undifferentiated MEL cells, the LCR might be required for differentiation-dependent recruitment of Ldb1 to the promoter mediated by LCR/promoter proximity. Alternatively, Ldb1 might be recruited to the promoter independent of the establishment of proximity between the LCR and gene, a step that might be required to establish the proximity. To distinguish between these possibilities, we determined β-globin promoter occupancy of Ldb1 in mice homozygous for a deletion of the LCR24 (Figure 4A). Homozygous deletion of the LCR is lethal because of dramatically reduced β-globin transcription, but mice remain viable because of the presence of a human β-globin locus YAC transgene.
The Ldb1 complex and Ser2P RNA pol II are enriched at the β-globin promoter in fetal liver erythroid cells of mice homozygous for a deletion of the LCR. (A) The mouse β-globin locus is diagrammed, and the positions of TaqMan probes used for real-time PCR are indicated below and named on the graphs. Endogenous (WT) and LCR-deleted (ΔLCR) murine β-globin loci are depicted. (B) Total RNA was isolated from E14.5 fetal liver of WT or ΔLCR mice, and globin expression was analyzed by quantitative real-time RT-PCR. α-Globin was used as a positive control of RNA expression. Each value was normalized with 18S ribosomal RNA. Error bars represent the SEM for several independent RNA preparations. (C-F) Chromatin was prepared from E14.5 fetal liver of WT (+) or ΔLCR (−) mice, and then ChIP and quantitative PCR were performed with (C) Ldb1, GATA-1, LMO2, and SCL, (D) Ser2P pol II, and (E) Cdk9, or (F) DSIF and FACT components as indicated on the graphs. Necdin served as a negative control. Error bars represent the SEM among independent chromatin preparations.
The Ldb1 complex and Ser2P RNA pol II are enriched at the β-globin promoter in fetal liver erythroid cells of mice homozygous for a deletion of the LCR. (A) The mouse β-globin locus is diagrammed, and the positions of TaqMan probes used for real-time PCR are indicated below and named on the graphs. Endogenous (WT) and LCR-deleted (ΔLCR) murine β-globin loci are depicted. (B) Total RNA was isolated from E14.5 fetal liver of WT or ΔLCR mice, and globin expression was analyzed by quantitative real-time RT-PCR. α-Globin was used as a positive control of RNA expression. Each value was normalized with 18S ribosomal RNA. Error bars represent the SEM for several independent RNA preparations. (C-F) Chromatin was prepared from E14.5 fetal liver of WT (+) or ΔLCR (−) mice, and then ChIP and quantitative PCR were performed with (C) Ldb1, GATA-1, LMO2, and SCL, (D) Ser2P pol II, and (E) Cdk9, or (F) DSIF and FACT components as indicated on the graphs. Necdin served as a negative control. Error bars represent the SEM among independent chromatin preparations.
Deletion of the LCR in E14.5 erythroid fetal liver cells reduced the level of β-globin mRNA to approximately 8% of wild-type levels (Figure 4B), consistent with earlier work.24 Notably, this is similar to the reduction in stable β-globin transcripts observed in MEL cells after shRNA-mediated reduction of Ldb1 (compare Figure 4B with Figure 3B). We observed no effect of the LCR deletion on transcription of the independently regulated α-globin gene, ruling out a generalized effect of the LCR deletion on gene transcription. In addition, primary RNA transcripts were reduced to 6% across the β-globin gene, similar to the reduction in spliced transcripts, and consistent with an overall reduced rate of transcription of the gene rather than transcript degradation (supplemental Figure 4).
A ChIP analysis in E14.5 fetal liver cells indicated that LCR deletion diminished but did not eliminate total pol II enrichment at the β-globin gene as expected17 (supplemental Figure 4). Notably, the Ldb1 complex occupied the β-globin promoter normally in the LCR deleted mice (Figure 4C), indicating that promoter binding is independent of the LCR and does not require proximity between these 2 distant regulatory elements. In contrast to results after shRNA-mediated Ldb1 reduction in MEL cells, the abundance of Ser2P pol II and Cdk9 in the β-globin gene appeared to be unaltered in the absence of the LCR (Figure 4D-E). However, although overall levels of enrichment were not greatly affected, Ser2P pol II and Cdk9 were redistributed such that peak enrichment now occurred at the promoter, with approximately a 2-fold reduction at the 3′ end of the gene, which is consistent with decreased or inefficient elongation.
In addition to phosphorylation of pol II, P-TEFb phosphorylates the Spt5 subunit of DRB-sensitivity inducing factor (DSIF), which releases this “promoter pausing factor” to advance through transcribed genes with elongating pol II.37 Spt5 and Ser2P pol II display an occupancy pattern overlapping that of the FACT components Spt16 and SSRP1 that facilitate removal of the H2A/H2B histone dimer during transcription elongation.38 To investigate whether chromatin occupancy by these elongation components is perturbed by loss of the LCR, we performed a ChIP analysis of E14.5 fetal liver cells with antibodies to the DSIF component Spt5 and the FACT components SSRP1 and SPT16. Figure 4F shows that FACT and DSIF are distributed over β-globin in actively transcribing fetal liver cells, but they are greatly reduced in the absence of the LCR. We favor the idea that the absence of these elongation components reflects the failure of the locus, absent the LCR, to relocalize from the nuclear periphery to active TFs.22 From these data, we conclude that a major event associated with loss of the LCR is the failure of FACT to associate with the β-globin gene.
Ldb1 is required for the relocalization of the β-globin locus to the nuclear interior
Several studies have shown that high-level transcription of the β-globin gene occurs after migration of the locus from the nuclear periphery to a more central position where it becomes associated with high levels of pol II in TFs.22,39 The LCR is required for this nuclear migration and only a low level of β-globin transcription can be achieved if the locus remains in the nuclear periphery.22 We wished to investigate the role of Ldb1 in nuclear relocalization of the globin locus. If globin loci migrate away from the nuclear periphery on MEL cell induction in the absence of Ldb1, we would conclude that the LCR, per se, is sufficient for such migration and, further, that residence in a TF can occur in the absence of proximity between the LCR and gene because this is dependent on Ldb1. On the other hand, if loss of Ldb1 compromised nuclear migration, it would support the idea that the LCR is not sufficient for nuclear relocalization and that Ldb1 provides an additional and necessary function.
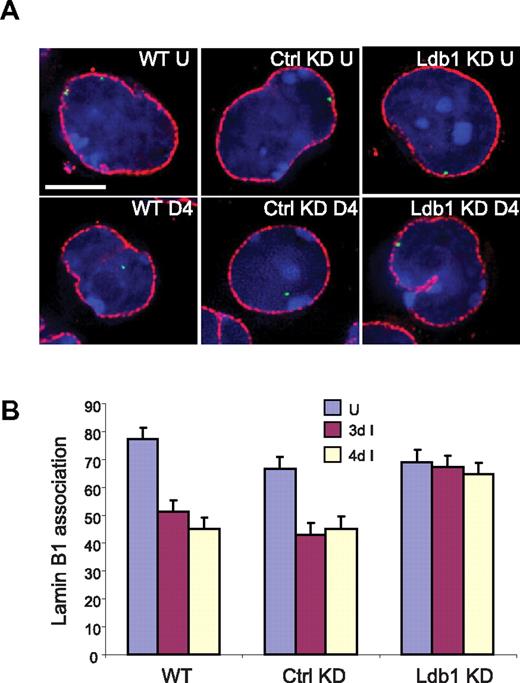
We performed immuno-FISH studies to determine the intranuclear position of the globin locus in Ldb1 knockdown MEL cells. Figure 5 reveals that approximately 75% of β-globin loci in WT MEL cells contact the lamina at the nuclear periphery before differentiation, and this is very similar in MEL cells with either control or Ldb1 shRNA expression. After differentiation, more than half the loci have moved away from the lamina in WT MEL cells and in cells with the control shRNA, consistent with earlier data40 (T.R. and A. Telling, unpublished data, March 2006). In contrast, in MEL cells in which Ldb1 was reduced there was no change in the association of globin loci with the lamina after differentiation. Thus, we conclude that Ldb1 is required for intranuclear migration of the β-globin locus on induction.
Ldb1 is required for localization of the β-globin locus away from the nuclear periphery during differentiation. (A) Three-dimensional immuno-FISH analysis of untreated cells and cells treated with DMSO. WT indicates MEL cells; Ctrl KD, MEL cells with control shRNA; KD, MEL cells with shRNA directed to Ldb1; U, uninduced; and 4D, treated with DMSO for 4 days. Red represents nuclear lamin immunofluorescence; and green, probe detecting the β-globin locus. Bar represents 5 μm. For details see “Image acquisition and analysis.” (B) Quantitation of association of globin loci with the nuclear lamina before and after DMSO treatment for 3 days (3d) or 4 days (4d). More than 100 cells were scored for each determination. Error bars represent the SE.
Ldb1 is required for localization of the β-globin locus away from the nuclear periphery during differentiation. (A) Three-dimensional immuno-FISH analysis of untreated cells and cells treated with DMSO. WT indicates MEL cells; Ctrl KD, MEL cells with control shRNA; KD, MEL cells with shRNA directed to Ldb1; U, uninduced; and 4D, treated with DMSO for 4 days. Red represents nuclear lamin immunofluorescence; and green, probe detecting the β-globin locus. Bar represents 5 μm. For details see “Image acquisition and analysis.” (B) Quantitation of association of globin loci with the nuclear lamina before and after DMSO treatment for 3 days (3d) or 4 days (4d). More than 100 cells were scored for each determination. Error bars represent the SE.
Broad role of Ldb1 in β-globin gene expression during erythroid differentiation
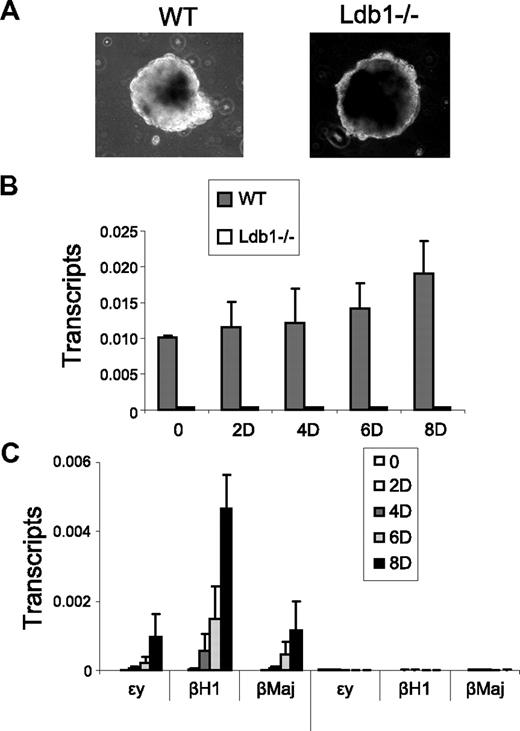
Ldb1 null mice die at embryonic day 8, and embryos fail to form blood islands in the yolk sac, indicating an important role for Ldb1 in primitive erythropoiesis.41 Ldb1 is also required for primitive erythropoiesis in zebrafish.42 Although Ldb1 was required for adult β-globin activation in MEL cells,11 we wished to examine definitive globin gene transcription in a more physiologic setting. We followed wild-type murine ES cells and ES cells that are homozygous null for Ldb1 (Ldb1−/−) during the course of differentiation along erythroid lines to determine the influence of Ldb1 in this system. We used a differentiation protocol that is expected to result in activation of both embryonic and adult globins during differentiation.27 Ldb1−/− ES cells differentiated more slowly than wild-type cells; however, they formed EBs that were indistinguishable from those of wild-type ES cells (Figure 6A).
Importance of Ldb1 to embryonic and adult β-globin gene expression during erythroid differentiation. Wild-type (WT) or Ldb1-null mutant (Ldb1−/−) ES cells were differentiated along erythroid lines into EBs with erythropoietin for 8 days as described under “EB differentiation.” (A) Morphology of WT or Ldb1−/− EB after 8 days of differentiation. (B-C) Total RNA was isolated at the indicated day of differentiation to determine expression of (B) Ldb1, or (C) ϵγ, βH1, and βmaj-globin. Each value was normalized with 18S ribosomal RNA. Error bars represent the SEM from independent RNA preparations.
Importance of Ldb1 to embryonic and adult β-globin gene expression during erythroid differentiation. Wild-type (WT) or Ldb1-null mutant (Ldb1−/−) ES cells were differentiated along erythroid lines into EBs with erythropoietin for 8 days as described under “EB differentiation.” (A) Morphology of WT or Ldb1−/− EB after 8 days of differentiation. (B-C) Total RNA was isolated at the indicated day of differentiation to determine expression of (B) Ldb1, or (C) ϵγ, βH1, and βmaj-globin. Each value was normalized with 18S ribosomal RNA. Error bars represent the SEM from independent RNA preparations.
Quantitative reverse-transcribed (RT)-PCR confirmed that Ldb1 mRNA was undetectable in the differentiated Ldb1−/− ES cells (Figure 6B). Furthermore, although wild-type ES cells produced embryonic ϵγ and βH1 globins and adult βmaj-globin, no detectable globin transcription was observed for the Ldb1 null ES cells (Figure 6C). This was not the result of cell death because normal levels of 18S RNA were produced by the differentiated Ldb1 null ES cells. Nor was this the result of failure of Ldb1 null ES cells to enter the erythroid differentiation pathway because important markers of that process, SCL, GATA-2, and RUNX1, were induced in Ldb1 null ES cells during differentiation (supplemental Figure 5). Transcription of α-globin was also abrogated in the null ES cells, indicating an important role for Ldb1 in its transcription (not shown).43 We conclude that Ldb1 is required for both primitive and definitive β-globin gene activation, most probably through mechanisms suggested here, including P-TEFb recruitment, establishment of proximity between the LCR and globin genes, and intranuclear migration of the locus.
Discussion
Long-range interactions between remote enhancers and their target genes underlie gene expression in many developmentally regulated gene families. The specificity of these interactions is thought to be mediated by tissue-specific DNA-binding proteins, although the prevalence of the interactions in different systems suggests the possibility of shared components. We explored the activity of Ldb1, a widely expressed nuclear factor with no known DNA binding or enzymatic activity that is a putative enhancer facilitator. Ldb1 and erythroid partners SCL, GATA-1, and LMO2 form a complex that occupies the β-globin LCR and gene and is required to establish spatial proximity between these elements, which is coincident with transcription activation, supporting the hypothesis that Ldb1 facilitates long-range gene activation.11 Here we have examined LCR-dependent and -independent functions of Ldb1 and find a role for this protein in migration of the β-globin locus away from the nuclear periphery during erythroid differentiation.
Reduction of Ldb1 suggests new roles for the Ldb1 complex
Reduction of Ldb1 by shRNA decreased β-globin transcription to 6% of the wild-type level. Further study of the knockdown cells revealed that Ldb1 stabilizes its erythroid complex partners on β-globin chromatin, even though it is not one of the DNA-binding components. We and others have observed that LMO2 is subject to degradation in the absence of Ldb1, but this is not the case for GATA-1.11,44 This observation might help to explain the differential sensitivity of LMO2 to Ldb1 knockdown, at least at some positions within the β-globin locus. The results of these experiments suggest that the essential role of GATA-1 in recruitment of RNA pol II to the β-globin gene during differentiation45 is carried out through participation in a chromatin-bound complex whose stabilization and function require Ldb1.
We further observed that the temporal-spatial pattern of recruitment of the important transcription component P-TEFb and of RNA pol II Ser2 phosphorylation at the β-globin locus parallel that of Ldb1 complex occupancy and establishment of spatial proximity between the LCR and gene during differentiation.11 Because the complex as a whole was diminished by the knockdown, we were unable to determine whether Ldb1 or another complex member is required for the association of P-TEFb. GATA-1 has been reported to interact with P-TEFb46 and could be important for P-TEFb recruitment to GATA-1 target genes. Others reported Ldb1 and Cdk9 interact directly.42 Thus, the role of Ldb1 in P-TEFb enrichment could be direct or indirect (or both). Notably, the shRNA reduction experiments indicate that, in the absence of Ldb1, a fully intact LCR, is insufficient for high-level transcription activation of β-globin, possibly because proximity between the LCR and gene cannot be established.
Ldb1 function in the absence of the LCR
In LCR-deleted fetal liver cells, Ldb1 complex recruitment to the β-globin promoter was not impaired. These data establish that the complete Ldb1 complex forms at the promoter independent of the presence of the LCR. Earlier work showed that GATA-1 occupancy of the β-globin promoter is likewise independent of the LCR, and our data are consistent with the idea that Ldb1 complex occupancy is dependent on GATA-1 (and possibly SCL) DNA binding.10,11 These results led us to conclude that promoter localization of the Ldb1 complex, in the absence of the LCR, insufficient for high-level transcription of the gene.
The continued presence of the Ldb1 complex in the absence of the LCR appears to be sufficient for recruitment of pTEF-b and for Ser2 pol II phosphorylation at the β-globin gene. Promoter occupancy by Ser2P pol II when transcription is reduced is surprising. However, we observed that the Ser2P pol II pattern of occupancy in ΔLCR cells was shifted toward the β-globin gene promoter with a decrease in signal of approximately 2-fold toward the 3′ end of the gene. Earlier work indicated that loss of the LCR was associated with a decrease in the Ser5P form of pol II across the β-globin gene.17 Both the decrease in Ser5P pol II and the redistribution of Ser2P pol II are consistent with blocked or inefficient elongation in ΔLCR erythroid cells.
It is unlikely that Pol II “promoter pausing” is the basis of the elongation block in ΔLCR cells, as Ser2P pol II is present at the β-globin promoter.37 Moreover, absent the LCR, DSIF and FACT are not observed over the body of the β-globin gene. FACT is necessary to remodel the template for efficient elongation by pol II. We propose that the failure of FACT to associate with β-globin is at least partially responsible for the lack of robust transcriptional elongation in the absence of the LCR but other mechanisms are possible. How FACT is recruited remains an open question. It seems likely that FACT and other pol II transcription components such as DSIF occupy TFs in the nuclear interior and that association of FACT with the locus might require LCR/β-globin proximity or migration away from the nuclear periphery or both.
Role of Ldb1 in intranuclear migration of the β-globin locus
Reduction of Ldb1 prevented relocalization of the β-globin locus from the nuclear periphery to the interior, a process that normally accompanies differentiation and robust transcription activation. This result indicates that the full LCR, per se, is insufficient for relocalization of the globin locus and supports the idea that a shared function of the LCR together with Ldb1, possibly the establishment of LCR/gene proximity, is the salient requirement for migration to TFs. This proposal is consistent with the hypothesis that LCR/promoter interaction occurs before stable association of the locus with a TF where high-level transcription takes place and might, indeed, poise the locus for TF association.10,22,39
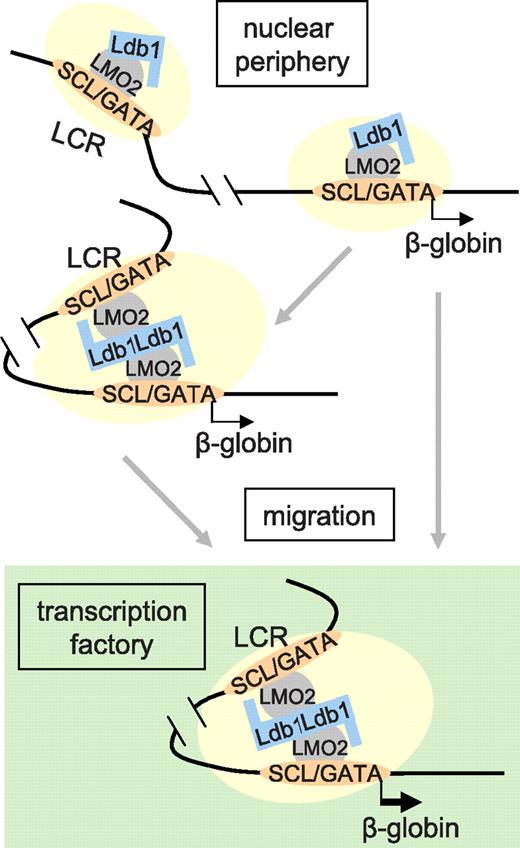
Taken together, our data contribute to elucidating the connection between the ubiquitous enhancer facilitator/chromatin factor Ldb1 and transcription activation of the β-globin locus by the LCR. Ldb1 is required to stabilize the GATA-1/SCL/LMO2 complex on β-globin chromatin and for association of P-TEFb and RNA pol II Ser2 phosphorylation in the locus. These functions are independent of the LCR and of interaction between the LCR and β-globin promoter. However, both Ldb1 and the LCR are required for intranuclear migration and the onset of high-level transcription. Two models could account for this observation. Ldb1 might be required to establish proximity between the LCR and β-globin gene through protein-protein interactions, which, in turn, is necessary for nuclear relocalization to a TF to achieve high-level transcription (Figure 7 left). This model is compatible with the observation that when transcription is interrupted and globin loci migrate out of a TF, proximity between the LCR and gene is retained.39
A model of dynamic long-range communication between the β-globin LCR and gene. Association of the Ldb1 complex (GATA-1/SCL/LMO2) and other activation components (shaded ovals), whose recruitment might be dependent on Ldb1, occupying the LCR and β-globin gene. Previous work comparing induced and uninduced MEL cells suggests that the Ldb1 complex binds first to the LCR.11 In the absence of the LCR, these components can co-occupy the β-globin gene, implying that promoter occupancy may normally occur independently; however, such occupancy is insufficient for high-level transcription. Full occupancy may be sufficient to establish LCR/β-globin proximity, before nuclear migration, via unknown mechanisms, and TF occupancy. Alternatively, the locus might migrate, again by unknown mechanisms, and establish LCR/β-globin proximity as a result of positioning within a TF. Proximity would then be stabilized by protein-protein interactions, possibly dependent on Ldb1.
A model of dynamic long-range communication between the β-globin LCR and gene. Association of the Ldb1 complex (GATA-1/SCL/LMO2) and other activation components (shaded ovals), whose recruitment might be dependent on Ldb1, occupying the LCR and β-globin gene. Previous work comparing induced and uninduced MEL cells suggests that the Ldb1 complex binds first to the LCR.11 In the absence of the LCR, these components can co-occupy the β-globin gene, implying that promoter occupancy may normally occur independently; however, such occupancy is insufficient for high-level transcription. Full occupancy may be sufficient to establish LCR/β-globin proximity, before nuclear migration, via unknown mechanisms, and TF occupancy. Alternatively, the locus might migrate, again by unknown mechanisms, and establish LCR/β-globin proximity as a result of positioning within a TF. Proximity would then be stabilized by protein-protein interactions, possibly dependent on Ldb1.
Alternatively, it is possible that the establishment of LCR/β-globin proximity may be the result of relocalization (Figure 7 right). In this model, Ldb1 might be required for the assembly and stabilization of an LCR-bound protein complex that facilitates nuclear relocalization to TFs or the nuclear interior, preceding high-level transcription. LCR/β-globin proximity may then potentially be mediated through tethering of these elements to a structural component. A more complete understanding of how loci destined to change their nuclear address acquire the ability to undergo movement within the nucleus should contribute to distinguishing these possibilities. Recent data suggest a role for actin-myosin motors in this process.47,48
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Catherine Porcher for SCL antibodies, Dr Paul Love for Ldb1−/− ES cells, Drs Qihui Gong and Indrani Som for assistance with animal and ES cell experiments, Agnes Telling for help with the FISH experiments, and Dr Elissa Lei for comments on the manuscript.
This work was supported by the Intra-mural program of the National Diabetes and Digestive and Kidney Diseases, National Institutes of Health (A.D.) and extramural National Institutes of Health (grants DK44746 and HL65440; M.G.).
National Institutes of Health
Authorship
Contribution: S.-H.S. and A.D. designed research; S.-H.S., A.K., M.A.B., and T.R. performed research; S.-H.S., A.K., A.D., T.R., M.A.B., and M.G. analyzed data; and S.-H.S., A.D., and M.G wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Sang-Hyun Song is Cancer Research Institute, Seoul National University College of Medicine, Seoul, South Korea.
Correspondence: Ann Dean, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bldg 50, Rm 3154, 50 South Dr, MSC 8028, Bethesda, MD 20892; e-mail: anndean@helix.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal