Abstract
Biopsies and cell lines of natural killer/T-cell lymphoma, nasal type (NKTCL) were subject to combined gene expression profiling and array-based comparative genomic hybridization analyses. Compared with peripheral T-cell lymphoma, not otherwise specified, NKTCL had greater transcript levels for NK-cell and cytotoxic molecules, especially granzyme H. Compared with normal NKcells, tumors were closer to activated than resting cells and overexpressed several genes related to vascular biology, Epstein-Barr Virus–induced genes, and PDGFRA. Notably, platelet-derived growth factor receptor α and its phosphorylated form were confirmed at the protein level, and in vitro the MEC04 NKTCL cell line was sensitive to imatinib. Deregulation of the AKT, Janus kinase–signal transducers and activators of transcription, and nuclear factor-κB pathways was corroborated by nuclear expression of phosphorylated AKT, signal transducers and activators of transcription 3, and RelA in NKTCL, and several deregulated genes in these pathways mapped to regions of recurrent copy number aberrations (AKT3 [1q44], IL6R [1q21.3], CCL2 [17q12], TNFRSF21 [6p12.3]). Several features of NKTCL uncovered by this analysis suggest perturbation of angiogenic pathways. Integrative analysis also evidenced deregulation of the tumor suppressor HACE1 in the frequently deleted 6q21 region. This study highlights emerging oncogenic pathways in NKTCL and identifies novel diagnostic and therapeutic targets.
Introduction
Several natural killer (NK)/T-cell lymphoma entities have a predilection for extranodal locations. One of the most common of these overall rare lymphoma entities is NK/T-cell lymphoma, nasal type (NKTCL). The disease, most prevalent in Asian and Central and South American populations, most commonly arises in the nasal cavity or adjacent structures but also can occur in other extranodal sites.1 This lymphoma exhibits an angiocentric and angiodestructive growth pattern characteristically associated with necrosis and ulceration. The tumor cells typically express CD2, cytoplasmic CD3 (CD3ϵ chain), and CD56; are negative for CD5, CD4, and CD8; and have a cytotoxic immunophenotype with expression of perforin, granzyme B (gzm B), and T cell–restricted intracellular antigen (TiA1). Most cases derive from NK cells and lack T-cell receptor (TCR) gene rearrangement, whereas a small proportion of cases have the phenotype and genotype of cytotoxic γδ or αβ T cells. Accordingly, the expression of killer immunoglobulin–like receptors has been documented in these tumors.2
In virtually all cases, most neoplastic cells harbor clonal episomal Epstein-Barr virus (EBV), suggesting the implication of the virus in tumor pathogenesis. The genetic alterations of NKTCL reported in a few studies include gain on chromosome 2q, losses of chromosomes 6q and 1p, and occasional presence of isochromosome 7q.3-6
Even with intensive therapies combining multiagent chemotherapy and involved-field radiotherapy, the prognosis of NKTCL remains poor, with 5-year overall survival ranging from 42% to 64% in 4 recent studies,7-10 and novel alternative approaches are needed. Several factors have been incriminated to account for aggressiveness or poor outcome, including Fas and p53 mutations,11,12 expression of the multidrug resistance 1 (MDR1) gene product P-glycoprotein,13 absence of CD94 transcript,14 and gzm B inhibitor PI9.9 However, the mechanisms involved in resistance to therapy remain poorly understood.
Genome-wide profiling studies of NKTCL are scarce,3,15 likely reflecting not only the rarity of the disease but also quantitative and qualitative restrictions imposed by small and often necrotic diagnostic samples. In one recent study, Iqbal et al5 focused on in-depth genomic and transcriptomic analysis of specific chromosomal regions, which led them to identify candidate suppressor genes mapping to 6q. In the current study, we analyzed a series of NKTCL samples in relation to normal cells and peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS), with the aims to (1) characterize the molecular signature of this lymphoma entity, (2) explore some signaling pathways implicated in its pathogenesis, and (3) search for novel markers potentially useful for diagnosis and/or targetable by therapeutic agents.
Methods
Patient characteristics and tumor samples
Nine newly diagnosed, previously untreated NKTCL patients with high-quality RNA and/or DNA extracted from frozen tumor biopsies containing more than 60% tumor cells were selected for this study. Their main clinical, phenotypic, and molecular characteristics are summarized in Table 1. Seven tumors originated in the nasopharyngeal area, 1 in the skin, and 1 in the hypophysis. All cases were reviewed by 3 hematopathologists (L.d.L., Y.H., and P.G.) and were diagnosed according to World Health Organization criteria.1 All cases had a CD3+ (cytoplasmic), CD2+, CD7+, CD5−, CD4−, CD8−, TiA1+, gzm B+ phenotype. Eight cases were positive for EBV by in situ hybridization with EBER probes. A high load of EBV DNA was demonstrated by polymerase chain reaction (PCR) in the remaining case. Specimens were also investigated for TCR-γ chain gene rearrangement by the use of a GC-clamp multiplex PCR-γ–denaturing gradient gel electrophoresis procedure. Seven cases without clonal T-cell population were regarded as of NK-cell origin, and a cytotoxic T-cell derivation was established in 2 cases with a clonal TCR gene rearrangement. For immunohistochemical validation, formalin-fixed, paraffin-embedded tumor samples from 16 NKTCL, including 7 of the cases described previously, and 17 PTCL, NOS were selected. The present study was approved by the institutional review board “Comité de Protection des Personnes Ile de France IX,” Créteil, France.
Summary of immunohistochemical and genotypic features of the NKTCL samples analyzed in the study
| Samples . | Age . | Sex . | Biopsy site . | CD3e . | CD2 . | CD5 . | CD7 . | CD4 . | CD8 . | CD56 . | TiA1 . | Gzm B . | EBV . | T clonality . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | 58 | M | Nasopharyngeal region | + | + | − | + | − | − | + | + | + | + | − |
| T2 | 25 | M | Nasopharyngeal region | + | + | − | − | − | − | + | + | + | + | − |
| T3 | 64 | M | Skin | + | − | − | ND | − | − | − | + | + | + | − |
| T4 | 62 | M | Nasopharyngeal region | − | + | − | − | − | − | + | + | + | + | − |
| T5 | 72 | M | Nasopharyngeal region | + | + | − | − | − | − | + | + | + | + | + |
| T6 | 52 | F | Nasopharyngeal region | + | + | − | − | ND | − | + | + | + | + | − |
| T7* | 42 | M | Hypophysis | + | + | − | − | − | − | + | + | + | + | − |
| T8† | 31 | F | Nasopharyngeal region | + | + | − | + | − | ND | + | + | + | + | − |
| T9† | 67 | M | Nasopharyngeal region | + | + | − | + | − | + | + | + | + | + | +‡ |
| SNK6 | NA | NA | NA | + | + | ND | ND | − | − | + | ND | + | + | − |
| SNT8 | NA | NA | NA | + | + | ND | ND | − | − | + | ND | ND | + | + |
| Samples . | Age . | Sex . | Biopsy site . | CD3e . | CD2 . | CD5 . | CD7 . | CD4 . | CD8 . | CD56 . | TiA1 . | Gzm B . | EBV . | T clonality . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | 58 | M | Nasopharyngeal region | + | + | − | + | − | − | + | + | + | + | − |
| T2 | 25 | M | Nasopharyngeal region | + | + | − | − | − | − | + | + | + | + | − |
| T3 | 64 | M | Skin | + | − | − | ND | − | − | − | + | + | + | − |
| T4 | 62 | M | Nasopharyngeal region | − | + | − | − | − | − | + | + | + | + | − |
| T5 | 72 | M | Nasopharyngeal region | + | + | − | − | − | − | + | + | + | + | + |
| T6 | 52 | F | Nasopharyngeal region | + | + | − | − | ND | − | + | + | + | + | − |
| T7* | 42 | M | Hypophysis | + | + | − | − | − | − | + | + | + | + | − |
| T8† | 31 | F | Nasopharyngeal region | + | + | − | + | − | ND | + | + | + | + | − |
| T9† | 67 | M | Nasopharyngeal region | + | + | − | + | − | + | + | + | + | + | +‡ |
| SNK6 | NA | NA | NA | + | + | ND | ND | − | − | + | ND | + | + | − |
| SNT8 | NA | NA | NA | + | + | ND | ND | − | − | + | ND | ND | + | + |
NA indicates not applicable; and ND, not done.
Sample used only for transcriptomic analysis.
Samples used only for genomic analysis.
This case showed βF1 immunoreactivity on formalin-fixed, paraffin-embedded tissue sections, and was negative for δTCR1 on frozen sections.
Cell lines and normal NK cells
Two NKTCL cell lines, SNK6 and SNT8 (Table 1),16 and 2 samples of normal CD56+ NK cells purified from peripheral blood, including resting and interleukin 2 (IL-2)–activated NK cells, also were subjected to gene expression profiling. The MEC04 NKTCL cell line also was used for in vitro proliferation assay and immunohistochemical validation.17 U937, a myelomonocytic cell line (ATCC LGC Standards), was used as control for in vitro assay. Cell lines were cultured in RPMI 1640 supplemented with 2mM l-glutamine and 10% heat-inactivated fetal bovine serum (Invitrogen) in the presence of 100 U/mL recombinant human IL-2, except for U937 cell line. Centrifugated pellets of SNK6 and MEC04 cells were fixed in ethanol to construct paraffin-embedded blocks.
RNA and DNA extraction
Total RNA was extracted with TRIzol reagent (Invitrogen), according to the manufacturer's instructions, and DNA was extracted with phenol-chloroform by the use of standard procedures. The integrity of the extracts was verified on an Agilent 2100 Bioanalyser (Agilent Technologies).
Microarray procedures
Microarray analyses were performed with 3 μg of total RNA as starting material and 10 μg of cRNA per hybridization (GeneChip Fluidics Station 400; Affymetrix). The total RNAs were amplified and labeled following the one-cycle target labeling protocol (http://www.affymetrix.com). The labeled cRNAs were hybridized to HG-U133 plus 2.0 Affymetrix GeneChip arrays (Affymetrix). The chips were scanned with an Affymetrix GeneChip Scanner 3000 and subsequent images analyzed by the use of GCOS 1.4 (Affymetrix).
Gene expression analyses
The gene expression analysis encompassed HG-U133 plus 2.0 Affymetrix array data from 7 NKTCL biopsies, 2 NKTCL cell lines (Table 1), 16 PTCL, NOS recently reported by our group under accession number E-TABM-702,18 6 normal NK-cell samples (including 4 previously published from GSE8059),19 18 recently published normal B-cell samples (8 from GSE15271 and 10 from GSE12195),20,21 and 15 activated B cell–like diffuse large B-cell lymphoma (ABC-DLBCL) samples from GSE12195.21 Two NK-cell samples without stimulation of IL-2 (1 each from ours and from GSE8059) represented the “resting NK-cell” group, whereas the 2 NK-cell samples stimulated by IL-2 for 24 hours (1 from our sample, the other from GSE8059) were regarded as the “activated NK-cell” group. Affymetrix raw data of the samples were normalized in batch by the use of the Robust Multichip Average method along with 58 other samples unrelated to this study. The clustering analysis of the 25 tumor samples was performed as already described.22
To identify genes differentially expressed between 2 groups of samples we used Welch t tests. To control for multiple testing, we measured the local false discovery rate by using kerfdr R package, an alternative approach to the more commonly used Bonferroni and Benjamini-Hochberg methods.23 The KEGG/Biocarta pathways enrichment scores were calculated by combining 4 methods, including Significance Analysis of Microarray to Gene-Set Analysis, Gene Set Analysis, and globaltest. The median score across the 4 methods was used to rank the pathways. Methodologic details are given in the supplemental Methods (available on the Blood website; see the Supplemental Materials link at the top of the online article). Raw gene expression data have been deposited to ArrayExpress under accession number E-TABM-702.
Array-based comparative genomic hybridization
Genome analyses of 8 NKTCLs, SNK6, and SNT8 cell lines were performed by the use of the human genome-wide CIT-CGH array (V6) containing 4434 sequence verified bacterial artificial chromosome (BAC) clones (with quadruplicate spots per clone) with a median gap between 2 successive clones of 600 kb. The characteristics of this array have been previously reported. Array-based comparative genomic hybridization raw data (deposited to ArrayExpress under accession number E-TABM-791; http://www.ebi.ac.uk/arrayexpress) were normalized by the use of the within print-tip group locally weighted scatterplot smoothing method; smoothing, breakpoints detection and status (gain/loss) determination were calculated by use of the Bioconductor package GLAD. Methodologic details are given in the supplemental Methods.
Immunohistochemistry
Immunohistochemistry was performed on deparaffinized tissue sections by the use of a standard indirect avidin-biotin immunoperoxidase method. After appropriate antigen retrieval, sections were stained for EBI3 (2G4H6)24 ; phosphorylated signal transducers and activators of transcription 3 (pSTAT3) at Tyr705 (D3A7), phosphorylated AKT (pAKT) at Ser473 (736E11; Cell Signaling Technology); VEGFA (C1), VCAM-1 (E10), cRel (B-6), PDGFRα, and pPDGFRα (Santa Cruz Biotechnology); E-cadherin (NCH-38; DakoCytomation); CD163 (10D6; Novocastra-Leica); clusterin (41D; Upstate-Millipore); CCND1 (SP4; Lab Vision-Thermo Fisher Scientific); epidermal growth factor receptor (EGFR, 31G7; Zymed-Invitrogen); β-catenin (14; BD Biosciences); and RelB (EP613Y; Epitomics). For granzyme H (gzm H; 4G5)25 and RelA (Santa Cruz Biotechnology), tyramide signal amplification system (CSAII kit; DakoCytomation) and peroxidase-labeled destran polymer (EnVision+; DakoCytomation) were applied, respectively. Adequate control tissues for specific antibodies were included.
Quantitative reverse-transcriptase PCR analysis of candidate genes
The expression of candidate genes (HACE1, CCL2, TNFRSF21, CCND3, MET) identified in altered chromosomal regions was determined by TaqMan quantitative reverse-transcriptase PCR (qRT-PCR; Applied Biosystems) in 6 primary tumors, 3 NKTCL cell lines, and in resting normal NK cells as control. The expression of HACE1 was compared with that of PRDM1, ATG5, and AIM1, 3 candidate tumor suppressor genes in NKTCL also mapping at 6q21.5 All primers and probes were purchased from Applied Biosystems, and the gene expression was measured by use of Mastercycler ep realplex2S system (Eppendorf). Quantifications were performed in duplicate, and mean values and SD were calculated for each transcript as previously described.26
PDGFRA gene analysis
PDGFRA gene analysis was performed in 6 primary tumors and in 3 cell lines. PDGFRA copy numbers in intron 1 (Syst: Hs05935655_cn,hg18.v7) and exons 7-8 (Syst: Hs04818823_cn,hg18.v7) were measured on a LightCycler 480 (Roche Diagnostics) by the use of TaqMan copy number assays according to the manufacturer's instructions (Applied Biosystems). ALB gene was used as the reference gene.27 The mutations of PDGFRA exons 12, 14, and 18 were searched by length analysis of PCR products and sequencing as described previously.28
To search for mutations and polymorphisms in the PDGFRA promoter region, genomic DNA was amplified by PCR by the use of primers 1651F and 727R as described by Toepoel et al.29 After purification (PCR purification kit; GE Healthcare), automated sequencing was performed on the ABI310 genetic analyzer with the Big Dye terminator sequencing kit (Applied Biosystems) with the primers 1562F, 1340R, 1110F, and 759R (see supplemental data). Sequences were compared with the reference sequence for PDGFRA promoter (GenBank accession number X80389).
3H thymidine–labeled proliferation assay
Cells were washed extensively to remove IL-2 and plated at 20 000 cells per well in the presence or absence of imatinib mesylate (Axon Medchem BV), at 3 different concentrations (1, 3, and 6μM), and incubated for 72 hours. Tritium thymidine (1 μCi) was added 6 hours before evaluating the proliferation by the use of Packard TopCount Liquid Scintillation Counter (GMI). Results were expressed as the percentage of proliferation measured in the absence of inhibitor. All experiments were performed in triplicate.
Results
The molecular signature of NKTCL differs from that of normal NK cells
The Affymetrix expression profiles of 7 NKTCL samples were compared with those of normal NK cells (n = 6). A total of 2447 and 2339 probe sets corresponding to 1721 and 1436 overexpressed and underexpressed genes, respectively, significantly distinguished NKTCL tissues from normal NK cells (P < .005; Table 2; supplemental Table 1). As expected, genes related to the microenvironment, notably macrophages (CLU, CD68, CD163) were among the most differentially overexpressed in NKTCL tissues. As shown in Table 2 and supplemental Table 1, the other genes overexpressed in NKTCL tissues included genes involved in the autophagy (ATG3, ATG7) and in the cell-cycle control (CCND1), related to cell-to-cell interactions (CDH1, ITGA7, ITGA9, ITGB4, VCAM1), chemokines (CX3CL1, CXCL9, CXCL10), cytokines (IL-8, IL-20, IL-33), extracellular matrix interactions (MMP11, MMP14, TIMP1, TIMP2, TIMP3), innate immunity (IL4I1, TLR4, TLR7, TLR8), local invasion and metastasis (COL1A1, COL1A2, FN1, LAMB1), angiogenesis (ANGPT2, VEGFA, VEGFB, VEGFC, KDR), growth factors and their receptors (PDGFRA, PDGFB, PDGFC, transforming growth factor-B2 [TGF-B2], TGF-B3), oncogenes (MAFB, MET, MYC), and genes reported to be induced by EBV in vitro (CDH1, EBI3, BASP1, DNASE1L3, HCK, HLA-DQA1, IFI30, IFI44L, IFITM3, LGALS9, THRA, TNFRSF10D).15,30 PCDH15, a gene previously shown to be specifically expressed in NKTCLs,31 also was overexpressed, albeit with less statistical significance. Some of these genes also were overexpressed in SNK6 and SNT8 cell lines compared with normal NK cells, for example, VCAM1, CXCL10, EBI3, TIMP1, MYC, PDGFRA, KDR, and VEGFA (Table 2; supplemental Table 1). Interestingly, NKTCL appeared to be more closely related to activated than to resting NK cells because the level of expression of about two-thirds of the NKTCL signature was closer to that of activated NK cells than that of resting NK cells.
Selection of genes overexpressed in NKTCL tissues compared to normal NK cells with P < .005
| Gene class and specific genes . | Fold change . |
|---|---|
| Cell-to-cell interactions | |
| CDH1 (E-cadherin)* | 1.54 |
| ITGA9 (Integrin alpha 9) | 2.37 |
| ITGB4 (Integrin beta 4)* | 2.15 |
| VCAM1 (Vascular cell adhesion molecule 1)* | 26.18 |
| Cell-cycle control | |
| CCND1 (Cyclin D1)* | 6.67 |
| Chemokines | |
| CCL18 | 10.50 |
| CCL19 | 80.67 |
| CCL2* | 88.11 |
| CCL8* | 51.61 |
| CXCL10 (IP10)* | 48.07 |
| CXCL12 (SDF-1) | 25.75 |
| CXCL9 (Mig) | 254.14 |
| Cytokines | |
| IL13RA1* | 9.19 |
| IL6R | 2.25 |
| IL33* | 2.75 |
| IL8* | 5.07 |
| Genes overexpressed in EBV-infected lymphoma cell lines | |
| EBI3 (EBV-induced gene 3)* | 2.76 |
| BASP1 (Brain abundant, membrane-attached signal protein 1)* | 13.21 |
| HCK (Hemopoietic cell kinase) | 16.01 |
| TNFRSF10D (Tumor necrosis factor receptor superfamily, member 10d)* | 1.53 |
| Extracellular matrix interactions | |
| COL1A1 (Collagen, type I alpha 1)* | 45.91 |
| COL1A2 (Collagen, type I alpha 2)* | 33.35 |
| COL3A1 (Collagen, type III alpha 1)* | 59.68 |
| FN1 (Fibronectin 1)* | 91.79 |
| TIMP1 (TIMP metallopeptidase inhibitor 1)* | 6.28 |
| TIMP2 (TIMP metallopeptidase inhibitor 2) | 10.79 |
| TIMP3 (TIMP metallopeptidase inhibitor 3) | 9.57 |
| Growth factors and receptors | |
| GHR (Growth hormone receptor)* | 1.35 |
| PDGFB (Platelet-derived growth factor beta)* | 2.16 |
| PDGFC (Platelet derived growth factor C)* | 7.92 |
| PDGFRA (Platelet-derived growth factor receptor, alpha polypeptide)* | 7.47 |
| Immunity and lymphocyte development | |
| CD86* | 6.04 |
| HLA-DOA* | 12.12 |
| HLA-DRA* | 12.18 |
| IL4I1 (Interleukin 4–induced 1)* | 6.40 |
| SLAMF8 (SLAM family member 8)* | 45.26 |
| TLR2 | 8.76 |
| TLR7* | 5.26 |
| TLR8 | 13.49 |
| Microenvironment | |
| CD163 | 134.47 |
| Oncogenes | |
| MAFB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog) | 37.94 |
| MET (Hepatocyte growth factor receptor)* | 2.00 |
| MYC* | 1.23 |
| Transcription factors | |
| STAT1* | 5.25 |
| STAT2* | 1.86 |
| Vascular biology | |
| KDR (VEGFR2)* | 1.47 |
| THBD (Thrombomodulin) | 4.33 |
| VEGFA* | 1.54 |
| VEGFC* | 1.68 |
| VWF* | 12.97 |
| Miscellaneous | |
| AIM2 (Absent in melanoma 2)* | 14.76 |
| CD40 (TNF receptor superfamily member 5)* | 3.95 |
| FZD1 (Frizzled homolog 1)* | 2.55 |
| Gene class and specific genes . | Fold change . |
|---|---|
| Cell-to-cell interactions | |
| CDH1 (E-cadherin)* | 1.54 |
| ITGA9 (Integrin alpha 9) | 2.37 |
| ITGB4 (Integrin beta 4)* | 2.15 |
| VCAM1 (Vascular cell adhesion molecule 1)* | 26.18 |
| Cell-cycle control | |
| CCND1 (Cyclin D1)* | 6.67 |
| Chemokines | |
| CCL18 | 10.50 |
| CCL19 | 80.67 |
| CCL2* | 88.11 |
| CCL8* | 51.61 |
| CXCL10 (IP10)* | 48.07 |
| CXCL12 (SDF-1) | 25.75 |
| CXCL9 (Mig) | 254.14 |
| Cytokines | |
| IL13RA1* | 9.19 |
| IL6R | 2.25 |
| IL33* | 2.75 |
| IL8* | 5.07 |
| Genes overexpressed in EBV-infected lymphoma cell lines | |
| EBI3 (EBV-induced gene 3)* | 2.76 |
| BASP1 (Brain abundant, membrane-attached signal protein 1)* | 13.21 |
| HCK (Hemopoietic cell kinase) | 16.01 |
| TNFRSF10D (Tumor necrosis factor receptor superfamily, member 10d)* | 1.53 |
| Extracellular matrix interactions | |
| COL1A1 (Collagen, type I alpha 1)* | 45.91 |
| COL1A2 (Collagen, type I alpha 2)* | 33.35 |
| COL3A1 (Collagen, type III alpha 1)* | 59.68 |
| FN1 (Fibronectin 1)* | 91.79 |
| TIMP1 (TIMP metallopeptidase inhibitor 1)* | 6.28 |
| TIMP2 (TIMP metallopeptidase inhibitor 2) | 10.79 |
| TIMP3 (TIMP metallopeptidase inhibitor 3) | 9.57 |
| Growth factors and receptors | |
| GHR (Growth hormone receptor)* | 1.35 |
| PDGFB (Platelet-derived growth factor beta)* | 2.16 |
| PDGFC (Platelet derived growth factor C)* | 7.92 |
| PDGFRA (Platelet-derived growth factor receptor, alpha polypeptide)* | 7.47 |
| Immunity and lymphocyte development | |
| CD86* | 6.04 |
| HLA-DOA* | 12.12 |
| HLA-DRA* | 12.18 |
| IL4I1 (Interleukin 4–induced 1)* | 6.40 |
| SLAMF8 (SLAM family member 8)* | 45.26 |
| TLR2 | 8.76 |
| TLR7* | 5.26 |
| TLR8 | 13.49 |
| Microenvironment | |
| CD163 | 134.47 |
| Oncogenes | |
| MAFB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog) | 37.94 |
| MET (Hepatocyte growth factor receptor)* | 2.00 |
| MYC* | 1.23 |
| Transcription factors | |
| STAT1* | 5.25 |
| STAT2* | 1.86 |
| Vascular biology | |
| KDR (VEGFR2)* | 1.47 |
| THBD (Thrombomodulin) | 4.33 |
| VEGFA* | 1.54 |
| VEGFC* | 1.68 |
| VWF* | 12.97 |
| Miscellaneous | |
| AIM2 (Absent in melanoma 2)* | 14.76 |
| CD40 (TNF receptor superfamily member 5)* | 3.95 |
| FZD1 (Frizzled homolog 1)* | 2.55 |
A complete list of genes is given in supplementary Table 1.
Genes are also overexpressed in NKTCL cell lines compared to normal NK cells.
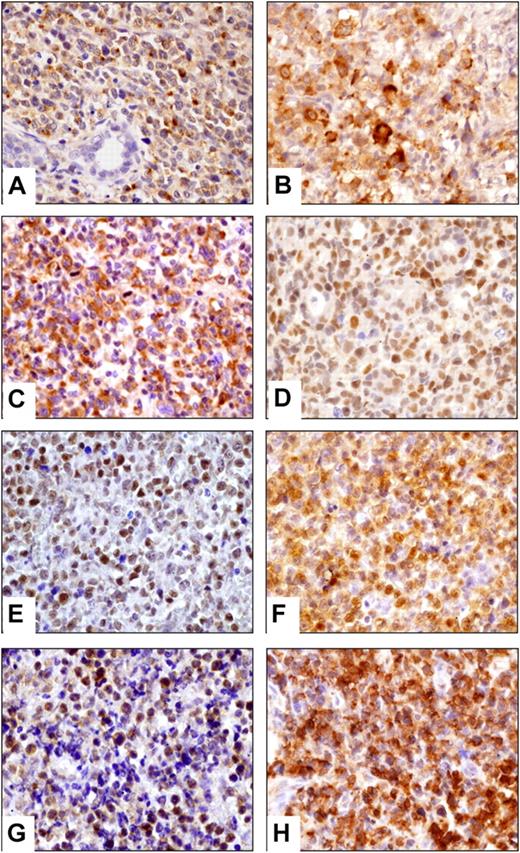
Among these overexpressed genes tested by immunohistochemistry, E-cadherin, CD163, clusterin, EGFR, and cyclin D1 expression was restricted to non-neoplastic cells, that is, epithelial cells (E-cadherin, clusterin, cyclin D1, EGFR), stromal cells (clusterin, cyclin D1), and numerous histiocytes (CD163; Table 3). In contrast, VCAM1 and the EBV-induced gene 3 (EBI3), a subunit of IL-27, labeled the neoplastic cells of 4 (31%) of 13 and 9 (75%) of 12 evaluable NKTCLs, respectively (Figure 1B). SNK6 and MEC04 cells were also positive for EBI3.
Summary of immunohistochemical results
| . | NKTCL (n = 16) . | NKTCL cell lines (n = 2) . | PTCL, NOS (n = 17) . |
|---|---|---|---|
| Clusterin | 0/4 (0%) | 0/1 | ND |
| E-cadherin | 0/4 (0%) | ND | ND |
| Cyclin D1 | 0/4 (0%) | ND | ND |
| EGFR | 0/4 (0%) | ND | ND |
| CD163 | 0/4 (0%) | 0/2 | ND |
| EBI3 | 9/12 (75%) | 2/2 | 1/13 (8%) |
| VCAM1 | 4/13 (31%) | 0/2 | 0/7 (0%) |
| Granzyme H | 16/16 (100%) | 2/2 | 2/16 (13%) |
| pSTAT3 | 13/14 (93%) | 2/2* | 2/17 (12%) |
| pAKT | 9/9 (100%) | NE | 0/16 (0%) |
| VEGFA | 9/9 (100%) | 2/2 | 5/13 (38%) |
| VEGFR2 | 4/5 (80%) | 2/2† | ND |
| PDGFRα | 11/13 (85%) | 1/1 | 10/10 (100%) |
| pPDGFRα | 13/13 (100%) | ND | 7/8 (88%) |
| RelA | 5/6 (83%)‡ | 2/2‡ | ND |
| RelB | 0/6 (0%) | ND | ND |
| cRel | 4/5 (80%)§ | 0/2 | ND |
| Beta-catenin | 0/8 (0%) | ND | ND |
| . | NKTCL (n = 16) . | NKTCL cell lines (n = 2) . | PTCL, NOS (n = 17) . |
|---|---|---|---|
| Clusterin | 0/4 (0%) | 0/1 | ND |
| E-cadherin | 0/4 (0%) | ND | ND |
| Cyclin D1 | 0/4 (0%) | ND | ND |
| EGFR | 0/4 (0%) | ND | ND |
| CD163 | 0/4 (0%) | 0/2 | ND |
| EBI3 | 9/12 (75%) | 2/2 | 1/13 (8%) |
| VCAM1 | 4/13 (31%) | 0/2 | 0/7 (0%) |
| Granzyme H | 16/16 (100%) | 2/2 | 2/16 (13%) |
| pSTAT3 | 13/14 (93%) | 2/2* | 2/17 (12%) |
| pAKT | 9/9 (100%) | NE | 0/16 (0%) |
| VEGFA | 9/9 (100%) | 2/2 | 5/13 (38%) |
| VEGFR2 | 4/5 (80%) | 2/2† | ND |
| PDGFRα | 11/13 (85%) | 1/1 | 10/10 (100%) |
| pPDGFRα | 13/13 (100%) | ND | 7/8 (88%) |
| RelA | 5/6 (83%)‡ | 2/2‡ | ND |
| RelB | 0/6 (0%) | ND | ND |
| cRel | 4/5 (80%)§ | 0/2 | ND |
| Beta-catenin | 0/8 (0%) | ND | ND |
Results refer to expression in neoplastic cells.
ND indicates not done; and NE, nonevaluable.
Phosphorylated STAT3 stained most MEC04 cells, but only a minority of SNK6 cells.
Performed by flow cytometric immunophenotyping.
Cytoplasmic and nuclear staining of neoplastic cells.
Cytoplasmic staining of most neoplastic cells.
Validation of gene expression profiling by immunohistochemistry. Representative NKTCLs disclosed cytoplasmic staining of neoplastic cells for (A) gzm H, (B) EBI3, (C) VEGFA, nuclear localization of (D) pSTAT3, (E) pAKT, cytoplasmic and nuclear staining of (F) RelA, and positivity for (G) PDGFRα and (H) pPDGFRα. Images were captured with a Zeiss Axioskop2 microscope (Zeiss). Photographs were taken with an Olympus DP70 camera. Image acquisition was performed with Olympus DP Controller 2002, and images were processed with Adobe Photoshop Version 7.0 (Adobe Systems). Original magnification, ×400 (A-H).
Validation of gene expression profiling by immunohistochemistry. Representative NKTCLs disclosed cytoplasmic staining of neoplastic cells for (A) gzm H, (B) EBI3, (C) VEGFA, nuclear localization of (D) pSTAT3, (E) pAKT, cytoplasmic and nuclear staining of (F) RelA, and positivity for (G) PDGFRα and (H) pPDGFRα. Images were captured with a Zeiss Axioskop2 microscope (Zeiss). Photographs were taken with an Olympus DP70 camera. Image acquisition was performed with Olympus DP Controller 2002, and images were processed with Adobe Photoshop Version 7.0 (Adobe Systems). Original magnification, ×400 (A-H).
NKTCL has a distinct molecular signature compared with PTCL, NOS
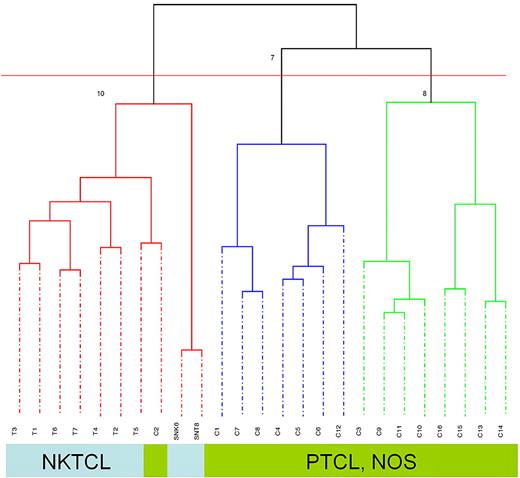
Unsupervised, consensus clustering applied to the molecular signatures of 9 NKTCL samples (7 biopsies and 2 cell lines) and 16 PTCL, NOS samples generated 2 main branches correlated to the pathologic diagnosis. Indeed, all NKTCL samples clustered in one branch, and the other branch comprised PTCL, NOS cases only (Figure 2). Interestingly, one case of nodal γδ PTCL, NOS with an activated cytotoxic profile (gzm B+) but negative for EBV (sample C2) clustered with NKTCL cases.
Unsupervised clustering of 7 NKTCLs, 2 NKTCL-derived cell lines, and 16 PTCLs, NOS. Dendrogram of 23 lymphoma tissues and 2 cell lines based on principal component analysis demonstrated 2 major clusters of NKTCL including tumor-derived cell lines and PTCL, NOS, respectively. T1-T7: 7 NKTCL samples, SNK6, SNT8; C1-16: 16 PTCL, NOS samples.
Unsupervised clustering of 7 NKTCLs, 2 NKTCL-derived cell lines, and 16 PTCLs, NOS. Dendrogram of 23 lymphoma tissues and 2 cell lines based on principal component analysis demonstrated 2 major clusters of NKTCL including tumor-derived cell lines and PTCL, NOS, respectively. T1-T7: 7 NKTCL samples, SNK6, SNT8; C1-16: 16 PTCL, NOS samples.
By supervised analysis, a total of 372 and 691 probe sets corresponding to 278 and 484 overexpressed and underexpressed genes, respectively, significantly distinguished tissues involved by NKTCLs versus PTCLs, NOS (P < .005; Table 4; supplemental Table 2). As expected, among the top overexpressed genes in NKTCLs were many genes encoding killer immunoglobulin–like receptors and others encoding NK cell–associated molecules (NCAM1, CD244) or related to cytotoxic functions (GZMB, GZMH, CTSW, PRF1…). Significantly, NKTCLs also overexpressed genes encoding cell adhesion molecules (ITGAM, ITGB6), chemokines and their receptors (CCL4, CCL5, CCR1), apoptosis-related molecules (FASLG, BCL2L2), cytidine deaminase (APOBEC3G), and oncogenes (MYC, RHOC).
Selection of genes overexpressed in NKTCL tissues compared to PTCL, NOS cells with P < .005
| Gene class and specific genes . | Fold change . |
|---|---|
| Apoptosis | |
| BCL2L2 (BCL-W) | 1.68 |
| FASLG (Fas ligand) | 5.99 |
| Cell-to-cell interactions | |
| ITGAM (Integrin, alpha M) | 2.30 |
| Chemokines | |
| CCL4 (MIP-1-beta) | 4.30 |
| CCL5 (RANTES) | 8.66 |
| Cytokines and receptors | |
| IL20 | 1.15 |
| IL2RB | 2.32 |
| Growth factors | |
| TGFB2 | 1.14 |
| Cytotoxic molecules | |
| CTSW (Cathepsin W) | 11.31 |
| GZMA (Granzyme A) | 8.03 |
| GZMB (Granzyme B) | 5.39 |
| GZMH (Granzyme H) | 13.66 |
| PRF1 (Perforin 1) | 3.34 |
| NK cell–associated molecules | |
| APOBEC3G (Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G) | 2.40 |
| CD244 (NK-cell receptor 2B4) | 6.75 |
| KIR2DL3 /// KIR2DS5 | 10.30 |
| KIR2DL4 | 11.77 |
| KIR2DL5A | 7.47 |
| KIR2DS4 | 4.94 |
| KIR3DL1 | 9.96 |
| KIR3DL2 | 14.02 |
| KIR3DL3 | 12.51 |
| KLRC1 /// KLRC2 | 25.75 |
| KLRC3 | 32.96 |
| KLRC4 /// KLRK1 | 10.33 |
| KLRD1 | 32.45 |
| NCAM1 (CD56) | 10.54 |
| Immune responses | |
| IRF6 (Interferon regulatory factor 6) | 1.54 |
| Oncogenes | |
| MYC | 1.18 |
| RHOC (Ras homolog gene family, member C) | 3.22 |
| Transcription factors | |
| ATF6 (Activating transcription factor 6) | 1.39 |
| Miscellaneous | |
| MAPK1 | 1.46 |
| PTPN22 (Protein tyrosine phosphatase, nonreceptor type 22) | 2.47 |
| YWHAH (Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide) | 1.62 |
| Gene class and specific genes . | Fold change . |
|---|---|
| Apoptosis | |
| BCL2L2 (BCL-W) | 1.68 |
| FASLG (Fas ligand) | 5.99 |
| Cell-to-cell interactions | |
| ITGAM (Integrin, alpha M) | 2.30 |
| Chemokines | |
| CCL4 (MIP-1-beta) | 4.30 |
| CCL5 (RANTES) | 8.66 |
| Cytokines and receptors | |
| IL20 | 1.15 |
| IL2RB | 2.32 |
| Growth factors | |
| TGFB2 | 1.14 |
| Cytotoxic molecules | |
| CTSW (Cathepsin W) | 11.31 |
| GZMA (Granzyme A) | 8.03 |
| GZMB (Granzyme B) | 5.39 |
| GZMH (Granzyme H) | 13.66 |
| PRF1 (Perforin 1) | 3.34 |
| NK cell–associated molecules | |
| APOBEC3G (Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G) | 2.40 |
| CD244 (NK-cell receptor 2B4) | 6.75 |
| KIR2DL3 /// KIR2DS5 | 10.30 |
| KIR2DL4 | 11.77 |
| KIR2DL5A | 7.47 |
| KIR2DS4 | 4.94 |
| KIR3DL1 | 9.96 |
| KIR3DL2 | 14.02 |
| KIR3DL3 | 12.51 |
| KLRC1 /// KLRC2 | 25.75 |
| KLRC3 | 32.96 |
| KLRC4 /// KLRK1 | 10.33 |
| KLRD1 | 32.45 |
| NCAM1 (CD56) | 10.54 |
| Immune responses | |
| IRF6 (Interferon regulatory factor 6) | 1.54 |
| Oncogenes | |
| MYC | 1.18 |
| RHOC (Ras homolog gene family, member C) | 3.22 |
| Transcription factors | |
| ATF6 (Activating transcription factor 6) | 1.39 |
| Miscellaneous | |
| MAPK1 | 1.46 |
| PTPN22 (Protein tyrosine phosphatase, nonreceptor type 22) | 2.47 |
| YWHAH (Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide) | 1.62 |
A complete list of genes is given in supplementary Table 2.
In addition to NCAM1 (CD56) and cytotoxic proteins such as TiA1 and gzm B, which were strongly expressed in all NKTCLs, we investigated the expression of another cytotoxic granule-associated protein, gzm H, of which transcript levels were 13.66 times greater than in PTCLs, NOS. As shown in Figure 1, a strong granular cytoplasmic staining was found in virtually all neoplastic cells of all NKTCL tumors tested (n = 16) as well as in the SNK6 and MEC04 cells, whereas gzm H was restricted to small lymphocytes in most PTCLs, NOS. Among PTCL, NOS, the γδ T-cell lymphoma was gzm H positive and another case disclosed heterogeneous partial staining.
Patterns of copy number aberrations and identification of genes relevant to the pathobiology of NKTCL
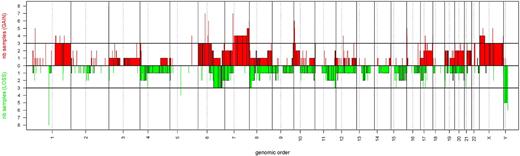
The array-based comparative genomic hybridization findings are summarized in Table 5 and in Figure 3. Extensive losses and gains of larger and smaller chromosomal regions were found in all NKTCL samples analyzed. Recurrent copy number aberrations observed in 3 or more (37.5%) of 8 NKTCL biopsy samples comprised 16 regions of chromosomal gains (on 10p15, 7q35-q36, 7p11.2-p12, 7q11.2-q34, 16p13.3, and 17q12 and on 1q21-q44, 4p16, 6p11.1-p25, 6q11.1-q14, 6q27, 8p23.3, 9q34, 10p14-p15, 11p15, and 22q11.21) and 4 regions of losses (on 6q16-q25, 11q24-q25, 17p13.3, and 8p22-p23).
Copy number alterations identified by aCGH in the NKTCL samples analyzed in this study
| Chromosomes . | Start position, Mb . | End position, Mb . | No. of clones . | NKTCL tissues and cell lines (n=10) . | NKTCL tissues (n=8) . | SNK6 . | SNT8 . |
|---|---|---|---|---|---|---|---|
| Gain | |||||||
| 1q21-q44 | 143 | 245 | 170 | 5 | 3 | 1 | 1 |
| 4p16 | 2 | 6 | 5 | 3 | 3 | 0 | 0 |
| 6p25-p11.1 | 0 | 58 | 93 | 4 | 3 | 1 | 0 |
| 6q11.1-q14 | 58 | 87 | 26 | 4 | 3 | 1 | 0 |
| 6q27 | 167 | 171 | 5 | 3 | 3 | 0 | 0 |
| 7p12-p11.2 | 53 | 55 | 4 | 5 | 4 | 0 | 1 |
| 7q11.2-q34 | 62 | 142 | 99 | 5 | 4 | 1 | 0 |
| 7q35-q36 | 143 | 156 | 11 | 6 | 5 | 1 | 0 |
| 8p23.3 | 0.13 | 1.96 | 5 | 4 | 3 | 1 | 0 |
| 9q34 | 134 | 138 | 7 | 4 | 3 | 1 | 0 |
| 10p15 | 0 | 4 | 5 | 6 | 6 | 0 | 0 |
| 10p15-p14 | 6 | 8 | 6 | 3 | 3 | 0 | 0 |
| 11p15 | 0 | 3 | 10 | 4 | 3 | 1 | 0 |
| 16p13.3 | 1 | 2 | 5 | 4 | 4 | 0 | 0 |
| 17q12 | 28.81 | 30.78 | 12 | 5 | 3 | 1 | 1 |
| 17q12 | 29.84 | 30.30 | 7 | 6 | 4 | 1 | 1 |
| 22q11.21 | 16.15 | 21.17 | 6 | 4 | 3 | 0 | 1 |
| Loss | |||||||
| 6q16-q25 | 100 | 154 | 75 | 4 | 3 | 1 | 0 |
| 8p23-p22 | 4 | 15 | 14 | 3 | 3 | 0 | 0 |
| 11q24-q25 | 128 | 133 | 8 | 4 | 3 | 0 | 1 |
| 17p13.3 | 0.34 | 0.96 | 6 | 4 | 3 | 1 | 0 |
| Chromosomes . | Start position, Mb . | End position, Mb . | No. of clones . | NKTCL tissues and cell lines (n=10) . | NKTCL tissues (n=8) . | SNK6 . | SNT8 . |
|---|---|---|---|---|---|---|---|
| Gain | |||||||
| 1q21-q44 | 143 | 245 | 170 | 5 | 3 | 1 | 1 |
| 4p16 | 2 | 6 | 5 | 3 | 3 | 0 | 0 |
| 6p25-p11.1 | 0 | 58 | 93 | 4 | 3 | 1 | 0 |
| 6q11.1-q14 | 58 | 87 | 26 | 4 | 3 | 1 | 0 |
| 6q27 | 167 | 171 | 5 | 3 | 3 | 0 | 0 |
| 7p12-p11.2 | 53 | 55 | 4 | 5 | 4 | 0 | 1 |
| 7q11.2-q34 | 62 | 142 | 99 | 5 | 4 | 1 | 0 |
| 7q35-q36 | 143 | 156 | 11 | 6 | 5 | 1 | 0 |
| 8p23.3 | 0.13 | 1.96 | 5 | 4 | 3 | 1 | 0 |
| 9q34 | 134 | 138 | 7 | 4 | 3 | 1 | 0 |
| 10p15 | 0 | 4 | 5 | 6 | 6 | 0 | 0 |
| 10p15-p14 | 6 | 8 | 6 | 3 | 3 | 0 | 0 |
| 11p15 | 0 | 3 | 10 | 4 | 3 | 1 | 0 |
| 16p13.3 | 1 | 2 | 5 | 4 | 4 | 0 | 0 |
| 17q12 | 28.81 | 30.78 | 12 | 5 | 3 | 1 | 1 |
| 17q12 | 29.84 | 30.30 | 7 | 6 | 4 | 1 | 1 |
| 22q11.21 | 16.15 | 21.17 | 6 | 4 | 3 | 0 | 1 |
| Loss | |||||||
| 6q16-q25 | 100 | 154 | 75 | 4 | 3 | 1 | 0 |
| 8p23-p22 | 4 | 15 | 14 | 3 | 3 | 0 | 0 |
| 11q24-q25 | 128 | 133 | 8 | 4 | 3 | 0 | 1 |
| 17p13.3 | 0.34 | 0.96 | 6 | 4 | 3 | 1 | 0 |
Genomic profiles of NKTCL. Pangenomic view of 8 NKTCL tissue samples. The horizontal axis represents the genomic order, and the vertical axis represents the number of samples with gains or losses. Gains are represented as red bars and losses are represented as green bars.
Genomic profiles of NKTCL. Pangenomic view of 8 NKTCL tissue samples. The horizontal axis represents the genomic order, and the vertical axis represents the number of samples with gains or losses. Gains are represented as red bars and losses are represented as green bars.
To identify candidate genes in the regions of copy number aberrations, we selected the genes with a Welch t test P value less than .001 and a fold average expression difference greater than 2.0 (or < 0.5) in NKTCL tissues compared with normal NK cells. The resulting list (supplemental Table 3) included genes related to malignant transformation and invasion (S100A16, LAMB1, LAMC1, COL1A2, CTSB), cell-cycle progression (CCND3), signal transduction (FYN), tumor suppressor genes (HACE1, CAV1, CAV2, DLC1), as well as members of important pathways which, according to GEP analysis, appear deregulated in NKTCL (see “Identification of distinct pathways in NKTCL”), such as angiogenesis (MET, S100A13), nuclear factor-κB (NF-κB; PRKCQ, TNFRSF21), WNT (CUL1, FZD1, SGK1), and Janus kinase–signal transducers and activators of transcription (JAK-STAT; AKT3, IL6R, CCL2) signaling pathways.
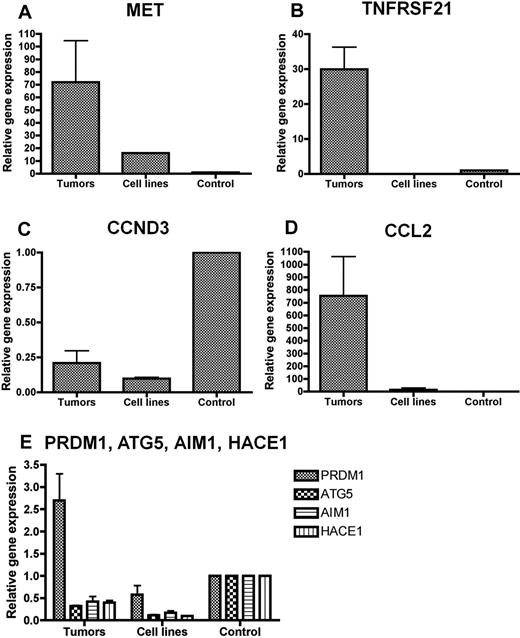
We selected for qRT-PCR correlation several biologically relevant deregulated candidate genes mapping to altered chromosomal regions at 6q21 (HACE1), 17q12 (CCL2), 6p12.3 (TNFRSF21), 6p21.1 (CCND3), and 7q31.2 (MET). We also evaluated ATG5, AIM1, and PRDM1 genes reported as candidate tumor suppressor genes at 6q21 in NKTCL cell lines, which were downexpressed in our microarray data albeit at less statistical significance. In agreement with the microarray results, we confirmed by qRT-PCR the overexpression of MET, CCL2, and TNFRSF21 in NKTCL samples compared with normal NK cells and the underexpression of CCND3 and HACE1 (Figure 4). ATG5 and AIM1 transcripts also were markedly reduced in both cell lines and primary tumors. In contrast, quantification of PRDM1 mRNA produced discrepant results in cell lines (underexpressed levels, in accordance with published data5 ) and primary tumors (increased transcript levels in 4 of 6 samples). These findings were independent of the presence or absence of 6q21 deletion.
Quantification of selected genes by qRT-PCR analysis. qRT-PCR analysis for (A) MET, (B) TNFRSF21, (C) CCND3, (D) CCL2, and (E) PRDM1, ATG5, AIM1, and HACE1 in NKTCL primary tumors and cell lines. The results are expressed as relative fold change compared with resting CD3−/CD56+ NK cells sorted from peripheral blood. Quantifications were performed in duplicate, and mean values and SD were calculated for each transcript. (A-D) In agreement with the microarray results, mRNA levels of MET, CCL2, and TNFRSF21 are increased in NKTCL primary tumors compared with normal NK cells, whereas CCND3 mRNA is reduced (E). The analysis of 4 putative tumor suppressors in 6q21 region show that the transcripts levels of HACE1, AIM1, and ATG5 are reduced in primary tumors and cell lines whereas the mRNA level of PRDM1 is increased in primary tumors, due to a wide variation from case to case (not shown).
Quantification of selected genes by qRT-PCR analysis. qRT-PCR analysis for (A) MET, (B) TNFRSF21, (C) CCND3, (D) CCL2, and (E) PRDM1, ATG5, AIM1, and HACE1 in NKTCL primary tumors and cell lines. The results are expressed as relative fold change compared with resting CD3−/CD56+ NK cells sorted from peripheral blood. Quantifications were performed in duplicate, and mean values and SD were calculated for each transcript. (A-D) In agreement with the microarray results, mRNA levels of MET, CCL2, and TNFRSF21 are increased in NKTCL primary tumors compared with normal NK cells, whereas CCND3 mRNA is reduced (E). The analysis of 4 putative tumor suppressors in 6q21 region show that the transcripts levels of HACE1, AIM1, and ATG5 are reduced in primary tumors and cell lines whereas the mRNA level of PRDM1 is increased in primary tumors, due to a wide variation from case to case (not shown).
Identification of distinct pathways in NKTCL
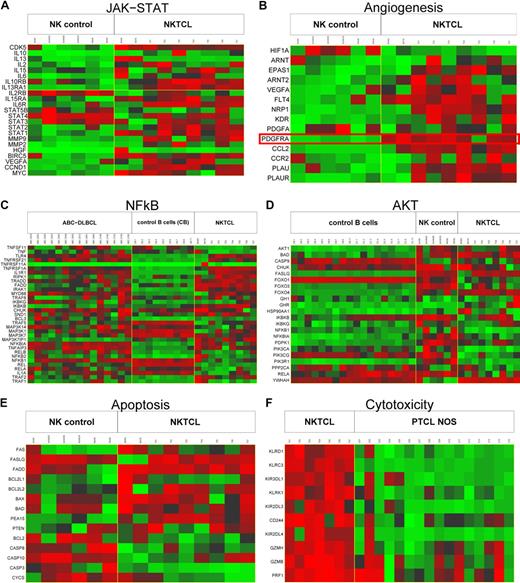
To identify potentially relevant biologic pathways in NKTCL, 4 gene sets analysis methods were applied. The most discriminatory pathways when comparing NKTCL and normal NK cells or PTCL, NOS, comprised those related to important cell functions such as apoptosis, cell adhesion, cell communication, extracellular matrix-receptor interaction, cell cycle (p27 regulation), cytokine-cytokine receptor interaction, as well as TGF-β, mitogen-activated protein kinase, WNT, and JAK-STAT signaling pathways (Table 6; Figure 5A,E). More specifically, angiogenesis-related (VEGF) and NF-κB also were discriminatory pathways between neoplastic samples and normal NK cells. FAS and AKT signaling genes pathways and especially genes related to NK cell–mediated cytotoxicity (Figure 5B,C,D,F) were differentially expressed between NKTCL and PTCL, NOS.
Selection of pathways differentially expressed in NKTCL tissues compared to normal NK cells or PTCL, NOS
| Pathway name . | Median P value, compared with normal NK cells* . | Median P value, compared with PTCL, NOS* . |
|---|---|---|
| Apoptosis | ||
| Apoptosis | .002 | < .002 |
| Regulation of BAD phosphorylation | .109 | < .018 |
| PTEN-dependent cell-cycle arrest and apoptosis | .246 | < .015 |
| FAS signaling pathway | .257 | < .006 |
| Cytotoxicity | ||
| NK cell–mediated cytotoxicity | .002 | < .0001 |
| Ras-independent pathway in NK cell–mediated cytotoxicity | .325 | < .0001 |
| Cell adhesion molecules/extracellular matrix receptor interaction | ||
| Cell communication | .001 | < .016 |
| ECM receptor interaction | .001 | < .016 |
| Focal adhesion | .001 | < .023 |
| Cell adhesion molecules | .002 | < .012 |
| Leukocyte transendothelial migration | .002 | .371 |
| Inhibition of matrix metalloproteinases | .003 | .018 |
| Integrin signaling pathway | .003 | .484 |
| Cell cycle | ||
| Regulation of p27 during phosphorylation | < .006 | .008 |
| Cyclin E destruction pathway | < .041 | .003 |
| Chemokines/cytokines | ||
| Cytokine cytokine receptor interaction | .002 | < .001 |
| Signal transduction | ||
| JAK-STAT signaling pathway | .002 | .01 |
| MAPK signaling pathway | .002 | .015 |
| mTOR signaling pathway | .002 | .06 |
| NF-κB signaling pathway | .002 | .06 |
| TGFβ signaling pathway | .002 | .019 |
| VEGF signaling pathway | .002 | .418 |
| Notch signaling pathway | < .005 | .025 |
| Wnt signaling pathway | < .005 | < .013 |
| AKT signaling pathway | .14 | < .001 |
| Pathway name . | Median P value, compared with normal NK cells* . | Median P value, compared with PTCL, NOS* . |
|---|---|---|
| Apoptosis | ||
| Apoptosis | .002 | < .002 |
| Regulation of BAD phosphorylation | .109 | < .018 |
| PTEN-dependent cell-cycle arrest and apoptosis | .246 | < .015 |
| FAS signaling pathway | .257 | < .006 |
| Cytotoxicity | ||
| NK cell–mediated cytotoxicity | .002 | < .0001 |
| Ras-independent pathway in NK cell–mediated cytotoxicity | .325 | < .0001 |
| Cell adhesion molecules/extracellular matrix receptor interaction | ||
| Cell communication | .001 | < .016 |
| ECM receptor interaction | .001 | < .016 |
| Focal adhesion | .001 | < .023 |
| Cell adhesion molecules | .002 | < .012 |
| Leukocyte transendothelial migration | .002 | .371 |
| Inhibition of matrix metalloproteinases | .003 | .018 |
| Integrin signaling pathway | .003 | .484 |
| Cell cycle | ||
| Regulation of p27 during phosphorylation | < .006 | .008 |
| Cyclin E destruction pathway | < .041 | .003 |
| Chemokines/cytokines | ||
| Cytokine cytokine receptor interaction | .002 | < .001 |
| Signal transduction | ||
| JAK-STAT signaling pathway | .002 | .01 |
| MAPK signaling pathway | .002 | .015 |
| mTOR signaling pathway | .002 | .06 |
| NF-κB signaling pathway | .002 | .06 |
| TGFβ signaling pathway | .002 | .019 |
| VEGF signaling pathway | .002 | .418 |
| Notch signaling pathway | < .005 | .025 |
| Wnt signaling pathway | < .005 | < .013 |
| AKT signaling pathway | .14 | < .001 |
With a median P value < .02 compared to either normal NK cells or PTCL, NOS.
Cellular programs deregulated in NKTCL. Representative molecular pathways differentially expressed in NKTCLs by comparison with either normal NK cells (JAK-STAT in panel A, angiogenesis in panel B, apoptosis in panel E), normal B cells and ABC-DLBCL (NF-κB in panel C), normal NK cells and normal B cells (AKT in panel D), or PTCL, NOS (cytotoxicity in panel F) were illustrated. For each line, green corresponds to the minimal intensity value (min), red corresponds to the maximal intensity value (max), and black corresponds to (min + max)/2. Several genes related to JAK-STAT, angiogenesis, and apoptosis-related pathways are overexpressed in NKTCLs compared with normal NK cells, and genes related to the cytotoxicity-related pathway also are overexpressed in NKTCLs compared with PTCL, NOS. Many genes in the NF-κB and AKT signaling pathways are overexpressed in NKTCLs compared with normal B cells, with a molecular signature similar to that of ABC-DLBCL in NK-κB pathway. The expression data of PDGFRA in NKTCLs, 2 NKTCL-derived cell lines, and normal NK cells is highlighted. T1-7: 7 NKTCL samples; C1-16: 16 PTCL, NOS samples; AcNK, AcNK02, AcNK08, AcNK24: activated NK cells; ReNK, ReNK: resting NK cells; CB.1-8 and CB.11: 9 centroblasts samples; CC.1-8 and CC.11: 9 centrocytes samples; and ABC.2024-2195: 15 ABC-DLBCL samples.
Cellular programs deregulated in NKTCL. Representative molecular pathways differentially expressed in NKTCLs by comparison with either normal NK cells (JAK-STAT in panel A, angiogenesis in panel B, apoptosis in panel E), normal B cells and ABC-DLBCL (NF-κB in panel C), normal NK cells and normal B cells (AKT in panel D), or PTCL, NOS (cytotoxicity in panel F) were illustrated. For each line, green corresponds to the minimal intensity value (min), red corresponds to the maximal intensity value (max), and black corresponds to (min + max)/2. Several genes related to JAK-STAT, angiogenesis, and apoptosis-related pathways are overexpressed in NKTCLs compared with normal NK cells, and genes related to the cytotoxicity-related pathway also are overexpressed in NKTCLs compared with PTCL, NOS. Many genes in the NF-κB and AKT signaling pathways are overexpressed in NKTCLs compared with normal B cells, with a molecular signature similar to that of ABC-DLBCL in NK-κB pathway. The expression data of PDGFRA in NKTCLs, 2 NKTCL-derived cell lines, and normal NK cells is highlighted. T1-7: 7 NKTCL samples; C1-16: 16 PTCL, NOS samples; AcNK, AcNK02, AcNK08, AcNK24: activated NK cells; ReNK, ReNK: resting NK cells; CB.1-8 and CB.11: 9 centroblasts samples; CC.1-8 and CC.11: 9 centrocytes samples; and ABC.2024-2195: 15 ABC-DLBCL samples.
For example, as illustrated in Figure 5A, several component genes of JAK-STAT pathway such as IL13RA1, IL6R, STAT1, STAT2, VEGFA, CCND1, and MYC were overexpressed in NKTCL compared with normal NK cells and IL12RB1, IL20, IL2RB, and MYC compared with PTCL, NOS. Altogether, the pathway analysis supports the involvement of several important pathways in NKTCL.
Activation of several oncogenic pathways (AKT, STAT3, NF-κB, VEGF) in NKTCL
To evaluate the possible activation of pathways identified by the use of statistical methods, the expression of several representative genes was assessed at the protein level (Table 3; Figure 1). Nuclear expression of pAKT (Ser473) and pSTAT3 (Tyr705) was found in 9 of 9 and 13 of 14 NKTCL cases, respectively. Strong expression of VEGFA, one of the important pSTAT3-regulated gene products related to angiogenesis and immune evasion,32 was demonstrated in 9 of 9 NKTCL, and expression of its receptor VEGFR2 was evidenced in the neoplastic cells of 4 of 5 tumors. Positivity for pSTAT3 and VEGFA was confirmed in MEC04 and SNK6 cell lines, and pAKT was demonstrated in MEC04 cells.
Differential expression of WNT and NF-κB pathway genes also were identified in pathway analyses. Nuclear expression of β-catenin, a hallmark of activation of WNT pathway, was not observed in neoplastic cells (n = 8), contrasting with cytoplasmic and/or membrane staining in surrounding epithelial and endothelial cells. However, RelA and cRel, 2 important molecules of the canonical NF-κB pathway, were expressed in the cytoplasm of most evaluable NKTCLs associated with nuclear localization of RelA (Figure 1). RelB, a molecule of the alternative NF-κB pathway, was negative in all investigated NKTCLs (n = 6).
Activation of PDGF pathway in NKTCL
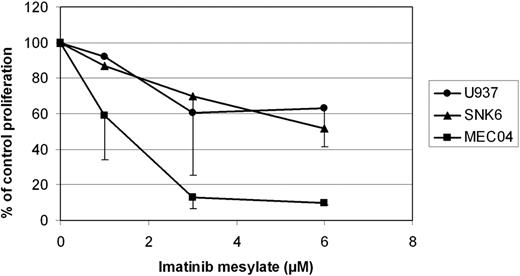
Because PDGFRα, a receptor tyrosine kinase mediating important cell functions such as migration, proliferation, and cell survival and known to interact with PI3K/AKT and STAT proteins,33 was highly expressed at the mRNA level (Table 2; Figure 5B), this factor was evaluated by immunohistochemistry. Eleven (85%) of 13 NKTCL cases demonstrated cytoplasmic expression of PDGFRα. In addition, its phosphorylated form (pPDGFRα) was found in all 13 NKTCLs (Table 3; Figure 1). In agreement with a recent report,34 expression of both PDGFRα and pPDGFRα also was demonstrated in most PTCLs, NOS (Table 3). In view of these findings, we investigated the potential implication of the PDGF signaling pathway in the proliferation of NKTCL cells. Imatinib mesylate induced dramatic concentration-dependent growth inhibition of PDGFRα-positive MEC04 cells, whereas the effect on SNK6 cells was minimal. As shown in Figure 6, a significant 50% growth inhibition was already observed when MECO4 cells were exposed to imatinib mesylate at 1μM that increased to 90% at 6μM. U937 used as a control cell line known to be resistant to imatinib mesylate maintained 63% proliferation with 6μM imatinib.
Proliferation of NKTCL-derived cell lines in the presence of imatinib mesylate. SNK6, MEC04, and U937 cells were incubated for 72 hours in media with or without imatinib mesylate at concentrations of 1, 3, and 6 μM. Imatinib induced concentration-dependent growth inhibition of MEC04 cells. The effect on SNK6 was not significantly different from that observed on the myelomonocytic U937 cell line. The horizontal axis is the concentration of imatinib mesylate. The vertical axis is the percentage of control proliferation. Bars indicate the SEM of triplicate experiments.
Proliferation of NKTCL-derived cell lines in the presence of imatinib mesylate. SNK6, MEC04, and U937 cells were incubated for 72 hours in media with or without imatinib mesylate at concentrations of 1, 3, and 6 μM. Imatinib induced concentration-dependent growth inhibition of MEC04 cells. The effect on SNK6 was not significantly different from that observed on the myelomonocytic U937 cell line. The horizontal axis is the concentration of imatinib mesylate. The vertical axis is the percentage of control proliferation. Bars indicate the SEM of triplicate experiments.
We further searched for potential genetic alterations underlying PDGFRA deregulation. The absence of copy number gain in PDGFRA locus validated by real-time PCR in 8 interpretable samples was concordant with the absence of genomic imbalance in 4q11-q13 observed in our BAC array CGH data. In addition, we did not evidence mutations in exons 12, 14, and 18 of PDGFRA, in agreement with previous findings.35
Finally, we performed mutations and polymorphisms analysis within the PDGFRA promoter region and in particular investigated the distribution of haplotypes, which have been reported in association with different transcriptional activities.36 Sequence analysis did not show any mutation in the 9 samples tested (compared with the reference sequence), and the distribution in the haplotypes—4 samples had a H1/H2α genotype, 3 H2α/H2α, 1 H2α/H2β, and 1 H2α/H2γ—corresponded to that reported in the Western European population (see supplemental Table 6).
Discussion
In the present study, we characterized the molecular signature of NKTCL in comparison with normal NK cells and to PTCL, NOS. This characterization led to the identification of deregulated genes and signaling pathways that might be relevant to the pathophysiology and clinicopathologic features of the disease and bring rationale for the development of new therapies.
Unsupervised clustering remarkably segregated NKTCL and PTCL, NOS samples. Notably, the NKTCL case with a T-cell cytotoxic phenotype (sample T5) clustered with those of NK-cell origin, providing another molecular argument for grouping nasal “true NK-” and cytotoxic T-cell lymphomas as a single entity as proposed in the current World Health Organization classification.1 Interestingly, the NKTCL case of T-cell derivation also showed the 6q16-q25 deletion. The only mismatch was represented by 1 PTCL, NOS in the NKTCL cluster. That particular case had a γδ activated cytotoxic phenotype, suggesting that derivation from the innate immune system might imprint a peculiar signature.37
By comparison with PTCL, NOS, the molecular signature of NKTCL was significantly contributed by an overexpression of genes associated with cytotoxic functions and NK cell–associated molecules. Interestingly, the greatest fold change of expression was observed for gzm H transcripts. gzm H, a gzm family member sharing a 90% amino acid sequence identity with gzm B, is constitutively expressed in NK cells irrespective of their activation status25 but acts differently from gzm B by inducing a caspase-independent cell death program.38 We confirmed a strong gzm H protein expression in all NKTCLs, contrasting with its negativity in most PTCLs, NOS, with the notable exception of the γδ T-cell lymphoma. Therefore, gzm H appears to be a novel sensitive marker for NKTCL, although its specificity needs to be delineated with respect to its possible expression in other lymphomas derived from the innate immune system.
Other aspects of the NKTCL signature could be related to some peculiar clinicopathologic features of the disease. Angio-invasion and angiocentricity, typical of NKTCL, might be accounted by the high expression of genes such as VCAM1, CXCL9, and CXCL10, encoding proteins involved in the interaction with endothelium or in the pathogenesis of tissue necrosis and vascular damage associated with EBV-positive lymphoproliferations.39 NKTCL, which in most instances arises in the nasal area, is also characterized by a strong tendency to disseminate to other extranodal distant sites. In view of the known roles of CCR7 and SELL/CD62L in peripheral lymph node homing and that of CCL27, CXCL12, in homing to the skin, intestine, and bone marrow, it is likely that the lower levels of CCR7 and SELL/CD62L and the overexpression of CXCL12 might explain the pattern of distribution of the disease.
We confirmed here recurrent genomic gains and losses previously reported in NKTCL. Deletion of chromosome 6q reported as the most characteristic but not specific genetic alteration in NKTCL,3-5,40-42 was present in 40% of our cases, including the SNK6 cell line. In line with the recent report by Iqbal et al,5 we also found recurrent gain of 1q21-q44 and loss of 17p13.3 in primary tumor samples. These authors found PRDM1, ATG5, and AIM1 as target genes in the region of del6q21 and reported both mutation and methylation in PRDM1. Here, we further extended these previous findings by showing low levels of ATG5 and AIM1 transcripts in primary tumors. Conversely, PRDM1 showed a wide range of expression from case to case. In addition, we also found marked reduction in transcripts of HACE1, a gene encoding a novel E3 ubiquitin ligase, which is the target of epigenetic inactivation in Wilms tumor and has been proposed as a tumor suppressor gene in multiple human cancers. Hace1−/− mice are spontaneously prone to developing multiple malignant tumors in various organs.43 Taken together, it is therefore tempting to speculate that HACE1 might be also involved in the pathogenesis of NKTCL.
The proto-oncogene MET mapping in 7q31, a region of recurrent gain in our series, was overexpressed in our samples. This receptor with tyrosine-kinase activity is a receptor to HGF, also overexpressed at the mRNA level in our NKTCL series. Interestingly, this pair of ligand-receptor appears to be linked to angiogenesis, tumor formation, invasion, and metastasis.44 These findings, together with the expression of VEGFA and its receptor VEGFR2, might reflect the implication of angiogenesis and/or VEGF signaling pathway in the pathophysiology of NKTCL.
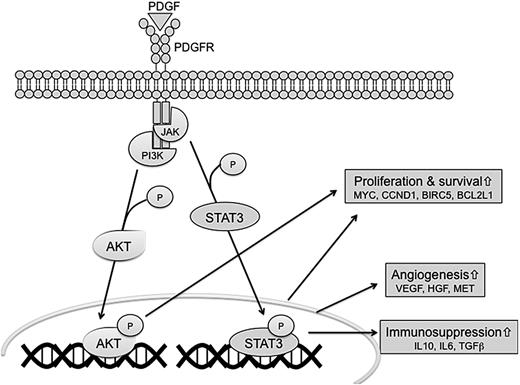
The molecular pathways involved in the pathogenesis of NKTCL are largely unknown. Our study identified deregulated pathways in NKTCL, in comparison with normal NK cells and PTCL, NOS. Among growth factor receptors, the receptor tyrosine kinase PDGFRα was expressed at a greater level than in normal NK cells, at both the mRNA and protein levels, in its activated phosphorylated form, a feature that appears to be shared with PTCL, NOS.34 PDGF signaling pathway is known to be associated with both JAK-STAT and AKT pathways. The AKT protein kinases play a critical role in cell proliferation, survival and programmed cell death, transcription, and cell migration via phosphorylation of a multitude of substrates. STATs are transcription factors activated in response to cytokines and growth factors. STAT3 in particular plays a crucial role in regulating cell growth and apoptosis.32 Several solid or hematologic malignancies, including ALK-positive anaplastic large cell lymphomas, show constitutive STAT3 activation. By using statistical methods, we showed here that AKT and JAK-STAT pathways were differentially expressed in comparison with normal NK cells and/or PTCL, NOS. In addition, we evidenced the nuclear expression of the phosphorylated forms of STAT3 and AKT in most NKTCLs, implying constitutive activation of these pathways in this disease. Our results expand recent findings that AKT was phosphorylated in NK-92 cell line and in a few NKTCL primary tumors, probably through involvement of IL-2 or IL-15.45 Many genes related to proliferation and survival, angiogenesis, and immunosuppression are known to be regulated by STAT3.32 The high transcript levels of several genes regulated by STAT3 in NKTCL compared with normal NK cells such as MYC, VEGFA, BCL2L1—and also less significantly BIRC5, HGF, IL6, MMP2, MMP9, IL10, and CDK5—suggests its implication in the pathogenesis of NKTCL. Among these, MMP2, MMP9, and IL-10 proteins have already been evidenced in NKTCL tumor cells, and we show here immunohistochemical expression of VEGFA.46,47 Recently, we demonstrated that the inhibition of STAT3 activation leads to underexpression of 2 STAT3 target gene products BCL-XL and MYC in MEC04 cells.17 Altogether, these data provide strong arguments supporting the involvement of JAK-STAT and AKT pathways in NKTCL (Figure 7).
Hypothetical representation of signaling pathways involved in NKTCL. The interactions of PDGF, AKT, and JAK-STAT signaling pathways may contribute to the angiogenesis, immunosuppression, proliferation, and survival of NKTCL.
Hypothetical representation of signaling pathways involved in NKTCL. The interactions of PDGF, AKT, and JAK-STAT signaling pathways may contribute to the angiogenesis, immunosuppression, proliferation, and survival of NKTCL.
EBV, constantly present in NKTCL, is suspected to play an important role in oncogenesis. Here we showed that, in comparison with EBV-negative normal NK cells, NKTCL overexpressed several EBV-induced genes.15,30 In agreement with a previous report,24 one of these genes, EBI3, was validated at the protein level. Among the various mechanisms involved in NF-κB pathway, EBV is known to activate NF-κB pathway through LMP-1 and/or TRAF,48 especially in Hodgkin lymphoma and in EBV-positive B-cell lymphoproliferative disorders. Here, we showed differential expression of this pathway in NKTCL and further demonstrate expression of RelA, supporting activation of NF-κB in this entity. Interestingly, the TNFAIP3 gene, encoding an inhibitor of the NF-κB pathway, maps to the region of recurrent loss in 6q16-q25 and was underexpressed in our study. Deletions and/or somatic mutations of this gene have been recently reported in classical Hodgkin lymphoma and primary mediastinal B-cell lymphoma as well as in MALT lymphoma,49,50 supporting the role of this key regulator of NF-κB activity as a novel tumor suppressor gene in these lymphomas. Altogether, these findings suggest the involvement of NF-κB pathway in the pathogenesis of NKTCL. The respective role of EBV and/or TNFAIP3 inactivation in NF-κB activation in NKTCL needs further investigation.
The demonstration of pPDGFRα by immunohistochemistry prompted us to test the sensitivity of NKTCL cell lines in vitro to imatinib mesylate. The effect of the drug already significant at a 1μM concentration with a 50% growth inhibition on MEC04 cells, was most prominent (90% inhibition of growth) at 6μM. Conversely, there was no substantial cytotoxic effect on the SNK6 cell line, which might be related to a lower expression of PDGFRA, as suggested at the RNA level (Figure 5). Although our results do not preclude the precise mechanism of action of imatinib and suggest heterogeneity in the sensitivity to the drug, the dramatic effect on MEC04 cells shed light on the possible use of tyrosine kinase inhibitors as a novel therapeutic option in some patients with NKTCL refractory to conventional therapies.
The cause of PDGFRA deregulation in NKTCL remains to be determined. We did not evidence either genomic imbalances or gene mutations. Furthermore, mutations in the promoter region were absent in NKTCL primary tumors and cell lines, and we did not find overrepresentation of H2α haplotype, known to result in up-regulation of PDGFRA in glioblastoma.29
In conclusion, this integrative genomic and transcriptomic study characterizes the molecular signature of NKTCL, highlights emerging oncogenic pathways in this disease entity, and offers a rationale for exploring new therapeutic options such as tyrosine kinase inhibitors in patients with this aggressive malignancy.
The online version of this article contains a data supplement.
Presented in part at the XIVth Meeting of the European Association for Hematopathology/Society for Hematopathology, Bordeaux, France, September 22, 2008.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are extremely thankful for the contributions made by the Carte d'Identité des Tumeurs (CIT) platforms (Affymetrix, IGBMC: Christelle Thibault, Philippe Kastner; BAC arrays, Institut Curie: Gaëlle Pierron, Olivier Delattre; RNA analysis, Saint-Louis Hospital: Daniela Geromin). We also thank Norio Shimizu for providing cell lines; Joe A. Trapani and Odile Devergne for providing antibodies; François Radvanyi for critical opinion in data interpretation; Marie-Laure Prunet from GELAP; and Virginie Fataccioli, Tony Noel, Yolaine Pothin, and Maryse Baia for technical assistance.
This work is part of the CIT program (http://cit.ligue-cancer.net/index.php/en) from the Ligue Nationale Contre le Cancer. This work was supported by Inserm, the Institut National du Cancer (INCa), the Belgian National Fund for Scientific Research (FNRS), and the Association pour la Recherche Thérapeutique, Génétique et Immunologique dans les Lymphomes (ARTGIL). Y.H. was supported by the Ligue Nationale Contre le Cancer.
Authorship
Contribution: L.d.L. and P.G. designed research; Y.H., A.d.R., B.G., N.M.-G., P.C., M.-H.D.-L., J.-F.E., M.T., and C.S. performed research; L.d.L., J. Bosq, J. Brière, B.P., and P.G. collected data; Y.H., A.d.R., and P.G. analyzed and interpreted the data; A.d.R., E.T., and T.M. contributed vital analytical tools; and Y.H., A.d.R., L.d.L., P.C., C.S., and P.G. drafted the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Philippe Gaulard, MD, Département de Pathologie and Inserm U955, Hôpital Henri Mondor, 94010 Créteil, France; e-mail: philippe.gaulard@hmn.aphp.fr.
References
Author notes
*Y.H. and A.d.R. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal