Abstract
The molecular pathway by which Fanconi anemia (FA) proteins function in oxidative stress response has not been defined. Here we report the functional interaction of the FA protein Fanconi anemia complementation group D2 (FANCD2) and the forkhead transcription factor forkhead box O 3a (FOXO3a). FOXO3a colocalized with FANCD2 foci in response to oxidative stress. The FANCD2-FOXO3a complex was not detected in cells deficient for the FA core complex component FANCA but could be restored in corrected cells. Consistent with this, a nonmonoubiquitinated FANCD2 mutant failed to bind FOXO3a. Although both mitomycin C and ionizing radiation induced FANCD2 monoubiquitination, neither could induce the association of FANCD2 and FOXO3a. Overexpression of FOXO3a reduced abnormal accumulation of reactive oxygen species, enhanced cellular resistance to oxidative stress, and increased antioxidant gene expression in corrected but not mutant FA-D2 cells. The novel oxidative stress response pathway identified in this study, in which FANCD2 and FOXO3a converge, probably contributes to cellular antioxidant defense.
Introduction
Fanconi anemia (FA) is a genetic disease characterized by genomic instability and cancer predisposition.1-3 Patients with FA have a pro-oxidant state that is associated with overproduction or impaired detoxification of reactive oxygen species (ROS).4-6 As a consequence, cells from FA patients demonstrate hypersensitivity to ambient oxygen and increased chromosomal aberrations.7-9 FA oxidant hypersensitivity has been documented in many studies using primary and immortalized cell cultures as well as ex vivo materials from patients.4-11 Significant evidence also suggests that excessive apoptosis of hematopoietic stem/progenitor cells induced by oxidative stress may be a critical factor in the pathogenesis of bone marrow failure and cancer progression in FA.5,12
Mammalian forkhead members of the class O (FOXO) transcription factors, including FOXO1, FOXO3a, FOXO4, and FOXO6, are implicated in the regulation of diverse physiologic processes, including cell-cycle arrest, apoptosis, DNA repair, stress resistance, and metabolism.13,14 Among these FOXO proteins, FOXO3a functions as a major regulator of oxidative stress.14,15 In this study, we identified a novel oxidative stress response pathway that converges Fanconi anemia complementation group D2 (FANCD2) and FOXO3a.
Methods
Cell culture and treatments
Human lymphoblast cell lines JY (normal), HSC72 (FA-A), HSC536 (FA-C), and PD20 (FA-D2) were cultured in 10% fetal bovine serum RPMI 1640 medium. HeLa cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum. Cells were treated with H2O2 (0.5mM for 6 hours), ionizing radiation (IR; 5 Gy), or mitomycin C (MMC; 0.5μM for 18 hours).
Constructs
The retroviral vectors encoding human pMMP-Puro, pMMP-wt-FANCD2, and pMMP-K561R-FANCD2 were generously provided by Dr Alan D'Andrea (Harvard Medical School, Boston, MA). The cDNA encoding the human FOXO3a was from Addgene Plasmid 8360 (Addgene). The retroviral vectors MIEG3, MIEG3-FANCA, and MIEG3-FANCC have been described elsewhere.16 The Flag-tagged FOXO3a was generated by polymerase chain reaction (PCR) using primers 5′-GGG GGA TCC ACC ATG GAT GGA CTA CAA GGA CGA TGA CGA TAA ACC-3′ (forward) and 5′-CCT CTA GAT CAG CCT GGC ACC CAG CTC TGA GAT G-3′ (reverse), and subcloned into the BamHI-XbaI of Lenti-X mammalian inducible vector (Clontech).
ROS production
Cells were incubated with CM-H2DCFDA (Invitrogen) in the dark for 15 minutes at 37°C. After washing, cells were analyzed by flow cytometry using a FACSCanto (BD Biosciences). Data were analyzed using the CellQuest program (BD Biosciences).17
Real-time PCR
Total RNA was prepared with RNeasy kit (QIAGEN) following the manufacturer's procedures. After treatment with RNase-free DNase, RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen). Real-time PCR was performed on an ABI PRISM 7700 sequence detection system (Applied Biosystems) with SYBR green PCR master mix (Applied Biosystems), according to the manufacturer's instructions. Samples were normalized to the level of GAPDH mRNA, and the relative expression levels were determined by the standard curve method. Primer sequences are shown in supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Immunofluorescent staining, immunoprecipitation and immunoblotting, cell viability (luminescent) assay, transduction, and subcellular fractionation
A detailed description of these assays is shown in supplemental data.
Results and discussion
FANCD2 forms a complex with FOXO3a in response to oxidative stress
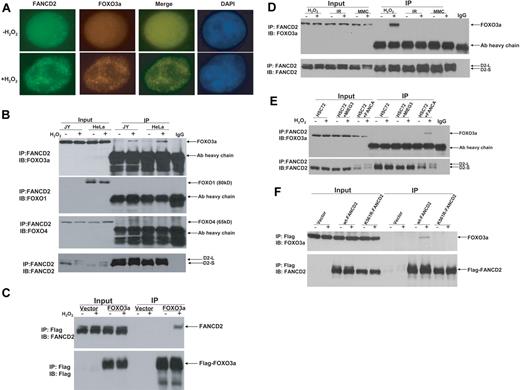
To investigate whether the FA pathway interplays with other cellular oxidative stress signaling pathways, we examined FANCD2 monoubiquitination and foci formation in response to oxidative stress. We found that H2O2 treatment induced both FANCD2 monoubiquitination and FANCD2 foci in normal human lymphoblasts as well as HeLa cells (supplemental Figure 1). Because FOXO3a is a major component in oxidative stress signaling,14,15 we determined whether FOXO3a was associated with the FANCD2 foci. Indeed, FOXO3a was found to colocalize with FANCD2 after H2O2 treatment in lymphoblasts (Figure 1A) and HeLa cells (supplemental Figure 2). To verify formation of a FANCD2-FOXO3a complex in H2O2-treated cells, we conducted FANCD2 immunoprecipitation (IP) of cell extracts from both normal lymphoblasts and HeLa cells treated with or without H2O2. The results showed that H2O2 treatment induced association of FANCD2 with FOXO3a but not FOXO1 or FOXO4 (Figure 1B). We then expressed FOXO3a as a Flag-tagged protein in HeLa cells and performed reciprocal IP with anti-Flag agarose. Again, the results showed that FANCD2 interacted with FOXO3a in cells subjected to oxidative stress (Figure 1C).
Oxidative stress–induced formation of the FANCD2-FOXO3a complex. (A) FOXO3a colocalizes with FANCD2 after H2O2 treatment in normal (JY) lymphoblasts. Cells were treated with H2O2 (0.5mM) for 6 hours and stained with antibodies against FOXO3a and FANCD2. Colocalization of nuclear FOXO3a with FANCD2 is shown as merged images. (B) Whole-cell lysates of normal lymphoblasts (JY) and HeLa cells treated with 0.5mM H2O2 for 6 hours were subjected to immunoprecipitation (IP) with an anti-FANCD2 antibody or an isotype IgG (negative control) followed by immunoblotting (IB) analysis with antibodies against FOXO3a, FOXO1, FOXO4, and FANCD2. (C) HeLa cells were transduced with vector or Flag-FOXO3a and treated with 0.5mM H2O2 for 6 hours. Cell lysates were prepared, immunoprecipitated using anti-Flag agarose, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblotted. Blots were probed with antibodies against FANCD2 or Flag. (D) Normal human lymphoblasts (JY) were treated with H2O2 (0.5mM for 6 hours), IR (5 Gy), or MMC (0.5μM for 18 hours), and whole-cell lysates were subjected to IP with an anti-FANCD2 antibody or an isotype IgG (negative control) followed by IB analysis with antibodies against FOXO3a and FANCD2. (E) HSC72 (human FA-A lymphoblast) cells were transduced with retrovirus carrying empty vector (MIEG3) or FANCA, and treated with H2O2 at 0.5mM for 6 hours. Cell extracts were subjected to IP with an anti-FANCD2 antibody or an isotype IgG (negative control) followed by IB analysis with antibodies against FOXO3a and FANCD2. (F) PD20 (human FA-D2 lymphoblast) cells transduced with retrovirus carrying empty vector wt-FANCD2 or K561R-FANCD2 were treated with H2O2 at 0.5mM for 6 hours. Cell extracts were prepared, immunoprecipitated using anti-Flag antibody conjugated to agarose, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblotted with antibodies against FANCD2 and FOXO3a.
Oxidative stress–induced formation of the FANCD2-FOXO3a complex. (A) FOXO3a colocalizes with FANCD2 after H2O2 treatment in normal (JY) lymphoblasts. Cells were treated with H2O2 (0.5mM) for 6 hours and stained with antibodies against FOXO3a and FANCD2. Colocalization of nuclear FOXO3a with FANCD2 is shown as merged images. (B) Whole-cell lysates of normal lymphoblasts (JY) and HeLa cells treated with 0.5mM H2O2 for 6 hours were subjected to immunoprecipitation (IP) with an anti-FANCD2 antibody or an isotype IgG (negative control) followed by immunoblotting (IB) analysis with antibodies against FOXO3a, FOXO1, FOXO4, and FANCD2. (C) HeLa cells were transduced with vector or Flag-FOXO3a and treated with 0.5mM H2O2 for 6 hours. Cell lysates were prepared, immunoprecipitated using anti-Flag agarose, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblotted. Blots were probed with antibodies against FANCD2 or Flag. (D) Normal human lymphoblasts (JY) were treated with H2O2 (0.5mM for 6 hours), IR (5 Gy), or MMC (0.5μM for 18 hours), and whole-cell lysates were subjected to IP with an anti-FANCD2 antibody or an isotype IgG (negative control) followed by IB analysis with antibodies against FOXO3a and FANCD2. (E) HSC72 (human FA-A lymphoblast) cells were transduced with retrovirus carrying empty vector (MIEG3) or FANCA, and treated with H2O2 at 0.5mM for 6 hours. Cell extracts were subjected to IP with an anti-FANCD2 antibody or an isotype IgG (negative control) followed by IB analysis with antibodies against FOXO3a and FANCD2. (F) PD20 (human FA-D2 lymphoblast) cells transduced with retrovirus carrying empty vector wt-FANCD2 or K561R-FANCD2 were treated with H2O2 at 0.5mM for 6 hours. Cell extracts were prepared, immunoprecipitated using anti-Flag antibody conjugated to agarose, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblotted with antibodies against FANCD2 and FOXO3a.
The interaction between FANCD2-FOXO3a is oxidative stress specific
Because we observed the FANCD2-FOXO3a interaction in cells subjected to oxidative stress, which induced FANCD2 monoubiquitination (supplemental Figure 1) and because other genotoxic agents such as MMC and IR can also induce FANCD2 monoubi-quitination,18,19 we next asked whether the FANCD2-FOXO3a interaction was oxidative stress specific. Co-IP experiments using cell extracts prepared from human lymphoblasts treated with H2O2, IR, or MMC showed that all agents could induce FANCD2 monoubiquitination. However, the FANCD2-FOXO3a complex was only detected in cells treated with H2O2 (Figure 1D). Further analysis by subcellular fractionation shows that, whereas FANCD2 was recruited to the chromatin by all 3 genotoxic treatments, FOXO3a was recruited to the chromatin only by H2O2 treatment (supplemental Figure 3). This result suggests that the interaction between FANCD2-FOXO3a was oxidative stress specific.
FANCD2 monoubiquitination is required for FANCD2-FOXO3a interaction
We next examined whether oxidative stress–induced FANCD2 monoubiquitination is required for the FANCD2-FOXO3a interaction. We performed 2 sets of experiments to address this. First, we examined whether FANCD2 could form a complex with FOXO3a in patient-derived lymphoblast cell lines deficient for the FA core complex component FANCA (HSC72) or FANCC (HSC536). The results showed that the FANCD2-FOXO3a complex was formed only in FANCA- (Figure 1E) or FANCC-corrected cells (supplemental Figure 4). Second, we expressed wild-type (WT) FANCD2 or a nonubiquitinated FANCD2 mutant (K561R) in cells deficient for FANCD2 and found that FOXO3a only interacted with WT FANCD2 (Figure 1F). These results collectively suggest that oxidative stress–induced FANCD2 monoubiquitination is required for the interaction.
The FANCD2-FOXO3a complex confers cellular resistance to oxidative stress
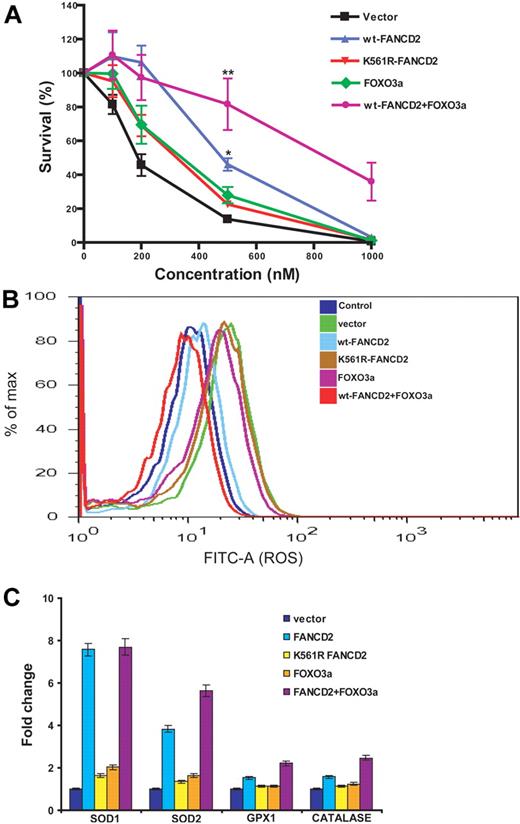
To investigate the functional consequence of the FANCD2-FOXO3a interaction, we coexpressed FANCD2 and FOXO3a in patient-derived lymphoblasts deficient for FANCD2. Vector-transduced cells served as a control. Expression of these proteins was verified by Western blot analysis (supplemental Figure 5). Cells were then analyzed for 3 biologic parameters in the context of oxidative stress response: sensitivity to H2O2 treatment, production of ROS, and expression of the FOXO3a-targeting genes encoding superoxide dismutases 1 and 2, glutathione peroxidase 1, and catalase involved in antioxidant defense.20,21 The results showed that coexpression of FOXO3a with WT FANCD2, but not with the mutant K561R-FANCD2, reduced abnormal accumulation of ROS, enhanced cellular resistance to oxidative stress, and increased expression of FOXO3a-controlled antioxidant genes (Figure 2; supplemental Figure 6).
Coexpression of FANCD2 and FOXO3a increases cellular resistance to oxidative stress. (A) PD20 cells were infected with retroviruses carrying vector, wt-FANCD2, K561R-FANCD2, FOXO3a, or both wt-FANCD2 and FOXO3a. The transduced cells were treated with H2O2 at the indicated doses for 16 hours. Cell survival was determined by luminescence and is shown as the percentage of the untreated cells. Mean values and SD from 3 independent determinations are shown. Statistical significance between vector and wt-FANCD2 + FOXO3a or wt-FANCD2 samples: **P < .01; *P < .05. (B) Transduced cells described in panel A were labeled with fluorescein isothiocyanate–conjugated CM-H2DCFDA, and levels of ROS were determined by flow cytometric analysis. (C) Expression of FOXO3a-targeted genes encoding antioxidant proteins. Transduced cells described in panel A were treated with H2O2 at 100μM for 12 hours. Total RNA was prepared, and gene expression was analyzed by real-time PCR. Data were presented as fold change in mRNA expression relative to glyceraldehyde-3-phosphate dehydrogenase in vector-transduced cells, which was given an arbitrary level of 1.0 for each gene. Results are mean ± SD of 3 independent experiments.
Coexpression of FANCD2 and FOXO3a increases cellular resistance to oxidative stress. (A) PD20 cells were infected with retroviruses carrying vector, wt-FANCD2, K561R-FANCD2, FOXO3a, or both wt-FANCD2 and FOXO3a. The transduced cells were treated with H2O2 at the indicated doses for 16 hours. Cell survival was determined by luminescence and is shown as the percentage of the untreated cells. Mean values and SD from 3 independent determinations are shown. Statistical significance between vector and wt-FANCD2 + FOXO3a or wt-FANCD2 samples: **P < .01; *P < .05. (B) Transduced cells described in panel A were labeled with fluorescein isothiocyanate–conjugated CM-H2DCFDA, and levels of ROS were determined by flow cytometric analysis. (C) Expression of FOXO3a-targeted genes encoding antioxidant proteins. Transduced cells described in panel A were treated with H2O2 at 100μM for 12 hours. Total RNA was prepared, and gene expression was analyzed by real-time PCR. Data were presented as fold change in mRNA expression relative to glyceraldehyde-3-phosphate dehydrogenase in vector-transduced cells, which was given an arbitrary level of 1.0 for each gene. Results are mean ± SD of 3 independent experiments.
In conclusion, we report functional interaction between FANCD2 and the major oxidative stress responsive transcription factor FOXO3a. The formation of the FANCD2-FOXO3a complex appears to be oxidative stress specific and require an intact FA core complex and, thus, oxidative stress–induced monoubiquitination of FANCD2. Functionally, we showed that overexpression of FOXO3a reduced abnormal accumulation of ROS, enhanced cellular resistance to oxidative stress, and increased antioxidant gene expression only in cells corrected by a FANCD2 protein capable of interacting with FOXO3a. Studies have shown that certain FA proteins interact with oxidative signaling pathways involving important cellular factors, such as glutathione S-transferase P1-1 and ASK1 that primarily function in redox-related processes.4,12 In this context, the novel oxidative stress response pathway converging FANCD2 and FOXO3a identified in this study probably contributes to cellular antioxidant defense.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Alan D'Andrea (Harvard Medical School) for the pMMP-Puro, pMMP-FANCD2, and pMMP-K561R-FANCD2 retroviral vectors; Jared Sipple for technical assistance; and the Vector Core of the Cincinnati Children's Research Foundation (Cincinnati Children's Hospital Medical Center) for the preparation of retroviruses and lentiviruses.
This work was supported in part by National Institutes of Health (grants R01 CA109641 and R01 HL076712). Q.P. is supported by a Leukemia & Lymphoma Scholar award. W.D. is supported by a Fanconi Anemia Research Fund grant.
National Institutes of Health
Authorship
Contribution: J.L. performed research, analyzed data, and wrote the paper; W.D. performed research and analyzed data; S.M. performed research; P.R.A. designed research; and Q.P. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qishen Pang, Division of Experimental Hematology and Cancer Biology, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: qishen.pang@cchmc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal