Abstract
We have identified a postentry CCR6-dependent mechanism of inhibition of HIV occurring at an early stage of infection mediated by the induction of the host restriction factor apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G). We observed induction of APOBEC3G expression only in CCR6+ cells but not in cells treated with the G inhibitory (Gi) pathway inhibitor pertussis toxin. CCR6 is highly expressed on peripheral blood CD4+CCR5+ memory T cells and by 2 populations of CD4+ T cells within the gut, α4β7+ and T helper type 17, that have been implicated in cell-to-cell spread of HIV and enhanced restoration of CD4+ T cells within gut-associated lymphoid tissue, respectively. This novel CCR6-mediated mechanism of inhibition allows the identification of pathways that induce intrinsic immunity to HIV, which could be useful in devising novel therapeutics that selectively target CCR6+ cells.
Introduction
The mucosal immune system is the initial physical and immunologic barrier that HIV encounters. During the acute phase of HIV infection, the gut-associated lymphoid tissue is a site of significant viral replication and rapid depletion of CD4+ T cells.1 Complete immune reconstitution does not occur in the gut even after initiation of highly active antiretroviral therapy.2,3 The chemokine receptor CCR6 is of crucial importance in mucosal immunity whereby it mediates mucosal homeostatic and inflammatory conditions.4 Lack of CCR6 in mice is associated with intestinal lymphoid structure defects, suggesting that it is required in the development of lymphoid tissues.5,6 The cognate ligand for CCR6, macrophage inflammatory protein (MIP)–3α (CCL20), is chemotactic for immature dendritic cells (DCs), effector/memory T cells, and B cells.4,7 In addition to MIP-3α, human β-defensin 2 (hBD2) is chemotactic for memory T cells, immature DCs, and tumor necrosis factor-α–treated neutrophils via CCR6.8,9 CCR6 is expressed on cell populations implicated in HIV infection, including DCs and effector/memory CD8+ and CD4+ T cells.10-12 CCR6+ memory T cells are selectively depleted from peripheral blood during HIV disease progression of which the majority of CD4+ T cells that express the HIV coreceptor CCR5 also express CCR6.13 The differential depletion of specific subsets of T cells may contribute to disease progression.1,2
CCR6 is also expressed on 2 populations of CD4+ T cells in the gut, CD4+α4β7+ and T helper type 17 (TH17) cells, that are highly relevant to HIV infection. CD4+ memory T cells within the lamina propria of the gut coexpress CCR6 and the gut homing receptor α4β7,11 which has been implicated in the spread of HIV in the gut.14 TH17 cells are an important component of mucosal immune defenses against bacteria and fungi, and CCR6 is expressed on all interleukin-17 (IL-17)–producing TH17 cells and is critical for TH17 cell homing to Peyer patches.15,16 TH17 cells are depleted from the gut in persons infected with HIV and in pathogenic simian immunodeficiency virus rhesus macaque models of infection.17 In HIV infection, the presence of mucosal TH17 cells is negatively correlated with blood viremia and positively correlated with enhanced restoration of CD4+ T cells in the gut.18,19 However, in nonpathogenic simian immunodeficiency virus sooty mangabey models of infection, TH17 cells within the gut are depleted during the acute phase of infection but are subsequently restored, suggesting this subset of cells may play a role in disease progression.17 Interestingly, IL-17 is a potent inducer of both MIP-3α and hBD2 in epithelial cells.20,21
The hBDs 1 to 3 are of particular interest in mucosal immunity because they are expressed by epithelial cells in mucosae, where they form an antimicrobial barrier and contribute, by their interaction with CCR6, to the homeostasis of lymphoid compartments.22-24 Defensins exert their antimicrobial activity on a broad range of Gram-positive and -negative bacteria, fungi, and enveloped and nonenveloped viruses.25,26 We and others have shown that hBD2 and hBD3 possess HIV-inhibitory activity.27,28 From a mechanistic viewpoint, our studies show that one component of the HIV-inhibitory activity of hBD2 results in lower infectivity of HIV virions, although we did not detect inhibition of env-mediated fusion.27,28 A second component of the HIV-inhibitory activity of hBD2 is imparted on viral target cells. In serum-free conditions, hBD2 and hBD3 have been reported to bind and down-regulate CXCR4, whereas we did not detect down-regulation in the presence of serum.27,28 This observation, however, does not account for the broad HIV-inhibitory activity of hBD2 and hBD3, which affects both X4 and R5 isolates, and for the intracellular activity of hBD2 observed in the presence of serum without modulation of CXCR4 or CCR5.28 We hypothesized that this intracellular component of inhibition by hBD2 could be due to an interaction with its known receptor, CCR6.
We report that CCR6 mediates an intracellular mechanism of inhibition of HIV and that the inhibitory activity depends on the induction of the host restriction factor apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G). APOBEC3G is a member of the apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like family of cytidine deaminases. Identified as a viral infectivity factor (vif)–sensitive anti-HIV factor,29 APOBEC3G inhibits HIV by lethal editing of nascent viral reverse transcripts30 and by the inhibition of reverse transcription.31,32 Thus, protecting CCR6+ cells from infection might protect cell populations that are highly susceptible to HIV infection, including TH17, memory, and α4β7 T cells, contributing to the preservation of an effective immune response.
Methods
Isolation and culture of primary cells and cell lines
Human peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of healthy human subjects with the use of Histopaque (Sigma-Aldrich). CD4+ T cells (purity of > 95% when analyzed by flow cytometry) were isolated from resting PBMCs with the use of the using CD4+ T Cell Isolation Kit II (Miltenyi Biotec). PBMCs and CD4+ T cells were maintained in complete RPMI-1640 media. Activated PBMCs and CD4+ T cells were obtained by stimulation with 10 ng/mL IL-2 and 5 μg/mL phytohemagglutinin for 48 hours. Activated PBMCs were maintained in complete RPMI-1640 media and 10 ng/mL IL-2 at a density of less than 2 × 106 cells/mL. JKT-FT7 and JKT-FT7 CCR6 GFP cells, derivatives of the CD4+ T lymphoblastoid cell line Jurkat, were maintained in complete RPMI-1640 media.33
Virus production
HIVIIIB (X4 strain) was prepared from acutely infected PM1 cells grown in complete RPMI-1640 media. HIVBaL (R5 isolate) was prepared from monocyte-derived macrophages in RPMI-1640 media/human AB serum. Pseudotyped virions were generated by cotransfection of 293T cells with pNL-4.3R−E− Luc plasmid and an amphotropic murine leukemia virus (AMLV) envelope-expressing plasmid, using Fugene 6 (Roche Diagnostics) and reagents kindly provided by Dan R. Littman (New York University).34
Infectivity assays
Activated PBMCs (105 PBMC/well) or JKT-FT7 and JKT-FT7 CCR6 GFP (0.5 × 106 cells/well) were infected for 2 hours with 100 TCID50 of HIVBaL, a R5 isolate, or HIVIIIB, an X4 isolate. After 2 hours, cells were washed with phosphate-buffered saline (PBS), and complete media was added with the appropriate treatment. Infection was monitored by p24 enzyme-linked immunoabsorbent assay (ELISA) with the use of a commercially available kit (PerkinElmer Life and Analytical Sciences). Single-round infections were performed with the use of AMLV envelope-pseudotyped HIV. PBMCs in complete media and 10 ng/mL IL-2 were untreated or pretreated with 500 ng/mL pertussis toxin (PTx; Calbiochem). Cells were incubated with pseudotype virus for 3 hours and subsequently washed with PBS and then incubated in the presence or absence of hBD2 or MIP-3α for 3 days at 37°C. Infected cells were collected, washed with PBS, and lysed with Reporter Lysis Buffer (Promega), and luciferase activity was measured in a Turner Luminometer. Percentage of inhibition in treated cells was calculated with the use of the following formula: [1 − (p24treated/p24control)] × 100, where p24treated and p24control are concentrations of HIV p24 measured in supernatants of treated and untreated cells, respectively. An analogous formula was used to calculate the percentage of inhibition in experiment measuring luciferase expression from HIV pseudovirions.
Quantitative PCR
Activated PBMCs (106 cells/time point) were untreated or pretreated with azidothymidine (AZT) or T20 and infected for 2 hours at 37°C with 300 TCID50 of DNase I–treated HIVIIIB or HIVBaL. Cells were trypsin treated, washed with PBS to remove extracellular virus, and resuspended in complete media and IL-2 (10 ng/mL), and infected untreated cells were incubated in the presence or absence of hBD2 (0.8-20 μg/mL). Total cellular DNA was extracted with the use of the DNeasy Blood and Tissue Kit (QIAGEN) at 4, 8, and 24 hours. DNA was analyzed by real-time quantitative polymerase chain reaction (PCR) with iQSYBR green supermix (Bio-Rad) with the use of primers specific for Strong Stop cDNA (5′-GGCTAACTAGGGAACCCACTG-3′ [sense] and 5′-CTGCTAGAGATTTTCCACACTGAC-3′ [antisense]). Reactions were performed in triplicate with the use of a Bio-Rad iQ5 Real-Time PCR machine with number of copies calculated with dilutions of HxB2 plasmid DNA.
Total chemical synthesis of defensins and MIP-3α
hBD2 and hBD3, human α-defensin 1 (HNP1), and MIP-3α were chemically synthesized by solid-phase peptide synthesis with the use of a custom-modified procedure tailored from the previously published in situ neutralization protocol developed for Boc chemistry.35 The β connectivity of 3 disulfide bonds (Cys1-Cys5, Cys2-Cys4, Cys3-Cys6) in highly pure synthetic hBD2 was independently verified by mass mapping of peptide fragments generated by enzymatic digestion and Edman degradation. Correct folding of synthetic MIP-3α was demonstrated by the solution of its high-resolution x-ray crystal structure. Protein concentrations were determined by absorbance measurements at 280 nm with the use of molar extinction coefficients.
Cell metabolism and proliferation assays
Cell metabolism was determined by MTS assay (Promega). Cells (105 cells/well) were cultured in triplicate in 96-wells plates with media alone as a control or experimental condition, in triplicate, and then the MTS/PMS mixture was added and incubated 1 to 4 hours before spectrophotometric absorbance reading at 490 nm. OD ratios of treated/control cells were calculated as percentages. Cell proliferation was determined by 3H-thymidine uptake as described.36
Immunoblotting
Cells were lysed with buffer containing 50mM HEPES, pH 7.4, 125mM NaCl, 0.2% NP-40, 0.1mM PMSF, and 1× EDTA-free protease inhibitor cocktail (Calbiochem). Total protein concentration was determined by DC Protein Assay (Bio-Rad), and equal amounts of total protein were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis. The primary antibodies included anti-APOBEC3G rabbit antisera (National Institutes of Health National Institutes of Health AIDS Research and Reference Reagent Program) and mouse anti–β-actin (Abcam Inc).
siRNA knockdown of APOBEC3G
Small interfering RNAs (siRNAs) targeting APOBEC3G messenger RNA, previously described37 (A3G, 5′-GCAUCGUGACCAGGAGUAUdTdT-3′), were chemically synthesized by QIAGEN. A mutant APOBEC3G siRNA (A3G mutant, 5′-GCAUCGUGCACAGGAGUAUdTdT-3′) containing a 2-nucleotide mismatch (italics) was used as a negative control. CD4+ T cells were transfected with the siRNA with the use of an AMAXA nucleofector apparatus (program V-024; 2 μg of siRNA per 5 × 106 cells).
Online supplemental material
Chemotaxis assay.
The in vitro chemotaxis was performed with the ChemoTx System (Neuro Probe no. 101-5) 96-well cell migration system. Thirty microliters of JKT-FT7 or JKT-FT7 CCR6 GFP cells (cells suspended in RPMI-1640 with 1% fetal bovine serum at 106 cells/mL) were added to the upper filter with a pore size of 5 μm and the indicated chemoattractants in the lower wells. The cells were incubated at 37°C and 5% CO2 for 2 hours. The results were presented as the mean chemotactic index, defined as the fold increase in the number of migrated cells in the presence of chemotactic factors over cell migration in the presence of media alone (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Quantitative PCR.
JKT-FT7 and JKT-FT7 CCR6 GFP cells (0.5 × 106 cells/time point) were untreated or pretreated with AZT and infected for 2 hours at 37°C with DNase I–treated HIVIIIB at a multiplicity of infection of 0.001. Cells were trypsin-treated, washed with PBS to remove extracellular virus, and resuspended in complete media, and infected untreated cells were incubated in the presence or absence of hBD2 (20 μg/mL) or MIP-3α (5 μg/mL). Total cellular DNA was extracted with the use of the DNeasy Blood and Tissue Kit (QIAGEN) at 4, 8, and 24 hours and analyzed by real-time quantitative PCR with iQSYBR green supermix (Bio-Rad). Reactions were performed in triplicate in parallel with sets of known quantitative standards for HIV DNA and albumin. HIV DNA was quantified with the use of the following primers: 5′-GGCTAACTAGGGAACCCACTG-3′ (sense) and 5′-CTGCTAGAGATTTTCCACACTGAC-3′ (antisense), and the number of copies were normalized to copies of albumin as a reference with the following primers: 5′-TGTTGCATGAGAAAACGCCA-3′ (sense) and 5′-GTCGCCTGTTCACCAAGGAT-3′ (antisense).
Results
hBD2 inhibits HIV replication after entry
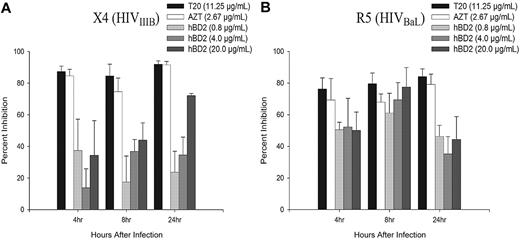
In addition to reduced infectivity, our previously published data obtained performing time-of-addition experiments suggested an intracellular mechanism of inhibition of HIV by hBD2.28 We did not observe down-regulation of CXCR4 or CCR5 in PBMCs treated with hBD228 or CD4+ T cells treated with hBD2 or MIP-3α (supplemental Figure 1). To gain insight into the intracellular mechanism of inhibition, the presence of early reverse transcription products in PBMCs infected with either X4 (IIIB) or R5 (BaL) HIV isolates were measured by quantitative PCR. PBMCs were infected, as described in “Infectivity assays,” and treated after infection with 0.8 to 20 μg/mL hBD2 (which is well within the range of concentrations of hBD2 measured in oral mucosa and epidermal tissue38,39 ) and compared with infected untreated samples and samples pretreated with the reverse transcriptase inhibitor AZT at 2.67 μg/mL or the fusion inhibitor T20 at 11.25 μg/mL. As shown in Figure 1, treatment with hBD2 after infection inhibited the accumulation of early (strong stop) reverse transcription products of both X4 (Figure 1A) and R5 (Figure 1B) HIV-infected cells at 4, 8, and 24 hours after infection. Pretreatment with AZT or T20 inhibited both X4 and R5 HIV.
hBD2 inhibits the accumulation of early reverse transcription products of HIV. PBMCs were infected with DNase I–treated X4 HIVIIIB (A) or R5 HIVBaL (B). After virus removal and washing, hBD2 (0.8-20 μg/mL) was added to the tissue culture media. Cells were pretreated with AZT (2.67 μg/mL) or T20 (11.25 μg/mL) as controls. Total cellular DNA was isolated at the indicated time points, and the presence of early reverse transcription products was determined by real-time PCR. Triplicate readings were averaged, and inhibition was determined as a percentage of the number of copies of early reverse transcription products in infected treated cells in reference to copies measured in untreated control infections. Shown here are mean percentage of inhibition (± SEM) of 4 independent experiments that used cells from different donors.
hBD2 inhibits the accumulation of early reverse transcription products of HIV. PBMCs were infected with DNase I–treated X4 HIVIIIB (A) or R5 HIVBaL (B). After virus removal and washing, hBD2 (0.8-20 μg/mL) was added to the tissue culture media. Cells were pretreated with AZT (2.67 μg/mL) or T20 (11.25 μg/mL) as controls. Total cellular DNA was isolated at the indicated time points, and the presence of early reverse transcription products was determined by real-time PCR. Triplicate readings were averaged, and inhibition was determined as a percentage of the number of copies of early reverse transcription products in infected treated cells in reference to copies measured in untreated control infections. Shown here are mean percentage of inhibition (± SEM) of 4 independent experiments that used cells from different donors.
CCR6 enhances the HIV-inhibitory activity of hBD2 on spreading infections in a CD4+ T lymphoblastoid cell line
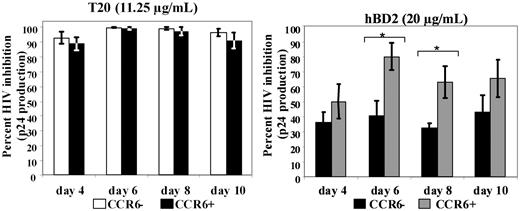
We hypothesized that the intracellular mechanism of inhibition could be due to receptor interaction. Because the only receptor known for hBD2 is CCR6, we conducted experiments to ascertain the role of this receptor in the inhibitory activity. We reasoned that the inhibitory effect of hBD2 would not be due exclusively to intracellular effects, because the spreading (budding) virus would also be subject to the intrinsic inhibitory activity that we and others observed on HIV virions.27,28 However, we expected that CCR6 expression would eventuate in more potent HIV inhibition by hBD2. To characterize the role of CCR6 in HIV inhibition, we used the Jurkat-derived CD4+ T lymphoblastoid cell line JKT-FT7, which expresses low or undetectable CCR6, and JKT-FT7 CCR6 GFP, which is stably transfected with an expression vector encoding for CCR6 and expresses high levels of this receptor.33 Both expression level and functionality of CCR6 were tested by flow cytometry and chemotaxis assays, confirming the expected pattern of CCR6 surface expression (supplemental Figure 2A) and chemotaxis to MIP-3α and hBD2 (supplemental Figure 2B-C). JKT-FT7 and JKT-FT7 CCR6 GFP were infected with X4 (IIIB) HIV, as described in “Infectivity assays.” Cells were pretreated with T20 as a control or treated after infection with 0.8 to 20 μg/mL hBD2, and the expression of HIV p24 in tissue culture supernatants was monitored by ELISA up to day 10 after infection. Percentage of inhibition with each treatment was calculated in reference to infected untreated controls. We did not observe any difference in HIV inhibition by T20 between CCR6+ and CCR6low/no JKT-FT7 cells (Figure 2). However, when hBD2 was used at the dose of 20 μg/mL, HIV inhibition was higher in CCR6+ cells than in CCR6low/no cells (Figure 2). The difference in the percentage of inhibition among the 2 variants of JKT-FT7 cells was statistically significant at day 6 and day 8 and approached significance on day 4. Differences in the percentage of inhibition of HIV were not statistically significant when hBD2 was used at lower doses (data not shown). These results are consistent with our dual mechanism model of the inhibitory activity of hBD2, targeting HIV by both an extracellular and a postentry mechanism. No significant difference in proliferation or cellular metabolism was observed among hBD2 treatment in both cell lines as determined by tritiated thymidine uptake and MTS assays (data not shown).
Role of CCR6 in mediating the antiviral activity of hBD2 in a spreading infection. CD4+ T lymphoblastoid cell lines JKT-FT7 (CCR6low/no) and JKT-FT7 CCR6 GFP (CCR6+) were infected with X4 (IIIB) HIV. After infection, cells were treated with T20 (11.25 μg/mL; left) or with hBD2 (20 μg/mL; right). At the indicated time points, HIV p24 in tissue culture supernatant was quantified by ELISA, and the percentage of inhibition was calculated in reference to measured p24 in untreated control infections. Shown here are average percentages of inhibition values (± SEMs) of 4 independent experiments. *P < .05 (2-tailed t test) between homologous treatment groups for CCR6low/no and CCR6+ cells.
Role of CCR6 in mediating the antiviral activity of hBD2 in a spreading infection. CD4+ T lymphoblastoid cell lines JKT-FT7 (CCR6low/no) and JKT-FT7 CCR6 GFP (CCR6+) were infected with X4 (IIIB) HIV. After infection, cells were treated with T20 (11.25 μg/mL; left) or with hBD2 (20 μg/mL; right). At the indicated time points, HIV p24 in tissue culture supernatant was quantified by ELISA, and the percentage of inhibition was calculated in reference to measured p24 in untreated control infections. Shown here are average percentages of inhibition values (± SEMs) of 4 independent experiments. *P < .05 (2-tailed t test) between homologous treatment groups for CCR6low/no and CCR6+ cells.
Postentry inhibition of HIV by hBD2 is mediated by CCR6
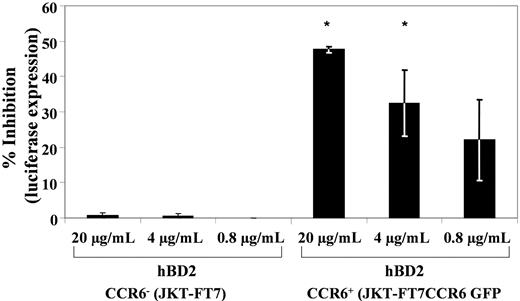
To show that the postentry inhibition is mediated by CCR6, 2 approaches were used to eliminate the effect of inactivation of budding virus. First, JKT-FT7 and JKT-FT7 CCR6 GFP cells were infected with X4 (IIIB) HIV, as described in “Infectivity assays,” and treated after infection with either hBD2 (20 μg/mL) or MIP-3α (5 μg/mL). Infections were measured at early time points (4, 8 and 24 hours after infection) by quantitative PCR for copies of early reverse transcription products normalized to copies of albumin. Shown in supplemental Figure 3, treatment with either hBD2 or MIP-3α resulted in reduced copies of early reverse transcription products in the JKT-FT7 CCR6+ cells but not in the CCR6low/no JKT-FT7 cells. Treatment with AZT (2.67 μg/mL) resulted in similar reduced levels of early reverse transcripts in both cell lines. Second, a single-cycle infection assay was used with the HIV luciferase virus pseudotyped with AMLV envelope that does not use CXCR4 or CCR5 for entry. JKT-FT7 and JKT-FT7 CCR6 GFP cells were infected in triplicate for 3 hours, and cells were washed 3 times and incubated 3 days with concentrations of hBD2 ranging from 0.8 to 20 μg/mL. Subsequently, luciferase activity was measured, relative light units were averaged, and the percentage of inhibition compared with infected untreated cells was calculated per each treatment group along with the statistical significance of differences among treatment groups (Figure 3). In the CCR6low/no JKT-FT7 cells, hBD2 treatment did not result in detectable inhibition of infection, as measured by luciferase activity. In contrast, hBD2 treatment of infections in the JKT-FT7 CCR6+ cells at doses of 20, 4, and 0.8 μg/mL resulted in an average inhibition of luciferase expression of 47.6%, 32.4%, and 22.1%, respectively. Treatment of AMLV pseudotypes under the same conditions with AZT (2.67 μg/mL) resulted in inhibitory activity greater than 75% (data not shown).
CCR6 is necessary to the intracellular HIV-inhibitory activity of hBD2. JKT-FT7 (CCR6low/no) and JKT-FT7 CCR6 GFP (CCR6+) cells were infected with HIV-luciferase pseudotyped with AMLV envelope. After infection, cells were incubated 3 days in complete RPMI, with or without hBD2 (20 μg/mL). Subsequently, cells were washed and lysed, and luciferase activity was measured. The percentage of inhibition was calculated in reference to luciferase activity as measured in untreated control infections. Shown here are the average percentage of inhibition values (± SEMs) of 3 independent experiments. *P < .05 (2-tailed t test) between homologous treatment groups for CCR6low/no and CCR6+ cells.
CCR6 is necessary to the intracellular HIV-inhibitory activity of hBD2. JKT-FT7 (CCR6low/no) and JKT-FT7 CCR6 GFP (CCR6+) cells were infected with HIV-luciferase pseudotyped with AMLV envelope. After infection, cells were incubated 3 days in complete RPMI, with or without hBD2 (20 μg/mL). Subsequently, cells were washed and lysed, and luciferase activity was measured. The percentage of inhibition was calculated in reference to luciferase activity as measured in untreated control infections. Shown here are the average percentage of inhibition values (± SEMs) of 3 independent experiments. *P < .05 (2-tailed t test) between homologous treatment groups for CCR6low/no and CCR6+ cells.
CCR6 ligand MIP-3α inhibits HIV replication
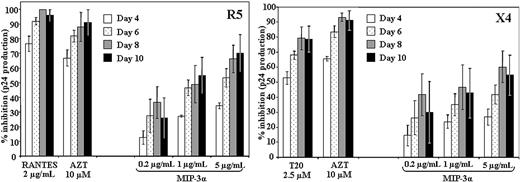
On the basis of the requirement of CCR6 in hBD2-mediated intracellular inhibition, we next tested whether MIP-3α, the cognate ligand of CCR6, has HIV-inhibitory activity.40 To this end, we tested concentrations between 1 and 5 μg/mL MIP-3α in PBMC infectivity assays, adding MIP-3α after infection. We observed a dose-dependent and significant inhibition of HIV replication with both R5 (BaL) and X4 (IIIB) HIV isolates (Figure 4). A lower degree of variability in the antiviral potency of MIP-3α among donors with the R5 HIV isolate BaL was observed, which is commensurate with the higher frequency of CCR6 expression on CD4+CCR5+ T cells. This could also contribute to the relatively higher potency in inhibiting R5 HIV in our PCR assay for early reverse transcription products (Figure 1). The postentry inhibitory activity of MIP-3α contributes another piece of evidence that CCR6 stimulation induces an antiviral response in HIV-infected cells.
MIP-3α inhibits HIV replication in PBMCs after infection. PBMCs were infected with R5 (BaL) HIV (left) or X4 (IIIB) HIV (right). After virus removal and washing, cells were treated with MIP-3α (0.2-5 μg/mL) or with T20 (11.25 μg/mL) or AZT (2.67 μg/mL) as controls. Shown here are the averages (± SEMs) of 3 experiments conducted in triplicate on cells from 3 separate donors.
MIP-3α inhibits HIV replication in PBMCs after infection. PBMCs were infected with R5 (BaL) HIV (left) or X4 (IIIB) HIV (right). After virus removal and washing, cells were treated with MIP-3α (0.2-5 μg/mL) or with T20 (11.25 μg/mL) or AZT (2.67 μg/mL) as controls. Shown here are the averages (± SEMs) of 3 experiments conducted in triplicate on cells from 3 separate donors.
Gi protein–induced signaling mediates the antiviral activity of hBD2 and MIP-3α
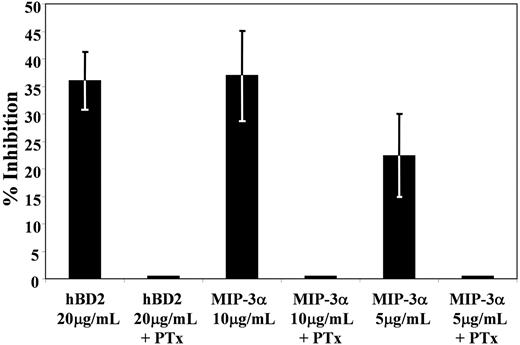
To gain insight on the intracellular pathways involved in the HIV-inhibitory activity of hBD2 and MIP-3α through CCR6 stimulation, we used PTx, a classical inhibitor of Gi proteins that are typically associated with chemokine receptors. PBMCs were pretreated with 500 ng/mL PTx and infected with an HIV luciferase virus pseudotyped with AMLV, and postinfection hBD2 (20 μg/mL) or MIP-3α (at 10 and 5 μg/mL) was tested. Treatment of single-cycle infections with PTx alone resulted in enhancement of infection (4.8 ± 1.9) over control, untreated infection (data not shown). We observed an abrogation of the HIV-inhibitory activity by both ligands in the presence of PTx (Figure 5). Thus, it is probable that Gi-activated intracellular pathways are involved in CCR6-mediated inhibition of HIV.
Treatment with Bordetella PTx abrogates HIV inhibition by CCR6 ligands hBD2 and MIP-3α. PBMCs were infected with HIV–luciferase pseudotype virus (AMLV envelope) as described in “Infectivity assays.” Three hours after infection, virus was washed 3 times with PBS, and cells were incubated 3 days in complete RPMI, with or without PTx, and with or without hBD2 (20 μg/mL) or MIP-3α (1 and 5 μg/mL). Subsequently, cells were washed and lysed with cell lysis reagent from Promega. Luciferase activity in cytoplasmic lysates was measured with a Turner luminometer. The percentage of inhibition was calculated in reference to luciferase activity as measured in untreated control infections. Shown here are averages (± SEMs) of 3 experiments. Treatment of single-cycle infections with PTx alone resulted in enhancement of infection (4.8 ± 1.9) over control, untreated infection.
Treatment with Bordetella PTx abrogates HIV inhibition by CCR6 ligands hBD2 and MIP-3α. PBMCs were infected with HIV–luciferase pseudotype virus (AMLV envelope) as described in “Infectivity assays.” Three hours after infection, virus was washed 3 times with PBS, and cells were incubated 3 days in complete RPMI, with or without PTx, and with or without hBD2 (20 μg/mL) or MIP-3α (1 and 5 μg/mL). Subsequently, cells were washed and lysed with cell lysis reagent from Promega. Luciferase activity in cytoplasmic lysates was measured with a Turner luminometer. The percentage of inhibition was calculated in reference to luciferase activity as measured in untreated control infections. Shown here are averages (± SEMs) of 3 experiments. Treatment of single-cycle infections with PTx alone resulted in enhancement of infection (4.8 ± 1.9) over control, untreated infection.
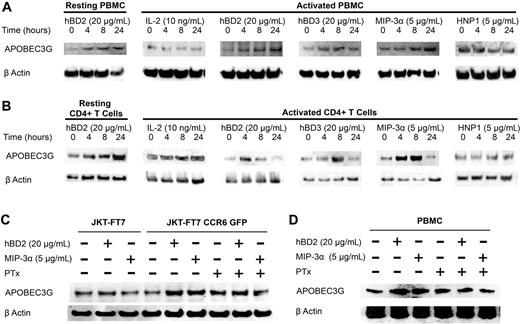
CCR6 ligands up-regulate APOBEC3G expression in both PBMCs and CD4+ T cells
Our results suggest that the postentry CCR6-mediated anti-HIV response occurs before or during early reverse transcription. APOBEC3G inhibits the accumulation of early reverse transcription products in the absence of hypermutation, probably by inhibition of nascent viral cDNA elongation.31,32 Therefore, we next investigated whether the CCR6 ligands hBD2, hBD3, and MIP-3α induce the expression of APOBEC3G. PBMCs or CD4+ T cells were treated with hBD2 (20 μg/mL), hBD3 (20 μg/mL), or MIP-3α (5 μg/mL), and lysates were prepared at 0, 4, 8, and 24 hours. Equal amounts of total protein were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting with anti–human APOBEC3G antibodies. Treatment of resting PBMCs or CD4+ T cells with hBD2 induced an up-regulation of APOBEC3G expression (Figure 6A-B). Further, treatment of both activated PBMCs and activated CD4+ T cells with hBD2, hBD3, or MIP-3α enhanced APOBEC3G expression (Figure 6A-B). The induction of APOBEC3G by CCR6 ligands was specific because postactivation maintenance in IL-2 (10 ng/mL) as well as treatment with HNP1 did not increase APOBEC3G expression (Figure 6A-B). Immunoblotting with anti–β-actin antibodies confirmed comparable protein loading.
Induction of APOBEC3G by CCR6 ligands. Resting or activated (A) PBMCs and (B) CD4+ T cells treated with hBD2 (20 μg/mL), hBD3 (20 μg/mL), MIP-3α (5 μg/mL), or HNP1 (5 μg/mL). (C) JKT-FT7 (CCR6low/no) and JKT-FT7 CCR6 GFP cells treated with hBD2 (20 μg/mL) or MIP-3α (5 μg/mL) for 4 hours or pretreated with 500 ng/mL PTx. (D) PBMCs treated with hBD2 (20 μg/mL) or MIP-3α (5 μg/mL) for 24 hours or pretreated with 500 ng/mL PTx. APOBEC3G and β-actin were detected at the indicated time points by Western blotting.
Induction of APOBEC3G by CCR6 ligands. Resting or activated (A) PBMCs and (B) CD4+ T cells treated with hBD2 (20 μg/mL), hBD3 (20 μg/mL), MIP-3α (5 μg/mL), or HNP1 (5 μg/mL). (C) JKT-FT7 (CCR6low/no) and JKT-FT7 CCR6 GFP cells treated with hBD2 (20 μg/mL) or MIP-3α (5 μg/mL) for 4 hours or pretreated with 500 ng/mL PTx. (D) PBMCs treated with hBD2 (20 μg/mL) or MIP-3α (5 μg/mL) for 24 hours or pretreated with 500 ng/mL PTx. APOBEC3G and β-actin were detected at the indicated time points by Western blotting.
Induction of APOBEC3G by hBD2 and MIP-3α is mediated by CCR6
Having shown that the intracellular inhibition of HIV by hBD2 and MIP-3α is mediated by CCR6 and both hBD2 and MIP-3α induce APOBEC3G, we assessed whether CCR6 is required for the induction of APOBEC3G. Treatment with hBD2 or MIP-3α increased the expression of APOBEC3G in JKT-FT7 CCR6+ cells but not in CCR6low/no cells (Figure 6C). Further, pretreatment of cells with PTx abrogated the induction of APOBEC3G by hBD2 and MIP-3α in JKT-FT7 CCR6+ (Figure 6C) and PBMCs (Figure 6D). No significant difference in cellular metabolism was observed between untreated and PTx-treated cells as determined by MTS assays (data not shown). These findings suggest Gi signaling through CCR6 induces the expression of APOBEC3G. Taken together with our findings that viral inhibition by hBD2 is restricted to CCR6+ cells (Figure 3), both hBD2 and MIP-3α increase the expression of APOBEC3G in PBMCs and CD4+ T cells (Figure 6A-B), and the induction is restricted to CCR6+ cells (Figure 6C) suggests that the postentry mechanism of inhibition is mediated by the induction of APOBEC3G.
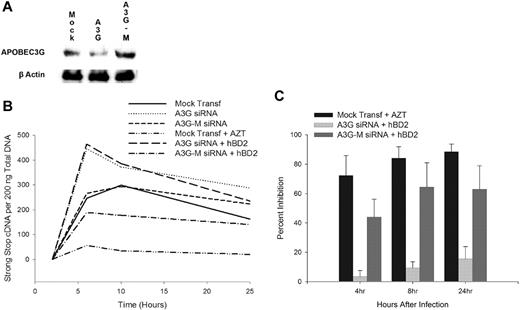
CCR6 intracellular inhibition of HIV is mediated by APOBEC3G
We next investigated whether the CCR6-mediated induction of APOBEC3G is the primary intracellular mechanism of inhibition. CD4+ T cells transfected with APOBEC3G-specific, but not mutant, siRNA reduced the expression of APOBEC3G (Figure 7A). Knockdown of APOBEC3G before infection with X4 (IIIB) HIV enhanced infection and resulted in a shift in kinetics with peak early reverse transcript synthesis occurring at 4 hours compared with 8 hours after infection in mock-transfected or mutant APOBEC3G siRNA-transfected CD4+ T cells (Figure 7B). The knockdown of APOBEC3G in CD4+ T cells before infection abrogated the inhibition by hBD2, whereas transfection with mutant APOBEC3G siRNA did not abrogate the postentry inhibition by hBD2 (Figure 7C). These findings suggest that the primary postentry intracellular mechanism of inhibition by hBD2 is mediated by the induction of APOBEC3G. No significant difference in cellular metabolism was observed among the mock- and siRNA-transfected cells as determined by MTS assays (data not shown).
Intracellular inhibition of HIV by hBD2 is mediated by the induction of APOBEC3G. (A) siRNA knockdown of APOBEC3G in CD4+ T cells. (B) CD4+ T cells transfected with APOBEC3G-specific siRNA or mutant APOBEC3G siRNA were infected with X4 (IIIB) HIV. After virus removal and washing, hBD2 (20 μg/mL) was added to the tissue culture media. Total cellular DNA was isolated at the indicated time points, and the presence of early reverse transcription products was determined by real-time PCR. (C) Triplicate readings were averaged, and inhibition was determined as a percentage of the number of copies of early reverse transcription products in infected treated cells in reference to copies measured in untreated control infections. Shown here are mean percentage of inhibition (± SEMs) of 4 independent experiments that used cells from different donors.
Intracellular inhibition of HIV by hBD2 is mediated by the induction of APOBEC3G. (A) siRNA knockdown of APOBEC3G in CD4+ T cells. (B) CD4+ T cells transfected with APOBEC3G-specific siRNA or mutant APOBEC3G siRNA were infected with X4 (IIIB) HIV. After virus removal and washing, hBD2 (20 μg/mL) was added to the tissue culture media. Total cellular DNA was isolated at the indicated time points, and the presence of early reverse transcription products was determined by real-time PCR. (C) Triplicate readings were averaged, and inhibition was determined as a percentage of the number of copies of early reverse transcription products in infected treated cells in reference to copies measured in untreated control infections. Shown here are mean percentage of inhibition (± SEMs) of 4 independent experiments that used cells from different donors.
Discussion
Mucosal immunity is the first line of defense against HIV infection, and a deeper understanding of its components and mechanisms will yield new strategies for prevention and therapy of HIV infection. Among the antimicrobial, and anti-HIV peptides that are expressed in mucosae, β-defensins, being expressed at high levels by epithelial cells, are probably the main players in the containment of HIV infection.22-24 In particular, we and others have shown that hBD2 has characteristics that further stress its potential importance in mucosal control of HIV.27,28 First, it has a broad HIV-inhibitory activity in the absence of any negative effect on cell metabolism, even at very high concentrations.27,28 Second, it is expressed at high levels in healthy human oral mucosa, which implies that this peptide does not induce inflammation.28,38 Third, its HIV-inhibitory activity is based on 2 mechanisms: an intrinsic component that results in a lower infectivity of hBD2-treated HIV and another component affecting HIV intracellularly.28 Because we did not detect inhibition of env-mediated fusion, it is possible that the intrinsic activity is due to defensin delivered to cells attached to virions. In this study, we set out to further investigate the intracellular component of the antiviral activity of hBD2. Among the possible explanations for this activity, we considered whether the intracellular inhibition of HIV could be due to an interaction of hBD2 with a cellular receptor. The known receptor for hBD2 is the chemokine receptor CCR6, although it is possible that hBD2 may use additional receptors.8 This interaction creates the potential for cross talk between a molecule that is part of innate immunity, hBD2, and adaptive immunity, that is, cells that express CCR6, which include memory T lymphocytes.11,41,42 Expression of CCR6 on DCs adds to the potential relevance of the hBD2–CCR6 axis to the establishment of immune responses against HIV.7,43
Our studies show that postentry treatment with hBD2 of PBMCs infected with either X4 (IIIB) or R5 (BaL) HIV inhibits HIV at an early stage of infection either before or during viral DNA synthesis. Because CCR6 ligands inhibit replication of both R5 and X4 isolates of HIV in primary cells, it is unlikely that the inhibition depends on coexpression or differential expression of CCR5 on CCR6+ and CCR6− cells; data obtained on the Jurkat-derived cell line, which does not express CCR5, are consistent with an independence of inhibition from CCR5 expression. The mechanism of inhibition is mediated by CCR6 and requires induction of the host restriction factor APOBEC3G. We observed an increase in APOBEC3G expression induced by hBD2 in resting PBMCs and CD4+ T cells. Our results also show a protective role in activated PBMCs and CD4+ T cells whereby the induction of APOBEC3G by hBD2 is required for inhibition. To our knowledge, this is the first time that activation of APOBEC3G has been shown in mitogen-activated cells. Previously, induction of APOBEC3G in CD4+ T cells by mitogens, IL-2, IL-7, IL-15, and interferon-α, had been observed only in resting CD4+ T cells.44,45 Further, the nonprotective high-molecular-mass form of APOBEC3G predominates in activated CD4+ T cells37 and in resting CD4+ T cells after induction by mitogens, IL-2 and IL-15 and to a lesser extent with IL-7.44 Our findings suggest that the CCR6-mediated induction of APOBEC3G may be beneficial to both resting and activated cells. We did not address in this study the expression of APOBEC3G and its high- or low-molecular forms in primary cells separated according to CCR6 status, but our data with siRNA inhibition show that APOBEC3G is necessary for the antiviral effects of CCR6 ligands. Therefore, studies to elucidate the mechanism and signaling pathways involved in the induction of an inhibitory form of APOBEC3G by CCR6 ligands in activated PBMCs and CD4+ T cells (separated in CCR6+ and CCR6− populations) are warranted.
The inhibition of HIV and induction of APOBEC3G by CCR6 ligands was abrogated by the pretreatment with PTx, suggesting Gi signaling. The infection-enhancing effect of PTx that we observed in our assays could be due to the abrogation of signaling by CCR6 and other Gi-linked receptors, that in turn eliminates an intracellular HIV-inhibitory pathway. Our experiments were performed in activated PBMCs, which produce significant MIP-3α, and perhaps other unknown HIV-inhibitory factors that bind to Gi-linked receptors. The inhibition of Gi proteins would therefore abrogate inhibition mediated by these receptors. Our findings are not at odds with those of Alfano et al,46 who reported HIV inhibition by PTx. Those studies used lower concentrations of PTx or the use of the B-oligomer of PTx, which suggest that ADP ribosylation is not required for the inhibitory effect. Therefore, the HIV-inhibitory effect of PTx observed by Alfano et al46 is not related to our findings.
The discovery of the role of CCR6 in mediating HIV inhibition is of importance to mucosal immunity. CCR6 is expressed on immature DCs,7,43 which are among the first cells to come in contact with HIV.47-49 Their role in HIV pathogenesis is crucial and complex, and studies are necessary to establish how hBD2 and MIP-3α affect these cells with regard to HIV. Furthermore, CCR6 is expressed on memory CD4+ T cells11,41,42 and is highly expressed on lymphocytes that infiltrate inflamed mucosa. In particular, CCR6 is coexpressed on approximately 70% of CD4+CCR5+ peripheral blood lymphocytes.13 There is growing evidence of the importance of 2 populations of CCR6+ T cells within the gut: CD4+CD45RO+α4β7+ T cells11,14 and TH17 cells.15,17-19 Preservation of IL-17–producing cells contributes to intestinal mucosal immunity through the induction of hBD2 and MIP-3α. Both hBD2 and MIP-3α exhibit antimicrobial activity, including the anti-HIV activity mediated by the induction of APOBEC3G described here, and in mice both hBD2 and MIP-3α are essential for intestinal homeostasis because the absence of either leads to defects in the formation of various organized lymphoid tissue.50 Thus, an HIV-inhibitory mechanism based on CCR6 is probably highly relevant to protect CD4+CCR5+ T cells as well as CD4+α4β7+ and TH17 cells within the gut-associated lymphoid tissue, and it provides a potential new target for the development of novel drugs that induce APOBEC3G for prevention and treatment of HIV infection.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Lai-Xi Wang for the gift of T20 peptide; Dr Sam Hwang for the gift of JKT-FT7 and JKT-FT7 CCR6 GFP cell lines; Drs Warner C. Greene and Ya-Lin Chiu for sharing of protocols; and Drs Marvin Reitz, Richard Koup, Ferenc Livak, and Nicholas Carbonetti for useful suggestions. The reagent anti-APOBEC3G from Dr Warner C. Greene was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
This work was supported by the National Institute of Neurological Disorders And Stroke (award R01NS066842), the National Institute of Dental and Craniofacial Research (award 1R21DE15508-01; A.G.-D.) and (award R01AI061482; W.L). M.K.L. was a trainee under Institutional Training Grant T32AI007540 from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institute of Neurological Disorders And Stroke, the National Institute of Dental and Craniofacial Research, or the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: M.K.L., L.S., and L.D. performed experiments; M.K.L., L.S., and A.G.-D. analyzed results and made figures; and M.K.L., W.L., and A.G.-D. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alfredo Garzino-Demo, Laboratory of Virus-Host Interactions, Division of Basic Science, Institute of Human Virology, University of Maryland School of Medicine, Rm S613, 725 W Lombard St, Baltimore, MD 21201; e-mail: agarz001@umaryland.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal