Abstract
A total of 143 adult acute myeloid leukemia (AML) patients with available karyotype (K) and FLT3 gene mutational status were assessed for minimal residual disease (MRD) by flow cytometry. Twenty-two (16%) patients had favorable, 115 (80%) intermediate, and 6 (4%) poor risk K; 19 of 129 (15%) carried FLT3-ITD mutation. Considering postconsolidation MRD status, patients with good/intermediate-risk K who were MRD− had 4-year relapse-free survival (RFS) of 70% and 63%, and overall survival (OS) of 84% and 67%, respectively. Patients with good- and intermediate-risk K who were MRD+ had 4-year RFS of 15% and 17%, and OS of 38% and 23%, respectively (P < .001 for all comparisons). FLT3 wild-type patients achieving an MRD− status, had a better outcome than those who remained MRD+ (4-year RFS, 54% vs 17% P < .001; OS, 60% vs 23%, P = .002). Such an approach redefined cytogenetic/genetic categories in 2 groups: (1) low-risk, including good/intermediate K-MRD− with 4-year RFS and OS of 58% and 73%, respectively; and (2) high risk, including poor-risk K, FLT3-ITD mutated cases, good/intermediate K-MRD+ categories, with RFS and OS of 22% and 17%, respectively (P < .001 for all comparisons). In AML, the integrated evaluation of baseline prognosticators and MRD improves risk-assessment and optimizes postremission therapy.
Introduction
In adult acute myeloid leukemia (AML), cytogenetic abnormalities detected at diagnosis represent the most relevant prognostic factors affecting complete remission (CR), overall survival (OS), and relapse-free survival (RFS) rates.1-6 However, there is consistent evidence that the presence of specific gene abnormalities, such as mutations in FLT3 and NPM1 genes, allows to identify, even within homogeneous cytogenetic groups, subsets of patients with distinct treatment outcome.7-10 This is critical in the large intermediate-risk category, which encompasses a very high proportion of cases with no karyotypic abnormalities (40%-50%)11 as well as a variety of different structural and numerical changes that are too infrequent to be reliably assigned a prognostic score and are not covered in favorable or unfavorable groups.1,2 Furthermore, the mutations in the c-KIT gene occurring in the context of the core-binding factor translocations confer a negative prognosis to these otherwise favorable karyotypic abnormalities.12-15
As a consequence of the molecular heterogeneity of cytogenetic categories, the therapeutic strategy for most patients still remains controversial, particularly in the postremission phase, with some authors advocating standard chemotherapy and others recommending autologous (AuSCT) or even allogeneic stem cell transplantation (ASCT).15-20 In light of this uncertainty, the search for alternative biologic parameters to better refine prognosis is warranted to allow clinicians to modulate the intensity of postremission therapy proportionally to disease aggressiveness. In this view, several gene mutations and gene profiling patterns have been described, showing variable impact on clinical outcome.21-26 Although some of these molecular signatures allow a reliable upfront prognostic evaluation in many patients (up to 60% for NPM mutations), the clinical impact of detecting these gene alterations for monitoring minimal residual disease (MRD) in the postremission phase is still under investigation.27-29
Multiparametric flow cytometry (MPFC) has been successfully used to quantify MRD in AML expressing leukemia-associated phenotypes (LAIPs).30-32 Our results in this field suggest that this technique is applicable to the majority of AML patients achieving morphologic CR and that the assessment of MRD status by MPFC after treatment predicts clinical outcome in a consistent manner, especially when measured at the postconsolidation time point.33-36
Based on these findings, it is conceivable that an improved outcome evaluation in AML may emerge from the combination of upfront and delayed prognosticators. As such, the Medical Research Council (MRC) published a robust prognostic scoring system that integrates baseline cytogenetics and the achievement of CR after 1 or 2 cycles of induction therapy.37 This study was designed to incorporate the speed and quality of response into a prognostic algorithm conventionally based on baseline prognostic parameters. Patients with poor-risk karyotype who entered CR after 2 induction courses had, by far, the worst outcome.
In the present study, we analyzed a large group of adult patients with newly diagnosed AML, in whom cytogenetics, molecular genetics, and serial MRD assessments were available. We aimed to verify whether the combination of these determinants would help to optimize risk stratification in this disease.
Methods
A total of 284 consecutive adult patients with de novo non-M3 AML were diagnosed at the Department of Hematology, University Tor Vergata, Rome, during the period 1998 to 2008. Patients were enrolled in the European Organization of Research and Treatment of Cancer/Gruppo Italiano Malattie EMatologiche dell'Adulto (EORTC/GIMEMA) protocols AML-10, AML-12, AML-13, and AML-17. Approval for this study was obtained from the University Tor Vergata Institutional Review Board. Informed consent was obtained from patients in accordance with the Declaration of Helsinki. At presentation, a LAIP was detected by MPFC in 245 of 284 (86%) patients. Of these, 158 of 245 (64%) achieved morphologic CR after induction and were therefore followed up by monitoring MRD. CR rates according to age were 73% and 50% for patients younger and older than 60 years, respectively. Of 158 patients, 143 (90%) had cytogenetic characterization. In this group of 143 patients, molecular status of FLT3 and NPM1 was available in 129 (90%) and 135 (94%) cases, respectively. The present series represents an extension of a cohort of patients already analyzed for different purposes and reported previously.34,36 The EORTC/GIMEMA AML-10/12 trials included patients 18 to 60 years of age. Induction treatment combined cytarabine, etoposide, and an anthracycline, as detailed elsewhere.38,39 As consolidation, patients received cytarabine and an anthracycline; thereafter, those with an human leukocyte antigen-compatible sibling were allografted. Patients without a donor were randomly assigned to peripheral or bone marrow (BM) AuSCT (AML-10) or underwent peripheral-blood AuSCT followed by no further therapy or subcutaneous interleukin-2 maintenance (AML-12). The EORTC/GIMEMA AML-13 and AML-17 included patients older than 61 years; the AML-13 protocol is detailed elsewhere.34,36,40 In the AML-17 protocol, patients received as induction mitoxantrone (7 mg/m2 on days 1, 3, and 5), cytarabine (100 mg/m2 on days 1-7), and etoposide (100 mg/m2 on days 1-3). On achievement of CR, patients received 2 cycles of a consolidation program, consisting of idarubicin (8 mg/m2 on days 1, 3, and 5), cytarabine (100 mg/m2 on days 1-5), and etoposide (100 mg/m2 day 1-3). All patients were randomized before induction, to receive or not, gemtuzumab ozogamicin as a single 2-hour infusion at 6 mg/m2 on days 1 and 15, repeated at 3 mg/m2 on day 1 of each consolidation cycle.
Cytogenetic analysis
A G-banded chromosome study was performed on diagnostic BM samples using standard cytogenetic techniques. Briefly, 2 unstimulated cultures were started in RPMI 1640 medium enriched with 20% fetal calf serum, l-glutamine, and antibiotics (penicillin and streptomycin). The cells were cultured for 24 and 48 hours in a 37°C incubator until harvest. Before harvesting, the cultures were treated with colcemid (0.05 μg/mL) for 16 to 18 hours. Soon after, the cells were exposed to hypotonic solution (0.075M KCl) and fixed with methanol/acetic acid (3:1). Slides of the cells were prepared and stained using a G-banding (trypsin-Giemsa-Wright) technique. Karyotyping was carried out on GTG-banded chromosomes and reported using the ISCN-1995 nomenclature, after analyzing a minimum of 20 metaphases for cases with no clonal aberrations.41 Karyotypic findings were classified according to the MRC criteria.1,2
Fluorescence in situ hybridization
For fluorescence in situ hybridization analysis, we used cytogenetic pellets from direct or overnight cultures of diagnostic BM samples. To screen the set of 8 probes, microscope slides were prepared for each case. The slides were aged for 20 minutes at 80°C on a hot plate and dehydrated at room temperature in 70%, 80%, and 100% ethanol (2 minutes each). The slides were then placed on a hot plate at 37°C, and 5 μL of each probe-buffer solution was applied inside the area of the slide. Probes were prepared following the manufacturer's instructions. The complete set of probes included LSI 5qEGR1/D5S23, LSI D7S486/Cep7, Cep8, LSI 20q13, LSI MLL, LSI BCR/ABL, LSI core-binding factor-β, LSI ETO/AML1, all purchased by Vysis (Olympus LifeScience). When the full probe set was applied, the slides were covered and placed in the Vysis Hybrite machine (Olympus LifeScience). Codenaturation was carried out at 68°C for 5 minutes and hybridization at 37°C overnight. Posthybridization washing was done at 71°C in 0.4 × saline sodium citrate for 2 minutes and at room temperature in 2 × saline sodium citrate for 1 minute. Slides were then counterstained with 4,6-diamidino-2-phenylindole 0.1 μg/mL and analyzed using an Olympus BX61 microscope (Olympus LifeScience) equipped with a 100-W lamp and a complete set of filters. The results of fluorescence in situ hybridization analysis were reported using the ISCN-1995 nomenclature.41
Molecular studies
Total RNA was extracted from Ficoll-Hypaque isolated BM mononuclear cells using standard procedures42 and reverse-transcription using random hexamers as primers. According to our routine laboratory protocol for AML genetic diagnosis, cDNA was used to amplify the most common AML gene fusions as described43 as well as for the mutational analysis of FLT3 and NPM1 genes. For the screening of the internal tandem duplications (ITDs) of FLT3, 2 μL of cDNA was amplified in a final volume of 50 μL of the reaction mixture containing 1 × polymerase chain reaction (PCR) buffer, 200μM of each deoxynucleoside triphosphate, 1.5 U of Taq-Gold DNA polymerase (PerkinElmer), and 30 pmol of each primer. Preheating of the mixture at 94°C for 5 minutes was followed by 35 cycles of 45 seconds at 56°C, 30 seconds at 72°C, and 30 seconds at 94°C. A final extension of 10 minutes was carried out at 72°C on a Gene Amp PCR System 2400 (PerkinElmer Life and Analytical Sciences). With the aim to search FLT3-ITD, we adopted a PCR strategy and primers reported elsewhere.44 The screening of NPM1 mutations was performed using a previously described reverse-transcribed PCR assay followed by capillary electrophoresis.45
Immunophenotypic studies and MRD detection
At diagnosis, immunophenotypic studies were performed with standard techniques, as detailed elsewhere.33-36 Cases with LAIP were selected and reanalyzed by staining with the relevant combinations of antibodies in multiple-color assays4-6 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Once the LAIP was established, it was used to track residual leukemic cells after each treatment step and during subsequent follow-up, as previously reported.33-36 At least 2 antibody combinations were selected in each case to minimize pitfalls because of “phenotypic switches” that have been reported to occur occasionally on relapse.46,47
Outcome definition and statistical analysis
Comparisons between groups were made using the χ2 test or Fisher exact test for categoric data. OS was calculated from the date of entry into the trial to the date of death or last follow-up, RFS was measured from achievement of CR until relapse or death from any cause, and remission duration was measured from the date of CR until the date of relapse.48 Probabilities of OS and RFS were calculated using the Kaplan-Meier method,49 whereas probabilities of cumulative incidence of relapse (CIR) were estimated using the cumulative incidence nonparametric estimator. Univariate comparisons were made using the log-rank test for OS and RFS and the Gray test for CIR, and variables found to be significant at the P less than .1 level were entered into a proportional hazards regression analysis. Multivariate analysis was carried out using the Cox proportional hazards model. Variables were checked for multicolinearity, proportional hazards, and influential or poorly fit subjects using R (http://www.r-project.org/). All quoted P values are from 2-sided tests. Quoted confidence intervals refer to 95% boundaries. The Spearman rank correlation (r) was used to assess the relationship between MRD level at the end of consolidation and time to relapse.
Results
Using the refined MRC cytogenetic classification,1,2 the large majority of patients were in the intermediate-risk group (115 of 143, 80%), whereas 22 of 143 (16%) and 6 of 143 (4%) belonged to good-risk and poor-risk categories, respectively. These 3 subsets were characterized by distinct survival outcomes (supplemental Figure 1A-C).
We evaluated the trend of standardized log-rank statistics50 using OS and RFS as dependent variables and the value of residual leukemic cells, determined at the postconsolidation (postcons) checkpoint, as an independent variable. The level of MRD at the end of the induction therapy was overpowered by the level of MRD at the end of consolidation, in line with our previous data.33-36 The postconsolidation experimental cut-off points, identified as the absolute peak in standardized log-rank statistics plots, were 3.5 × 10−4 and 3.4 × 10−4 residual leukemic cells for OS and RFS, respectively (data not shown). Such a previously established threshold was independently validated in this updated cohort of patients; therefore, 3.5 × 10−4 residual leukemic cells at the end of consolidation therapy was identified as the reference cut-off, in accordance with our previous results.33-36 Finally, the selected threshold was also validated in different age groups (> 60 and < 60 years), retaining in both of them its prognostic value (data not shown).
For the purpose of this study, we stratified the patients with good and intermediate karyotype (K) into 2 categories: those with a level of residual leukemic cells less than 3.5 × 10−4 being referred to as good K-MRD− and intermediate K-MRD−, respectively. Those with levels more than or equal to 3.5 × 10−4 were labeled as good K-MRD+ and intermediate K-MRD+, respectively. At the end of consolidation therapy, 14 of 22 (63%) good-risk and 30 of 115 (26%) intermediate-risk patients tested MRD−, respectively. None of poor-risk patients reached an MRD− status and experienced an early relapse before being addressed to transplantation or additional consolidation.
Table 1 summarizes the clinicobiologic characteristics of patients according to age. A total of 129 and 135 cases were studied for FLT3-ITD and NPM1 exon 12 mutations, respectively. Nineteen of 129 (15%) carried a FLT3-ITD, and 40 of 135 (30%) tested positive for NPM1 exon 12 mutations.
Clinical characteristics of the 143 patients entering the study*
| . | Age > 60 y (n = 40) . | Age < 60 y (n = 103) . | Total (n = 143) . | P . |
|---|---|---|---|---|
| Sex | ||||
| Male | 21 | 59 | 80 | NS |
| Female | 19 | 44 | 63 | |
| WBC | ||||
| Less than 50 × 109/L | 37 | 73 | 110 | NS |
| 50-100 × 109/L | 1 | 21 | 22 | |
| More than 100 × 109/L | 2 | 9 | 11 | |
| FAB | ||||
| M0 | 2 | 11 | 13 | NS |
| M1 | 9 | 21 | 30 | |
| M2 | 13 | 31 | 44 | |
| M4 | 9 | 13 | 22 | |
| M5 | 5 | 26 | 31 | |
| M6 | 2 | 1 | 3 | |
| FLT3 status (n = 129) | ||||
| Wild-type | 31 | 79 | 110 | NS |
| ITD | 5 | 14 | 19 | |
| NPM1 status (n = 135) | ||||
| Wild-type | 27 | 68 | 95 | NS |
| Mutated | 13 | 27 | 40 | |
| Cytogenetics | .014 | |||
| Good-risk | 2 | 20 | 22 | |
| Intermediate-risk | 8 | 24 | 32 | |
| Normal karyotype | 26 | 57 | 83 | |
| Poor-risk | 4 | 2 | 6 |
| . | Age > 60 y (n = 40) . | Age < 60 y (n = 103) . | Total (n = 143) . | P . |
|---|---|---|---|---|
| Sex | ||||
| Male | 21 | 59 | 80 | NS |
| Female | 19 | 44 | 63 | |
| WBC | ||||
| Less than 50 × 109/L | 37 | 73 | 110 | NS |
| 50-100 × 109/L | 1 | 21 | 22 | |
| More than 100 × 109/L | 2 | 9 | 11 | |
| FAB | ||||
| M0 | 2 | 11 | 13 | NS |
| M1 | 9 | 21 | 30 | |
| M2 | 13 | 31 | 44 | |
| M4 | 9 | 13 | 22 | |
| M5 | 5 | 26 | 31 | |
| M6 | 2 | 1 | 3 | |
| FLT3 status (n = 129) | ||||
| Wild-type | 31 | 79 | 110 | NS |
| ITD | 5 | 14 | 19 | |
| NPM1 status (n = 135) | ||||
| Wild-type | 27 | 68 | 95 | NS |
| Mutated | 13 | 27 | 40 | |
| Cytogenetics | .014 | |||
| Good-risk | 2 | 20 | 22 | |
| Intermediate-risk | 8 | 24 | 32 | |
| Normal karyotype | 26 | 57 | 83 | |
| Poor-risk | 4 | 2 | 6 |
Patients were stratified according to refined MRC classification of cytogenetic risk, as follows: “favorable” risk, cases with t(8;21), t(15;17), or inv(16)/t(16;16); “adverse” risk, cases with complex cytogenetic changes (> 3 unrelated abnormalities), −5, add(5q)/del(5q), −7/add(7q), t(6;11), t(10;11), t(9;22), −17, abn(17p) with other changes, 3q abnormalities excluding t(3;5), inv(3)/t(3;3); and “intermediate” risk, cases with normal karyotype and other noncomplex.
WBC indicates white blood cell; FAB, French-American-British; and NS, not significant.
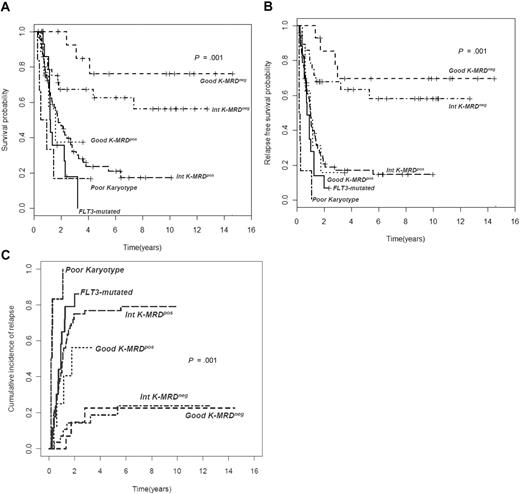
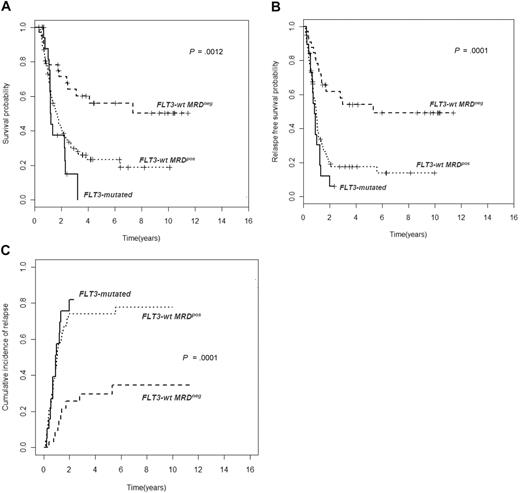
Overall, relapse rate was significantly higher among MRD+ patients (74% vs 27% of MRD−, P < .001); this was also confirmed in continuous variable analysis showing the inverse correlation between the level of MRD after consolidation and time to relapse (Spearman r = −0.5, P < .001). The prognostic role of MRD status at the end of consolidation persisted unaltered when the relapse rate was calculated by breaking down the series according to cytogenetic subsets (Table 2). The distinct outcome of intermediate K-MRD− and intermediate K-MRD+ patients was also clearly outlined in terms of 4-year RFS (63% vs 17%; P < .001), OS (67% vs 23%; P = .002), and CIR (18% vs 77%; P < .001). Similarly, good K-MRD− and good K-MRD+ patients had discrete RFS (70% vs 15%; P = .001), OS (84% vs 38%; P = .006), and CIR (23% vs 56%; P = .07). Consequently, good K-MRD− and intermediate K-MRD− categories shared a very favorable prognosis, whereas good K-MRD+ and intermediate K-MRD+ patients fared as badly as those with poor-risk karyotype (poor-risk K) or FLT3-ITD (Figure 1). The impact of MRD status at the end of consolidation was evident even in specific molecular subsets. Indeed (Figure 2), FLT3 wild-type patients achieving an MRD− status after consolidation experienced a significantly superior outcome compared with those who did not reach MRD negativity in terms of 4-year RFS (54% vs 17%, P < .001), OS (60% vs 23%, P = .002), and CIR (30% vs 74%, P < .001). Likewise, mutated and unmutated NPM1 cases (supplemental Figure 2A-C) had a significantly different OS, RFS, and CIR on the basis of MRD status at the end of consolidation. CIR in NPM1 mutated MRD− was 43% versus 77% of NPM1 mutated MRD+ (P = .09), whereas in NPM1 unmutated MRD− was 28% versus 77% of NPM1 unmutated MRD+ (P < .001). Indeed, the 3 categories of FLT3 wild-type MRD+, NPM1 mutated MRD+, and NPM1 unmutated MRD+ fared as poorly as FLT3-ITD patients (Figure 2; supplemental Figure 2).
Relapse rate in different cytogenetic groups according to the status of MRD at the end of consolidation
| Cytogenetic group . | Relapse . | P . | |
|---|---|---|---|
| No, no. (%) . | Yes, no. (%) . | ||
| Good-risk K-MRD+ | 4 (50) | 4 (50) | |
| Good-risk K-MRD− | 11 (79) | 3 (21) | |
| Intermediate-risk K-MRD+ | 22 (26) | 64 (74) | |
| Intermediate-risk K-MRD− | 23 (79) | 6 (21) | |
| Poor-risk K | 0 (0.0) | 6 (100) | < .001 |
| Cytogenetic group . | Relapse . | P . | |
|---|---|---|---|
| No, no. (%) . | Yes, no. (%) . | ||
| Good-risk K-MRD+ | 4 (50) | 4 (50) | |
| Good-risk K-MRD− | 11 (79) | 3 (21) | |
| Intermediate-risk K-MRD+ | 22 (26) | 64 (74) | |
| Intermediate-risk K-MRD− | 23 (79) | 6 (21) | |
| Poor-risk K | 0 (0.0) | 6 (100) | < .001 |
Clinical outcome in different cytogenetic groups according to MRD status after consolidation. (A) Overall survival (OS), (B) relapse-free survival (RFS), and (C) cumulative incidence of relapse (CIR) of 143 AML patients stratified according to levels of MRD after consolidation. Those with a level of residual leukemic cells less than 3.5 × 10−4 are referred to as intermediate K-MRD− or good K-MRD−, whereas those with levels more than or equal to 3.5 × 10−4 are categorized as intermediate K-MRD+ or good K-MRD+. Survival outcomes of these subsets and those of recognized unfavorable categories, such as poor-risk cytogenetics and FLT3-ITD mutation, are shown (P < .001 for all comparisons).
Clinical outcome in different cytogenetic groups according to MRD status after consolidation. (A) Overall survival (OS), (B) relapse-free survival (RFS), and (C) cumulative incidence of relapse (CIR) of 143 AML patients stratified according to levels of MRD after consolidation. Those with a level of residual leukemic cells less than 3.5 × 10−4 are referred to as intermediate K-MRD− or good K-MRD−, whereas those with levels more than or equal to 3.5 × 10−4 are categorized as intermediate K-MRD+ or good K-MRD+. Survival outcomes of these subsets and those of recognized unfavorable categories, such as poor-risk cytogenetics and FLT3-ITD mutation, are shown (P < .001 for all comparisons).
Assessment of MRD at the end of consolidation splits FLT3 wild-type in 2 categories with different prognoses. Patients carrying FLT3 wild-type who achieve an MRD− status have a significantly better OS (A), RFS (B), and CIR (C) than those who remain MRD+ (OS, P > .001; RFS and CIR, P < .001). The outcome of FLT3 wild-type MRD+ patients is such to replicate the one of those FLT3-ITD+.
Assessment of MRD at the end of consolidation splits FLT3 wild-type in 2 categories with different prognoses. Patients carrying FLT3 wild-type who achieve an MRD− status have a significantly better OS (A), RFS (B), and CIR (C) than those who remain MRD+ (OS, P > .001; RFS and CIR, P < .001). The outcome of FLT3 wild-type MRD+ patients is such to replicate the one of those FLT3-ITD+.
The prognostic variables achieving a statistical significance in univariate analysis (karyotype, FLT3-ITD/poor-risk K, MRD status after induction and consolidation, postremission treatments) were challenged in a multivariate model to determine to what extent they independently affected treatment outcome. The small patient number in the poor-risk K category led us to combine this group together with the intermediate-risk K one, for this analysis. Similarly, we pooled together poor-risk K and FLT3-ITD. Finally, postremission treatment type was included in the analysis to address the role of MRD status after consolidation in predicting outcome in the 3 different therapeutic contexts (chemotherapy vs AuSCT vs ASCT). In this analysis (Table 3), postconsolidation negative MRD status was found to be independently and significantly associated with a longer duration of RFS (P < .001) and OS (P = .001).
Impact of prognostic factors on OS and RFS by univariate and multivariate analysis
| . | Univariate analysis: P . | Multivariate analysis . | ||
|---|---|---|---|---|
| HR . | 95% CI . | P . | ||
| OS | ||||
| MRD postconsolidation | ||||
| Positive vs negative | < .001 | 2.38 | 1.03-5.45 | .04 |
| MRD postinduction | ||||
| Positive vs negative | .014 | 0.96 | 0.43-1.98 | NS |
| Karyotype/FLT3 risk | ||||
| Poor K/FLT3-ITD vs others | .006 | 1.19 | 0.65-1.17 | NS |
| Karyotype (MRC) | ||||
| Good vs intermediate/poor | .006 | 0.56 | 0.30-1.67 | NS |
| Postconsolidation treatment | ||||
| AuSCT vs chemotherapy | .006 | 2.58 | 0.69-9.59 | NS |
| ASCT vs chemotherapy | .035 | 4.16 | 1.25-13.8 | .02 |
| RFS | ||||
| MRD postconsolidation | ||||
| Positive vs negative | < .001 | 2.68 | 1.27-5.67 | .009 |
| MRD postinduction | ||||
| Positive vs negative | < .001 | 1.31 | 0.65-2.66 | NS |
| Karyotype/FLT3 risk | ||||
| Poor K/FLT3-ITD vs others | < .001 | 1.78 | 1.03-3.08 | NS |
| Karyotype (MRC) | ||||
| Good vs intermediate/poor | .007 | 0.79 | 0.37-1.69 | .038 |
| Postconsolidation treatment | ||||
| AuSCT vs chemotherapy | .13 | 2.44 | 1.06-5.64 | .036 |
| ASCT vs chemotherapy | .17 | 0.89 | 0.33-2.42 | NS |
| . | Univariate analysis: P . | Multivariate analysis . | ||
|---|---|---|---|---|
| HR . | 95% CI . | P . | ||
| OS | ||||
| MRD postconsolidation | ||||
| Positive vs negative | < .001 | 2.38 | 1.03-5.45 | .04 |
| MRD postinduction | ||||
| Positive vs negative | .014 | 0.96 | 0.43-1.98 | NS |
| Karyotype/FLT3 risk | ||||
| Poor K/FLT3-ITD vs others | .006 | 1.19 | 0.65-1.17 | NS |
| Karyotype (MRC) | ||||
| Good vs intermediate/poor | .006 | 0.56 | 0.30-1.67 | NS |
| Postconsolidation treatment | ||||
| AuSCT vs chemotherapy | .006 | 2.58 | 0.69-9.59 | NS |
| ASCT vs chemotherapy | .035 | 4.16 | 1.25-13.8 | .02 |
| RFS | ||||
| MRD postconsolidation | ||||
| Positive vs negative | < .001 | 2.68 | 1.27-5.67 | .009 |
| MRD postinduction | ||||
| Positive vs negative | < .001 | 1.31 | 0.65-2.66 | NS |
| Karyotype/FLT3 risk | ||||
| Poor K/FLT3-ITD vs others | < .001 | 1.78 | 1.03-3.08 | NS |
| Karyotype (MRC) | ||||
| Good vs intermediate/poor | .007 | 0.79 | 0.37-1.69 | .038 |
| Postconsolidation treatment | ||||
| AuSCT vs chemotherapy | .13 | 2.44 | 1.06-5.64 | .036 |
| ASCT vs chemotherapy | .17 | 0.89 | 0.33-2.42 | NS |
OS indicates overall survival; RFS, relapse-free survival; HR, hazard ratio; CI, confidence interval; and NS, not significant.
Based on these results, we identified 2 categories of risk with distinct prognosis: (1) low-risk, including good K-MRD− and intermediate K-MRD− with 4-year RFS, OS, and CIR of 58%, 73%, and 15%; and (2) high-risk, including poor-risk cytogenetics, FLT3-ITD mutated cases, good K-MRD+ and intermediate K-MRD+ categories, with RFS, OS, and CIR of 22%, 17%, and 77% (P < .001 for all comparisons; Figure 3). Clinical characteristics and treatment course of the 2 risk groups are summarized in Table 4.
AML risk stratification may be simplified integrating determination of MRD at the end of consolidation, with conventional cytogenetic/genetic classification. Two prognostically distinct groups can be identified: (1) low-risk, including good K-MRD− and intermediate K-MRD− patients; and (2) high-risk, including good K-MRD+, intermediate K-MRD+, poor-risk cytogenetics, and FLT3-ITD cases. The 2 groups stand for significantly different OS (A), RFS (B), and CIR (C) than those who remain MRD+ (P < .001 for all comparisons).
AML risk stratification may be simplified integrating determination of MRD at the end of consolidation, with conventional cytogenetic/genetic classification. Two prognostically distinct groups can be identified: (1) low-risk, including good K-MRD− and intermediate K-MRD− patients; and (2) high-risk, including good K-MRD+, intermediate K-MRD+, poor-risk cytogenetics, and FLT3-ITD cases. The 2 groups stand for significantly different OS (A), RFS (B), and CIR (C) than those who remain MRD+ (P < .001 for all comparisons).
Clinical characteristics of patients classified as high- or low-risk based on a comprehensive risk assessment, including baseline cytogenetics/genetics and status of MRD after consolidation*
| . | High-risk (n = 97) . | Low-risk (n = 32) . | P . |
|---|---|---|---|
| Postconsolidation MRD | |||
| Positive | 96 | 0 | < .001 |
| Negative | 1 | 32 | |
| Age | |||
| Older than 60 y | 32 | 5 | .059 |
| Younger than 60 y | 65 | 27 | |
| FLT3 status | |||
| Wild-type | 78 | 32 | .006 |
| ITD | 19 | 0 | |
| Cytogenetics | |||
| Good-risk | 7 | 10 | .001 |
| Intermediate-risk | 75 | 22 | |
| Poor-risk | 5 | 0 | |
| Postconsolidation therapy | |||
| Chemotherapy | 15 | 2 | .039 |
| AuSCT | 30 | 22 | |
| ASCT | 17 (1 MRD−) | 4 | |
| Reason for not giving postconsolidation therapy | |||
| Relapse | 31 | 3 | .025 |
| Toxicity | 0 | 1 | |
| Medical decision | 1 | 0 | |
| Too early | 3 | 0 |
| . | High-risk (n = 97) . | Low-risk (n = 32) . | P . |
|---|---|---|---|
| Postconsolidation MRD | |||
| Positive | 96 | 0 | < .001 |
| Negative | 1 | 32 | |
| Age | |||
| Older than 60 y | 32 | 5 | .059 |
| Younger than 60 y | 65 | 27 | |
| FLT3 status | |||
| Wild-type | 78 | 32 | .006 |
| ITD | 19 | 0 | |
| Cytogenetics | |||
| Good-risk | 7 | 10 | .001 |
| Intermediate-risk | 75 | 22 | |
| Poor-risk | 5 | 0 | |
| Postconsolidation therapy | |||
| Chemotherapy | 15 | 2 | .039 |
| AuSCT | 30 | 22 | |
| ASCT | 17 (1 MRD−) | 4 | |
| Reason for not giving postconsolidation therapy | |||
| Relapse | 31 | 3 | .025 |
| Toxicity | 0 | 1 | |
| Medical decision | 1 | 0 | |
| Too early | 3 | 0 |
Patients were stratified according to refined MRC classification of cytogenetic risk, as follows: “favorable” risk, cases with t(8;21), t(15;17), or inv(16)/t(16;16); “adverse” risk, cases with complex cytogenetic changes (> 3 unrelated abnormalities), −5, add(5q)/del(5q), −7/add(7q), t(6;11), t(10;11), t(9;22), −17, abn(17p) with other changes, 3q abnormalities excluding t(3;5), inv(3)/t(3;3); and “intermediate” risk, cases with normal karyotype and other noncomplex.
MRD indicates minimal residual disease; ITD, internal tandem duplication; AuSCT, autologous stem cell transplantation; and ASCT, allogeneic stem cell transplantation.
As a further step of analysis, we focused on patients younger than 60 years, submitted to AuSCT or ASCT (56 and 23 patients, respectively) who were followed up for MRD assessment over the posttransplantation period. This analysis was performed with the aim to determine the impact of stem cell transplantation on the outcome of the 2 newly identified risk categories.
Twenty-six of 56 (46%) and 6 of 23 (26%) patients who received AuSCT and ASCT belonged to the low-risk group, respectively. The outcome of these low-risk patients was equally favorable with AuSCT or ASCT (RFS 69% vs 50%, P = .052). Conversely, 30 of 56 (54%) and 17 of 23 (74%) of those who received AuSCT and ASCT belonged to the high-risk group, respectively. In this high-risk category, ASCT conferred a superior outcome than AuSCT (RFS 47% vs 13%, P = .029; supplemental Figure 3). Analysis of transplantation-related mortality demonstrated that 1 of 56 (2%) patients treated by AuSCT died in continuous complete remission because of toxicity, whereas of 23 undergoing ASCT, 5 (22%) died of transplantation-related mortality (P = .003). Overall, 5 of these 6 transplantation-related deaths occurred within the low-risk category.
Discussion
The main practical implication of our approach, which uses MRD assessment by MPFC to implement risk stratification in AML, is that the conventional cytogenetic/genetic classification might be simplified into 2 prognostically defined categories (Figure 3): a low-risk group, including good K-MRD− and intermediate K-MRD− patients; and a high-risk group, including good K-MRD+, intermediate K-MRD+, poor-risk cytogenetics, and FLT3-ITD patients. In this scenario, MRD status at the end of consolidation appears to alter substantially the initial prognosis as dictated by the sole genetic allocation.
Among some new insights in AML prognostic assessment provided by our investigation, we remark the potential to differentiate the large group of patients with intermediate karyotype and wild-type FLT3. This group, conventionally recognized as a good-risk population, shows a very different course of disease depending on patient MRD status at the end of consolidation (Figure 1). Of note, data on the intermediate group and particularly in patients with wild-type FLT3 were available for a substantial number of patients in our study.
Differently from what we observed for good and intermediate karyotype, the role of MRD detection might be of minor impact in categories, such as poor-risk cytogenetics and FLT3-ITD groups for which any further prognostic stratification appears meaningless because of the urgency to address them as early as possible to transplantation procedures or investigational treatment.20,36,51,52 However, it is worth recognizing that, in the present investigation, data related to the poor-risk group, particularly as far as karyotype only is concerned, and those related to the good-risk category were obtained in quite a low number of patients. Therefore, a similar study reproducing our results in larger series is warranted before drawing firm conclusions.
Whether confirmed in further studies, our data might lead to recommend ASCT not only for poor-risk karyotype or FLT3-ITD AML, but also for good, intermediate, and FLT3 unmutated patients not gaining MRD negativity after consolidation, this option being able to provide a superior chance of prolonged RFS.36 On the other hand, based on our findings, the ASCT option seems inappropriate for patients who can experience a long-term survival approaching 70% to 80%, such as those belonging to good K-MRD−, intermediate K-MRD−, and FLT3 unmutated-MRD− categories, who may have their life expectancy jeopardized by the choice of a therapeutic strategy with a disadvantageous risk/benefit ratio. Whereas in poor-risk karyotype or FLT3-ITD AML the choice of very intensive front-line approach is mandatory regardless of MRD status, in good- and intermediate-risk karyotype, measurement of MRD allows to deliver an intensity of cure proportional to the individual risk of relapse and could save additional lives, even without any major advance in chemotherapy or transplantation technologies.20 In this view, whereas baseline cytogenetic/genetic features dictate the intensity of the induction therapy, MRD detection helps to modulate appropriately the intensity of postremissional strategies.
In conclusion, our study suggests that, in AML, the combination of baseline biologic parameters (cytogenetics, molecular biology) and the assessment of the quality of response (MRD determination) enables a better definition of discrete prognostic categories. This approach may potentially allow improved tailoring of postconsolidation therapy aimed at avoiding undertreatment or overtreatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors thank Dr Luca Battistini (Laboratorio di Neuro-Immunologia, Fondazione Santa Lucia, Rome, Italy) for his excellent technical support and critical reading of the manuscript.
Authorship
Contribution: F.B., L.M., S.A., and A.V. conceived and designed the study; D.F., M.I.C., S.L., T.O., E.A., S.C., L.O., and C.S. provided study materials or patients; F.B., L.M., M.I.D.P., P.P., S.C., L.O., and C.S. collected and assembled data; F.B., L.M., A.S., P.B., M.I.D.P., D.F.A., A.D., G.D.P., F.L.-C., V.G., W.A., and A.V. analyzed and interpreted data; F.B., L.M., F.L.-C., and A.V. wrote the manuscript; and A.V. gave final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adriano Venditti, Ematologia, Fondazione Policlinico Tor Vergata, Viale Oxford 81, 00133, Rome, Italy; e-mail: adriano.venditti@uniroma2.it.