Abstract
Posttranscriptional modifications of histones play important roles in the control of chromatin structure and transcription. H3K4 (histone H3 lysine 4) methylation by the SET domain of the trithorax-group protein MLL (mixed-lineage leukemia) is important for the control of homeobox (HOX) gene expression during development. MLL is tethered to the HOXA locus through interaction of its amino-terminus with menin. MLL fusion proteins associated with human leukemia contain the menin interaction peptide and frequently recruit H3K79 (histone H3 lysine 79) methylation activity. This allows sustained expression of HOXA genes important for cellular transformation. We have characterized a novel recurrent chromosomal aberration, inv(11)(p15q23), as an isolated chromosomal abnormality in 2 patients with acute myeloid leukemia. This aberration is predicted to result in the expression of an NUP98 (nucleoporin 98 kDa)–MLL fusion protein that is unable to interact with menin. As expected, low levels of HOXA gene expression were observed in the patients' samples. This fusion protein is predicted to participate in cellular transformation by activating MLL targets other than HOXA genes.
Introduction
Chromatin structure is important for the epigenetic control of gene transcription. It is modulated through posttranslational modifications of histones. The polycomb group of proteins catalyzes the methylation of H3K27 (histone H3 lysine 27), which is an inactive mark. In contrast, the trithorax group of proteins catalyzes H3K4 (histone H3 lysine 4) methylation, a mark of activation. The balance between these activities is tightly regulated during embryogenesis and is known to be important for the correct regulation of homeobox (HOX) gene expression.1
The MLL (mixed-lineage leukemia) protein is a member of the trithorax group and is involved in the maintenance of HOX gene expression. MLL harbors a SET domain at its C-terminus endowed with H3K4 methyl transferase activity.2,3 The MLL gene is frequently rearranged in human leukemia. All MLL-fusion proteins that result from chromosomal translocations lack the C-terminus part of MLL and frequently acquire, directly or indirectly, H3K79 (histone H3 lysine 79) or H4R3 (histone H4 arginine 3) methyltransferase or histone acetyltransferase activity. These activities are essential for cellular transformation because they mediate a high level of HOXA gene expression.4,5 Menin is an essential partner of MLL wild-type and MLL fusion proteins. It interacts with the first 15 amino acids of MLL, which are included in all MLL-fusion proteins known to date, and targets MLL wild-type and MLL fusion proteins to the HOXA and MEIS (myeloid ecotropic viral integration site) gene promoters. MLL fusion proteins lacking the menin interaction peptide do not activate Meis and HoxA expression and fail to transform murine primary hematopoietic cells in in vivo models.6,7
The NUP98 (nucleoporin 98 kDa) protein is a mobile component of the nuclear pore.8,9 It is involved in nucleocytoplasmic trafficking and is associated with transcriptionally active chromatin within the nucleoplasm in eukaryote cells.10-12 As MLL is the NUP98 gene is rearranged with many partner genes in human hematologic malignancies.13 All rearrangements reported to date result in fusion proteins that contain the amino-terminal moiety of NUP98, which includes the nucleoporin FG/GLFG repeat motif. NUP98 fusion proteins may interfere with transport and with the remaining wild-type NUP98.14,15 Some NUP98 fusion proteins are recruited to the chromatin through DNA binding or interaction with methylated histones, which results in the aberrant recruitment of transcriptional coregulators.16
We report on a novel recurrent chromosomal aberration that fuses the NUP98 and MLL genes. The predicted NUP98-MLL fusion protein lacks the menin interaction peptide and is predicted to be unable to activate HOXA gene transcription, as confirmed by the low level of HOXA gene expression observed in the patients' samples.
Methods
Patient 1 was a 79-year-old male diagnosed with acute myeloid leukemia–M1. Blood parameters were white blood cell count 132.5 × 109/L, red blood cell count 1.5 × 1012/L, hemoglobin 56 g/L (5.6 g/dL), hematocrit 0.14 (14.4%), platelets 14 × 109/L, and blasts 98%. Karyotype analyses showed 46,XY,inv(11)(p15q23)[18]/46,XY[8]. The patient died of his leukemia.
Patient 2 was a 30-year-old female diagnosed with acute myeloid leukemia–M2 with 29% of blasts in the bone marrow. Blood parameters were white blood cell count 2.0 × 109/L, red blood cell count 2.11 × 109/L, hemoglobin 67 g/L (6.7 g/dL), hematocrit 0.19 (19%), platelets 126 × 109/L, and blasts 27%. Karyotype analyses showed 46,XX,inv(11)(p15q23)[21]. She achieved remission but died of secondary acute lymphoblastic leukemia 3 years later.
Other patients bore the indicated genetic abnormality: NUP, NUP98 fusion gene (see Romana et al9 for details); MLL, MLL-AF10 fusion gene; ITD-MLL, internal tandem duplication; amplification, amplification of the MLL locus; and RUNX1-ETO resulting from the t(8;21)(q22;q22). Signed informed consent was obtained from all patients.
Cytogenetic and molecular methods were as described previously.9 Images were visualized under a Leica DM RXA microscope equipped with a fluorescence epi-illumination 100×/130-0.60 oil-immersion objective lens (Leica). Leica QFISH software was used to digitally acquire images after capturing them with a Photometrics Sensys camera (Roper Scientific). Primers were as follows: MLLex1F, TCGTCTTCGTCATCGTCCTCA; MLLex3R, TCGGTCAGAGCCACTTCTAG; NUP98ex13F, CGGAATCCGATGTCAGACCCTA; and NUP98ex16R, GTATCATCCACACTGTTGCTGT. The identity of amplified fragments was verified by DNA sequence analysis. Expression was normalized with respect to ABL expression.17
Probes were from Applied Biosystems as follows: MEIS1 (Hs00180020_m1), HOXA5 (Hs00430330_m1), HOXA7 (Hs00600844_m1), HOXA9 (Hs00365956_m1), HOXA10 (Hs00538183_m1), and MLL (Hs00610538_m1). All research was approved by the Inserm institutional review board.
Results and discussion
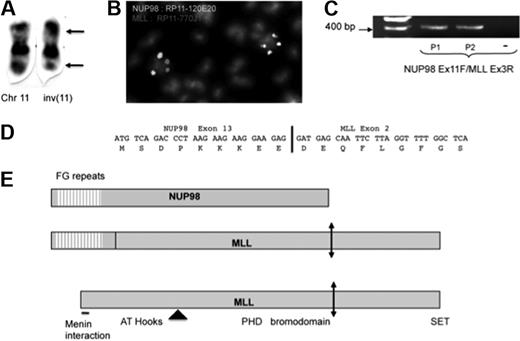
We have identified inv(11)(p15;q23) as a recurrent and isolated chromosomal abnormality in human acute myeloblastic leukemia. Both patients were adults and harbored the inverted chromosome 11 as a unique cytogenetic abnormality (Figure 1A). The location of the breakpoints suggested involvement of the NUP98 and MLL genes, and fluorescence in situ hybridization with specific bacterial artificial chromosome probes confirmed their involvement in both patient 1 (Figure 1B) and patient 2 (data not shown). Using fosmid-derived probes, we next narrowed the location of the breakpoints in patient 1's metaphase chromosomes to the common breakpoint region for NUP98 and to the first or second intron of MLL (data not shown). No gross genomic deletion was observed at either of the chromosomal breakpoints. Reverse transcription–polymerase chain reaction with primers from NUP98 exon 11 and MLL exon 3 allowed detection of a fusion transcript in the cDNA of both patients (Figure 1C). Sequence analyses of the amplified fragments demonstrated an in-frame fusion transcript between exon 13 of NUP98 and exon 2 of MLL in both patients (Figure 1D). The predicted fusion protein, which includes the first 154 amino acids of NUP989,13 and all but the first 142 amino acids of MLL, contains 4340 amino acids (Figure 1E). The predicted reciprocal fusion transcript, between the first exon of MLL and exon 14 of NUP98, was detected in patient 1 but not in patient 2 (data not shown).
Analysis of the inv(11)(p15q23) in 2 patients with acute myeloid leukemia. (A) Partial karyotype of patient 1. Breakpoints are indicated by arrows. Chr indicates chromosome. (B) Fluorescence in situ hybridization on metaphase chromosomes of patient 1 shows split of the NUP98 (green) and MLL (red) loci. The bacterial artificial chromosomes used as probes are indicated on the Figure. (C) Reverse transcription–polymerase chain reaction with primers corresponding to NUP98 exon 11 and MLL exon 3 amplified an NUP98-MLL fusion transcript in samples with inv(11) but not in those from control. (D) Sequence analysis of amplified cDNA showed an in-frame fusion between exon 13 of NUP98 and exon 2 of MLL. (E) Schematic of the wild-type and fusion proteins. Some identified domains are indicated. The location of the majority of the MLL breakpoints is indicated by an arrowhead, and the taspase cleavage site is shown by a double-headed arrow.
Analysis of the inv(11)(p15q23) in 2 patients with acute myeloid leukemia. (A) Partial karyotype of patient 1. Breakpoints are indicated by arrows. Chr indicates chromosome. (B) Fluorescence in situ hybridization on metaphase chromosomes of patient 1 shows split of the NUP98 (green) and MLL (red) loci. The bacterial artificial chromosomes used as probes are indicated on the Figure. (C) Reverse transcription–polymerase chain reaction with primers corresponding to NUP98 exon 11 and MLL exon 3 amplified an NUP98-MLL fusion transcript in samples with inv(11) but not in those from control. (D) Sequence analysis of amplified cDNA showed an in-frame fusion between exon 13 of NUP98 and exon 2 of MLL. (E) Schematic of the wild-type and fusion proteins. Some identified domains are indicated. The location of the majority of the MLL breakpoints is indicated by an arrowhead, and the taspase cleavage site is shown by a double-headed arrow.
We used quantitative reverse transcription–polymerase chain reaction to compare MLL expression in the NUP98-MLL and other MLL-rearranged samples. As shown in Figure 2, fusion to the NUP98 promoter did not result in overexpression of MLL compared with other samples with or without MLL abnormalities. We also evaluated the expression of a subset of HOXA genes and of the MEIS1 gene (Figure 2). Both NUP98-MLL samples showed low expression levels of HOXA genes, comparable with those seen in RUNX1-ETO–positive samples.4
Expression of MLL and HOXA genes. Quantitative reverse transcription–polymerase chain reaction analyses from RNA samples of various acute myeloid leukemia samples with the indicated oncogenic events. Expression was normalized with respect to ABL expression. Note the logarithmic nature of the scales. Patients bore the indicated genetic abnormality. NUP indicates NUP98 fusion gene (see Romana et al9 for details); MLL, MLL-AF10 fusion gene; ITD-MLL, internal tandem duplication; amplification, amplification of the MLL locus; and RUNX1-ETO, runt-related transcription factor 1–821 gene. Because the MLL assay maps at the 3′ end of the transcript, only expression of the untranslocated copy of MLL was detected in samples with MLL fusion.
Expression of MLL and HOXA genes. Quantitative reverse transcription–polymerase chain reaction analyses from RNA samples of various acute myeloid leukemia samples with the indicated oncogenic events. Expression was normalized with respect to ABL expression. Note the logarithmic nature of the scales. Patients bore the indicated genetic abnormality. NUP indicates NUP98 fusion gene (see Romana et al9 for details); MLL, MLL-AF10 fusion gene; ITD-MLL, internal tandem duplication; amplification, amplification of the MLL locus; and RUNX1-ETO, runt-related transcription factor 1–821 gene. Because the MLL assay maps at the 3′ end of the transcript, only expression of the untranslocated copy of MLL was detected in samples with MLL fusion.
Taken together, the present results show that a naturally occurring MLL mutant protein lacking the menin interaction domain is less able than wild-type MLL oncoproteins to maintain active transcription of the HOXA locus but still participates in the transformation of hematopoietic progenitors. The size of the open reading frame in the fusion transcript, 13 kb, did not allow a formal test of the oncogenic properties of this fusion protein with the bone marrow transplantation assay. In addition to MLL amplification and ITD,18 a recent report concerning the oncogenic properties of AF4-MLL, however, supports the notion that the C-terminus of MLL is able to participate in hematopoietic transformation.19
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank N. Dastugue and E. A. Macintyre for some RNA samples, H. Drabkin and T. Mercher for critical reading of the manuscript, and P. D. Aplan for discussion.
This work was supported by grants from Inserm, Association pour la Recherche sur le Cancer, and Institut National du Cancer.
Authorship
Contribution: S.K. performed research, interpreted data, and drafted the manuscript; G.S. performed research and interpreted data; C.B. and C.G. provided vital material; O.A.B. interpreted data and wrote the manuscript; and V.P.-L. and S.P.R. interpreted data and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Olivier Bernard, Inserm U985, Institut Gustave Roussy, PR1, 39 rue Camille Desmoulins, 94805 Villejuif, France; e-mail: olivier.bernard@inserm.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal