Abstract
Active immunization with the idiotype of follicular lymphoma induces tumor-specific immunity. T cells induced in vivo by idiotype vaccination recognize human leukocyte antigen (HLA)–restricted hypervariable but not conserved idiotype peptides. We hypothesized that idiotype-directed T-cell immunity occurs naturally and performed a reverse immunology analysis of idiotype HLA binding in 39 follicular lymphoma patients. For every idiotype, the sum of HLA-A or -B binding scores of the 20 highest-scoring peptides was calculated for all 39 HLA types through the BIMAS algorithm. The idiotype sum score of every patient's lymphoma was compared on the respective patient's HLA type to the mean of the sum scores of the remaining 38 idiotypes. Autologous idiotypes had lower immunogenicity than allogeneic idiotypes. Dif ferential immunogenicity resided predominantly in all 3 complementarity-determining regions rather than in framework peptides. Idiotype immunogenicity was not changed by somatic hypermutation. These findings indicate T cell–mediated immunosurveillance of follicular lymphoma directed specifically against individual idiotype epitopes.
Introduction
The individual composition of potentially immunogenic epitopes of any immunoglobulin is designated as “idiotype.” The idiotype is a unique feature for every B-cell clone and a tumor-specific antigen in B-cell lymphoma. Immunization of patients with follicular lymphoma (FL) against their lymphoma idiotype induces tumor-specific immunity.1-3 When idiotype vaccination is administered as remission consolidation after conventional chemotherapy, humoral immune responses correlate with superior outcome.4-6
Idiotype-specific, major histocompatibility complex (MHC) class I–restricted cytotoxic T cells circulate in lymphoma-bearing patients.7 Immunization with recombinant idiotype Fab fragments induces idiotype-directed T cells in the majority of patients.8,9 In the first trial testing idiotype vaccination as upfront intervention without prior cytoreductive therapy, anti-idiotype T-cell responses correlated with superior progression-free survival and with durable lymphoma remissions.9 Peptide mapping experiments revealed that these T-cell responses are consistently directed against human leukocyte antigen (HLA) class I–restricted peptides from the complementarity-determining regions (CDRs) or against hypermutated idiotype epitopes.8,9 These findings are in accordance with animal studies demonstrating T-cell tolerance for germline-encoded immunoglobulin sequences but responsiveness to novel idiotopes arising from junctional diversity or somatic mutations.10 Because mouse models have also shown T cell–mediated immunosurveillance against hypervariable idiotypic peptides,11 we sought to obtain evidence that spontaneous idiotype-directed T-cell immunity is operational in nonimmunized lymphoma patients and impairs expansion of malignant B-cell clones that express an idiotype with high immunogenicity. To test this hypothesis, we performed a “reverse immunology” bioinformatics study in 39 FL patients.
Methods
Patients
The study was approved by the institutional ethics committee of University Medical Center Freiburg. All patients provided informed consent for biopsies and immunologic studies in accordance with the Declaration of Helsinki. HLA types were identified by serotyping and high-resolution genotyping (Olerup SSP kit; QIAGEN).
Analyses of idiotype sequences
Immunoglobulin heavy- and light-chain cDNAs from FL biopsies were cloned by anchored polymerase chain reaction and sequenced.12 Identification of variable regions, mutational status, and assignment to CDR and framework regions (FRs) were performed by the ImMunoGeneTics/V-QUEST algorithm (www.imgt.cines.fr). Peptides predicted to bind to HLA-A and -B alleles were identified by the BIMAS algorithm (www.bimas.cit.nih.gov/molbio/hla_bind). The sum of the binding scores of the 20 highest ranking peptides served as a measure for the theoretical idiotype HLA-binding capacity and was calculated for all idiotypes and all 39 HLA types. For every patient's HLA type, the idiotype sum scores of his/her lymphoma (“autologous” score) was compared with the mean of the sum scores on the remaining 38 idiotypes (“allogeneic” scores) in a matched-pair analysis with Wilcoxon signed-rank test (Prism 5.02; GraphPad Software). For region-specific analyses, peptides containing at least 2 amino acids assigned to a CDR were defined as CDR epitopes, the remaining as FR peptides.
Results and discussion
The VH usage (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) did not differ significantly from reported series.13 Except for an overrepresentation of the rare allele HLA-A68 (P < .01; 2-sided χ2 test with Bonferroni adjustment), the HLA allele distribution was representative for the German population.14,15
A median of 3 (range, 1-4) HLA alleles per patient could be analyzed in BIMAS. The median score of the highest-ranking peptide of each idiotype was 69.6 (range, 4-2000). Scores more than 320 occurred exclusively for HLA-B2705 (2 patients). Median scores for peptides with rank 15 and 20 were 2.0 and 1.1, respectively. Therefore, calculation of sum scores from the 20 highest ranking peptides reliably included all relevant epitopes. Most of these bona fide peptides are expected to physically bind to HLA and to serve as functional T-cell epitopes.8,9,16-18
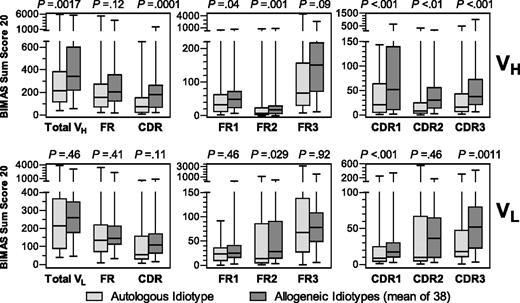
In a matched-pair comparison of the idiotype BIMAS sum scores, the VH region of an affected patient's own lymphoma had lower HLA-binding scores than allogeneic idiotypes (Figure 1). No such global difference was observed for idiotype VL regions.
BIMAS sum scores of the 20 highest-scoring peptides located in the indicated parts of VH and VL sequences from idiotypes of 39 follicular lymphomas. Horizontal bars represent median. Boxes represent 25%/75% quartiles. Whiskers represent range. Sum scores of autologous and the mean of 38 allogeneic idiotypes were compared by Wilcoxon signed-rank test.
BIMAS sum scores of the 20 highest-scoring peptides located in the indicated parts of VH and VL sequences from idiotypes of 39 follicular lymphomas. Horizontal bars represent median. Boxes represent 25%/75% quartiles. Whiskers represent range. Sum scores of autologous and the mean of 38 allogeneic idiotypes were compared by Wilcoxon signed-rank test.
Additional analyses were performed to assign differential idiotype immunogenicity to FR and CDR. Because immunogenicity of HLA-presented peptides may depend on a single nonanchor residue, we considered peptides as CDR-derived if at least 2 amino acids were assigned to a CDR. With this definition, the cumulative sequence length that could harbor CDR or FR epitopes was relatively balanced (VH: FR, 56%; CDR, 44%; VL: FR, 58%; CDR, 42%). As described before,18 the majority of peptides with high HLA-binding scores were located in FR: only 35% and 25% of all peptides with scores more than 10 and more than 50, respectively, were CDR-derived. Despite higher FR sum scores, weaker HLA binding of autologous idiotypes was dependent on hypervariable sequences, and each CDR contributed significantly to this difference (Figure 1). The same phenomenon was observed for VL CDR 1 and 3. Regarding FR, a highly significant difference between autologous and allogeneic sequences was found only for VH FR2.
Animal studies have demonstrated immunosurveillance against MHC class II–restricted idiotype epitopes.10,11 The available in silico tools permit only limited analyses for human MHC class II. Therefore, our study had to focus on HLA class I. The results indicate natural selection against FL idiotypes with relatively high HLA-binding capacity by T cell–mediated immunosurveillance.
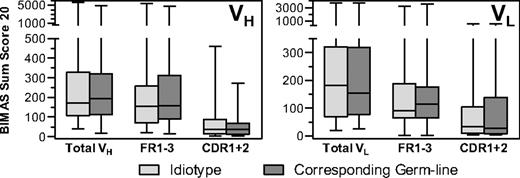
Hypermutated FL idiotypes and corresponding germline VH and VL segments had similar HLA-binding sum scores (Figure 2). We cannot ascertain absolute identity of the germline sequences deposited in the ImMunoGeneTics database with the individual patients' V genes. Despite this caveat, the dominant influence of the hypervariable regions on differential HLA binding scores was apparently unaltered by somatic hypermutation, a process constitutively activated in FL.19,20 Therefore, immunosurveillance appears to take effect in suppressing FL cells with idiotypes that acquire increased immunogenicity by random mutations.
BIMAS sum scores of the 20 highest-scoring peptides located in the indicated parts of VH and VL sequences (excluding CDR3) from idiotypes of 39 follicular lymphomas and their corresponding germline sequences. Horizontal bars represent median. Boxes represent 25%/75% quartiles. Whiskers represent range. Sum scores of idiotypes and corresponding germline sequences were compared by Wilcoxon signed-rank test. No significant differences were observed.
BIMAS sum scores of the 20 highest-scoring peptides located in the indicated parts of VH and VL sequences (excluding CDR3) from idiotypes of 39 follicular lymphomas and their corresponding germline sequences. Horizontal bars represent median. Boxes represent 25%/75% quartiles. Whiskers represent range. Sum scores of idiotypes and corresponding germline sequences were compared by Wilcoxon signed-rank test. No significant differences were observed.
If the immune system suppresses FL cells expressing idiotypes with relatively high immunogenicity, the idiotype of a clinically manifest lymphoma is apparently ignored. Cells carrying the FL-associated t(14;18) circulate in healthy persons over extended time periods.21,22 Our results permit speculation that idiotype-specific T cells contribute to prevent malignant outgrowth of these B-cell clones.
Immunosurveillance of idiotypes appears to be predominantly dependent on individual, CDR-derived epitopes despite their weaker predicted HLA binding compared with FR peptides. Consistent with superior HLA binding, FR peptides that are shared by numerous B-cell clones have been shown to function as epitopes that are efficiently recognized by cytotoxic T cells in vitro.18 However, these experiments were performed with isolated CD8+ T cells. When we compared the functional immunogenicity of a unique MHC class I–restricted CDR3 peptide with a widely shared FR4 epitope after immunization of syngeneic mice in vivo, only the unique CDR3 peptide induced a robust T-cell response.23 In contrast, the FR4 peptide elicited a strong regulatory T-cell response that systemically suppressed antigen-specific responses of T-cell receptor–transgenic cytotoxic T cells in vivo.
The combined available evidence therefore indicates that T-cell responses to potentially immunogenic FR peptides are controlled by peripheral tolerance in vivo. FL cells themselves can induce a regulatory phenotype on conventional T cells in situ.24,25 This effect could potentially enhance peripheral tolerance to shared and individual idiotype epitopes. Therefore, the efficacy of idiotype-directed T-cell immunity presumably depends on the balance between the immunogenicity of individual idiotype epitopes and on the extent of regulatory T-cell activity in the local immune environment.
This scenario suggests therapeutic potential of idiotype vaccination beyond the widely accepted role of humoral anti-idiotype immunity.4,5 Therapeutic idiotype vaccination should be tailored to shift the balance of idiotype-directed T-cell immunity from ignorance to effective recognition of tumor-individual epitopes, as we have demonstrated for T-cell responses to recombinant idiotype vaccination as upfront intervention in FL patients.9
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
Due to a software malfunction, some P values in Figure 1 were incorrect in the First Edition version of this article. These errors have been corrected in the print version.
Acknowledgment
The authors thank Heide Dierbach for expert technical assistance.
Authorship
Contribution: A.-M.S., D.P., M.D.-v.M., and M.N. performed idiotype analysis, polymerase chain reaction HLA typing, and statistical analyses; K.Z. and K.H.-M. recruited patients and provided clinical data and serologic HLA data; and H.V. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hendrik Veelken, Department of Hematology, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: j.h.veelken@lumc.nl.