Abstract
PRKAR1A (R1A)–retinoic acid receptor-α (R1A-RARα) is the sixth RARα–containing fusion protein in acute promyelocytic leukemia (APL). Using the murine bone-marrow retroviral transduction/transformation assay, we showed that R1A-RARα fusion protein could transform bone-marrow progenitor/stem cells. In gel-shift assays, R1A-RARα was able to bind to a panel of retinoic acid response elements both as a homodimer and as a heterodimer with RXRα, and demonstrated distinct DNA-binding characteristics compared with wild-type RARα/RXRα or other X-RARα chimeric proteins. The ratio of R1A-RARα to RXRα proteins affected the retinoic acid response element interaction pattern of R1A-RARα/RXRα complexes. Studies comparing R1A-RARα with R1A-RARα(ΔRIIa) demonstrated that the RIIa protein interaction domain located within R1A was responsible for R1A-RARα homodimeric DNA binding and interaction with wild-type R1A protein. However, the RIIa domain was not required for R1A-RARα–mediated transformation because its deletion in R1A-RARα(ΔRIIa) did not compromise its transformation capability. In contrast, introduction of point mutations within the RARα portion of either R1A-RARα or R1A-RARα(ΔRIIa), previously demonstrated to eliminate RXRα interaction or treatment of transduced cells with RXRα shRNA or a RXRα agonist, reduced transformation capability. Thus, leukemic transformation by APL fusion protein PRKAR1A-RARα is critically dependent on RXRα, which suggests RXRα is a promising target for APL.
Introduction
Acute promyelocytic leukemia (APL) accounts for 10% of cases of acute myeloid leukemia (AML) and is characterized by blast cells whose morphology reveals a block in myeloid cell differentiation at the promyelocyte stage.1 With the successful introduction of all-trans retinoic acid (ATRA) differentiation therapy, APL is the most curable AML subtype. The t(15;17)(q22;q21) chromosomal translocation, which generates PML-RARα fusion protein, is detected in the majority of APL patients.1,2 In addition to PML-RARα, 4 other APL-associated RARα–containing fusion proteins, PLZF-RARα, NPM-RARα, NuMA-RARα, and STAT5b-RARα, have been identified and characterized at the molecular level.3,4 Recently, a sixth APL-fusion protein, PRKAR1A-RARα (R1A-RARα), was identified in a 66-year-old man with APL who became disease-free after ATRA and arsenic treatment.5
PRKAR1A-RARα (R1A-RARα) resulted from a cytogenetically cryptic recombination event between the RARα and PRKAR1A (R1A) genes on chromosome 17.5 R1A encodes the regulatory subunit of protein kinase A (PKA RIα) and was initially described as the tissue-specific extinguisher 1. The cyclic adenosine monophosphate/protein kinase A (cAMP/PKA) signaling pathway plays a central role in the regulation of metabolism, cell proliferation, differentiation, and apoptosis.6 The tetrameric PKA holoenzyme, a serine/threonine kinase and the major mediator of cAMP signaling, is composed of a homodimer of regulatory (R) subunits that bind 2 catalytic (C) subunits that are inactive in the absence of cAMP ligand. The best documented function of the R subunits is inhibition of C subunit kinase activity. In the presence of cAMP, the C subunits are released from the inactive tetrameric complex and, in turn, are free to phosphorylate a wide variety of substrate proteins on serine or threonine residues.6 Inactivating mutations of the R1A gene have been found in sporadic tumors and in the germline of the majority of patients with the Carney complex syndrome, a form of multiple endocrine neoplasia.7-9
The R1A-RARα chimeric fusion protein consists of a putative dimerization domain (RIIa domain) within R1A at its N-terminus along with the DNA-binding and ligand-binding domains contributed by RARα, which are identical to portions of RARα found in all other APL-fusion proteins.1,3 It has recently been demonstrated that homo-oligomerization domains within the RARα fusion partners, including PML,10 PLZF,11,12 and Stat5,13 are essential for the oncogenic activity of their respective APL fusion proteins. The molecular basis for R1A-RARα–mediated bone marrow transformation is not known. In particular, the contribution of the dimerization domain of R1A-RARα to its oncogenic functions has not been evaluated.
In this study, we showed that both R1A-RARα and R1A-RARα(ΔRIIa), in which the RIIa dimerization domain was deleted, transformed murine bone marrow progenitors. We also demonstrated that R1A-RARα bound to retinoic acid response elements (RAREs) either as a homodimer or as a heterodimer with RXRα, formed a complex with wild-type R1A and displayed a nuclear localization pattern distinct from that of R1A but similar to the one of RARα. Finally, we demonstrated that RXRα and its interaction with R1A-RARα, but not R1A-RARα homodimeric DNA binding, are essential for transformation by this novel APL fusion protein.
Methods
Cell lines and reagents
The 293T and GP2-293T cells were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum (FBS; Invitrogen). HeLa cells were maintained in Opti-MEM I media with 4% FBS.13 The A03_1 cell line, which contains an integrated lac operator heterochromatic array arranged in a globular structure, was cultured in F-12 Ham medium (without hypoxanthine and thymidine; Invitrogen) with 0.3μM methotrexate and 10% dialyzed FBS (HyClone) as described.13 ATRA was purchased from Sigma-Aldrich, and SR11237, a pan-RXR agonist, has been reported in our former study.14 G418/neomycin and puromycin were purchased from Invitrogen and Sigma-Aldrich, respectively.
Constructs
Wild-type RARα, RXRα, PML-RARα, NPM-RARα, YFP-RARα, and YFP-RXRα expression vectors as well as murine RXRα shRNA sh563 (Open Biosystems) and sh131 (in pSUPER.retro.puro vector; OligoEngine) have been described previously.13-16 R1A-RARα was constructed in the pSG5 expression vector (Stratagene) by fusing human R1A (from GFP-R1A, provided by Dr M. Mavrakis, National Institutes of Health) and RARα using a polymerase chain reaction (PCR) approach based on the published sequence.5,8,15,17 A similar PCR approach was also applied to construct pSG5-R1A-RARα(ΔRIIa) in which the dimerization domain (RIIa) of R1A is deleted (Figure 1A). R1A (in pSG5) was cloned from the GFP-R1A vector.8 M2-R1A-RARα and M2-R1A in pSG5 were PCR-generated from their pSG5-version counterpart by inserting the M2 FLAG epitope sequence into the N-terminus of the R1A-RARα (in pSG5) or R1A (in pSG5) sequence, respectively. The Myc and HA epitope sequences were also cloned into a series of pSG5-based expression vectors to produce Myc-R1A-RARα, Myc-R1A-RARα(ΔRIIa), Myc-R1A, Myc-PLZF-RARα, Myc-PAX5, HA-R1A-RARα, HA-R1A-RARα(ΔRIIa), and HA-R1A vectors. The R1A-RARα, R1A-RARα(ΔRIIa), or R1A open-reading sequence was subcloned into pBD-S5 vector, a modified pSG5 vector containing Gal4-DNA binding sequence, or pAD-S5 vector, a modified pSG5 vector with addition of the AD (activation-domain) sequence (from pCMV-AD vector; Stratagene), to make BD-R1A-RARα, BD-R1A-RARα(ΔRIIa), BD-R1A, AD-R1A-RARα, AD-R1A-RARα(ΔRIIa), and AD-R1A vector constructs for the mammalian 2-hybrid assay. The cDNA sequences for R1A-RARα, R1A-RARα(ΔRIIa), or R1A from its pSG5 expression vectors were also cloned into pEYFP-S5 vector (a modified pSG5 vector containing EYFP sequence) or pECFPL-S5 vector (a modified pSG5 vector containing ECFP plus lac repressor sequence13 ) to make YFP-R1A-RARα, YFP-R1A-RARα(ΔRIIa), YFP-R1A, CFP-LacR-R1A-RARα, and CFP-LacR-R1A-RARα(ΔRIIa) expression vectors by standard methods,17 respectively. R1A-RARα (MSCV-neo) and R1A-RARα(ΔRIIa) (MSCV-neo) were made by transferring the inserts from the aforementioned expression plasmids into MSCV-neo vector (Clontech). PML-RARα (MSCV-neo), PLZF-RARα (MSCV-neo), MLL-ENL (MSCV-neo), and MLL-GAS7 (MSCV-neo) have been described previously.12,14,18 Vector constructs containing mutations within the RXRα-binding domain of RARα, R1A-RARα-M412R/T415R (MSCV-neo), R1A-RARα(ΔRIIa)-M374R/T377R (MSCV-neo), PML-RARα-M872R/T875R (MSCV-neo), PLZF-RARα-M775R/T778R (MSCV-neo), as well as PML-RARα(ΔCC) (MSCV-neo),19 were made from their parent form using a PCR-based mutagenesis approach described previously.15,20 All constructs were confirmed by DNA sequencing and immunoblotting.
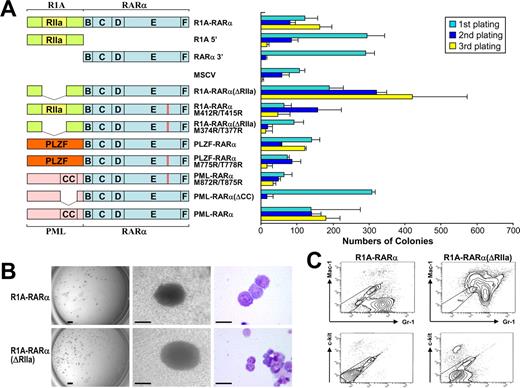
Persistent self-renewal of primary hematopoietic progenitor/stem cells by R1A-RARα. (A) Schematic diagram of R1A-RARα, PLZF-RARα, PML-RARα, and their mutant forms used in RTTA (left). The RIIa homodimerization domain of R1A and the B, C, D, E, and F domains of RARα are indicated. The bar chart (right) represents the corresponding numbers of colonies after each round of plating in methylcellulose culture medium. Data are mean ± SD of 4 independent experiments. (B) Typical third-round colony (left and middle panels) and cell morphology after May-Grunwald-Giemsa staining (right panel) of primary bone marrow cells transduced with the indicated constructs. Scale bars from left to right represent 1 mm, 100 μm, and 10 μm, respectively. (C) Flow cytogram of bone marrow cells transduced by R1A-RARα or R1A-RARα(ΔRIIa) and stained for Mac-1 or Gr-1 as indicated. The ellipse frames unstained negative control cells.
Persistent self-renewal of primary hematopoietic progenitor/stem cells by R1A-RARα. (A) Schematic diagram of R1A-RARα, PLZF-RARα, PML-RARα, and their mutant forms used in RTTA (left). The RIIa homodimerization domain of R1A and the B, C, D, E, and F domains of RARα are indicated. The bar chart (right) represents the corresponding numbers of colonies after each round of plating in methylcellulose culture medium. Data are mean ± SD of 4 independent experiments. (B) Typical third-round colony (left and middle panels) and cell morphology after May-Grunwald-Giemsa staining (right panel) of primary bone marrow cells transduced with the indicated constructs. Scale bars from left to right represent 1 mm, 100 μm, and 10 μm, respectively. (C) Flow cytogram of bone marrow cells transduced by R1A-RARα or R1A-RARα(ΔRIIa) and stained for Mac-1 or Gr-1 as indicated. The ellipse frames unstained negative control cells.
RTTA
Retrovirus transduction/transformation assay (RTTA) was performed as previously described.12,14,21 Briefly, viral supernatants were collected 48 hours after transfection of GP2-293 cells (Clontech) and used to infect the purified murine hematopoietic progenitor/stems cells that were positively selected for c-Kit expression by magnetic activated cell sorting (MACS; Miltenyi Biotec).14 Donor cells were harvested from the bone marrows of 4- to 10-week-old wild-type C57BL/6 mice. All animal studies were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. After spinoculation by centrifugation at 800g for 2 hours at 32°C, transduced bone marrow cells were plated in 1% myeloid-conditioned methylcellulose that contained an Iscove modified Dulbecco medium–based Methocult (Methocult M3231; StemCell Technologies) supplemented with 20 ng/mL stem cell factor and 10 ng/mL each of interleukin-3, interleukin-6, and granulocyte-macrophage colony-stimulating factor (PeproTech) in the presence or absence of appropriate drug selection (1 mg/mL G418/neomycin, Invitrogen; 1 μg/mL puromycin, Sigma-Aldrich). Replating was repeated every 7 days. For knockdown experiments, spinoculation was repeated 3 times before the plating. For drug studies, SR11237 (1μM) and ATRA (1μM) were added to the transduced cells after the second plating. Transformation results were determined from at least 3 independent experiments.
Phenotype analysis
Immunophenotypic analysis was performed by fluorescence-activated cell sorter (FACS) using fluorophore-conjugated monoclonal antibodies to c-Kit (2B8 clone), Mac-1 (M1/70 clone), and Gr-1 (RB6-8C5 clone; BD Biosciences PharMingen). After staining, which was performed in the dark on ice for 30 minutes, the cells were washed twice in staining medium and resuspended in 1 μg/mL propidium iodine before analysis using a BD LSR II flow cytometry system (BD Biosciences). Dead cells were gated out by high propidium iodine staining and forward light scatter.
Electrophoretic mobility shift assay
Whole-cell extracts were prepared from 293T cells after transient expression of R1A-RARα, R1A-RARα(ΔRIIa), PML-RARα, NPM-RARα, RARα, or RXRα using lysis buffer.19,22 Gel shift assay using in vivo–expressed whole-cell extract proteins or in vitro–translated proteins (TNT Quick Coupled Transcription/Translation Systems; Promega) has been described elsewhere.11,13,15,22 For supershift experiment, 1 μg of antibody was included in the reaction system when needed. Protein-DNA complexes were run on 5% polyacrylamide gels equilibrated in 0.25× Tris borate ethylenediaminetetraacetic acid and analyzed by PhosphorImager using ImageQuant software (GE Healthcare).
Immunoblotting, coimmunoprecipitation assay, and mammalian 2-hybrid assay
Immunoblotting and coimmunoprecipitation assay were essentially performed as previously described.14,15,22,23 The antibodies used in the immunoprecipitation assay were mouse anti-FLAG M2-agarose and Myc-Tag rabbit polyclonal antibody (Sigma-Aldrich) or HA-Tag mouse monoclonal antibody (6E2; Cell Signaling Technology).
For mammalian 2-hybrid assay, the GeneJuice transfection reagent was used for transfection of HeLa cells in 24-well tissue-culture plates according to the manufacturer's instructions.13,15 The amounts of plasmid DNA used per well were 250 ng of reporter vector, 0.5 to 1.0 μg of expression vector, and 200 ng of β-galactosidase vector, which was used as transfection control. Luciferase activity was measured in a luminometer (Thermo Fisher Scientific), expressed in arbitrary units and divided by the activities of β-galactosidase.13,15,23 Each point is the mean of at least 3 independent experiments.
Fluorescence and deconvolution microscopy
HeLa cells and A03_1 cells were plated onto poly-D-lysine–coated coverslips in 24-well plate at a concentration of 104 cells/well at 24 hours before transfection. Cells with transient expression of YFP-R1A-RARα, YFP-R1A-RARα(RIIa), YFP-R1A, YFP-RARα (in HeLa cell), and CFP-LacR–tagged plus YFP-tagged constructs (in A03_1 cell) were fixed, and deconvolution microscopy was performed with a DeltaVision Restoration Microscopy System (Applied Precision) as described previously.13,22
Results
R1A-RARα–mediated transformation of primary hematopoietic cells is not dependent on its dimerization domain
R1A-RARα, which was initially identified in an APL patient,5 is a newly described member of the X-RARα fusion gene family. To assess the oncogenic ability of this sixth APL fusion gene product, we preformed RTTAs in which the R1A-RARα chimeric protein was expressed under the control of long terminal repeat of the murine stem cell virus (MSCV) after transduction into c-Kit+ murine hematopoietic progenitor/stem cells. RTTA has been used successfully to examine the activity of several oncogenic transcription factors reported in human leukemia.12,14,21,24 Whereas hematopoietic progenitor/stem cells transduced with empty vector (MSCV) exhausted their colony-forming ability after third-round plating, hematopoietic progenitor/stem cells expressing R1A-RARα continued to form compact GM-CFU–like colonies after third-round plating (Figure 1A-B) similar to cells transduced with PML-RARα.12 On their own, neither the truncated form of R1A nor RARα exhibited any detectable transformation ability (Figure 1A)12,14 ; cells transduced with either vector construct exhausted their self-renewal potential after the third round of plating. Therefore, like PLZF-RARα12 and STAT5b-RARα,14 R1A-RARα–mediated transformation of primary hematopoietic cells is dependent on the contribution of both the R1A and the RARα portions of the chimeric protein. Interestingly, R1A-RARα–transduced primary hematopoietic progenitor/stem cells demonstrated many red granules in the cytoplasm after third-round plating (Figure 1B), which has been reported in APL patient cells.25 Immunophenotypic analyses of R1A-RARα-mediated transformed cells revealed the phenotype of myeloid precursors (c-kitlo, Mac-1lo, Gr-1+; Figure 1C).
It is well established that dimerization or oligomerization is required for the functional activities of RARα fusion proteins, including cellular transformation.1,12,14,15,26 We performed a domain structure analysis using the SMART program (http://smart.embl-heidelberg.de), which has been widely applied for the protein functional-domain structure analysis,27,28 and revealed the presence of a RIIa domain within the R1A moiety of R1A-RARα protein. This domain also is called the dimerization/docking or D/D domain because it contains the putative dimerization interface and docking site for A-kinase-anchoring proteins.6 To evaluate whether dimerization of R1A-RARα is required for increasing self-renewal of transduced cells, we generated a mutant R1A-RARα construct, R1A-RARα(ΔRIIa), in which the RIIa domain was deleted (Figure 1A), and performed RTTA. Unexpectedly, R1A-RARα(ΔRIIa) transduction yielded transformed cells that made compact colonies after third-round plating (Figure 1A) with colony morphology and cytoplasmic granularity very similar to cells transduced with intact R1A-RARα (Figure 1B). FACS analysis of R1A-RARα(ΔRIIa)–transformed cells revealed an immunophenotype of myeloid precursors (c-kitlo, Mac-1+, Gr-1+; Figure 1C). Moreover, unlike PML-RARα(ΔCC), which exhausted its colony formation ability after third-round plating, R1A-RARα(ΔRIIa) transduced cells consistently yielded an even increased number of colonies compared with transduction with R1A-RARα (Figure 1A), suggesting that deletion of the RIIa dimerization domain did not compromise its transformation ability.
R1A-RARα forms a homodimer that binds RAREs
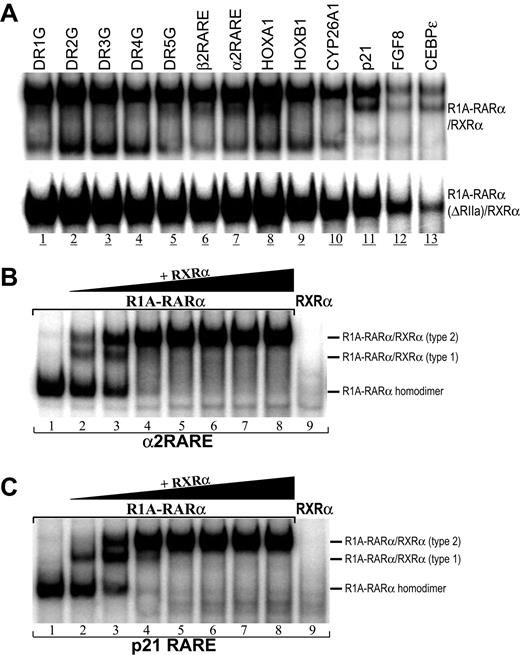
Aberrant transcriptional regulation of retinoic acid (RA)–responsive genes is thought to be a major underlying cellular transformation mechanism of X-RARα chimeric proteins in APL.1-3 To determine whether the transformation activity by R1A-RARα is the result of its ability to directly bind RAREs, in vivo–expressed R1A-RARα proteins were examined in gel-shift assays using a series of RARE duplex oligonucleotides.15 As expected, R1A-RARα chimeric protein could form homodimers capable of binding all RAREs tested (Figure 2A top panel). This binding of the R1A-RARα homodimer could be competitively inhibited by a 100-fold excess of unlabeled RARE, indicating that binding is specific (Figure 2B). The shifted band corresponded to a homodimer of R1A-RARα (Figure 2C lane 2), judging from its migration characteristics compared with the heterodimers R1A-RARα/RXRα and R1A-RARα(ΔRIIa)/RXRα or homodimers of PML-RARα (Figure 2C). These putative R1A-RARα homodimer or heterodimer bands could be supershifted by the addition of the antibody against RARα or RXRα (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Like other X-RARα,14,20 the formation of R1A-RARα/RXRα complex suggested that RXRα may also play an important role in the transformation mediated by R1A-RARα.29 However, as shown in Figure 2A, whereas PML-RARα and NPM-RARα proteins bound to RAREs, DR2G to DR5G and HOXA1, more efficiently than to other RAREs tested, R1A-RARα fusion protein bound efficiently to all RAREs tested except FGF8 and CEBPϵ and, in this regard, resembled the pattern of RARE binding of wild-type RARα/RXRα (Figure 2A). Moreover, R1A-RARα and NPM-RARα proteins bound to α2RARE and p21 RARE more efficiently than PML-RARα protein. We also observed strong CEBPϵ RARE binding by NPM-RARα protein but not by PML-RARα and R1A-RARα proteins. These results indicated that R1A-RARα protein bound to RARE as a homodimer and demonstrated a binding pattern distinct from wild-type RARα/RXRα as well as other X-RARα proteins.
Binding of RAREs by R1A-RARα fusion protein homodimer. (A) Binding of R1A-RARα homodimer to a series of RAREs assayed by gel-shift assays. Equivalent amounts of in vivo–expressed R1A-RARα, PML-RARα, NPM-RARα, or RARα/RXRα proteins were incubated with the following radiolabeled RARE probes: the synthetic RARE sequences, DR1G to DR5G, and the RAREs from the natural enhancer/promoter regions of the human RARβ2 (β2RARE), RARα2 (α2RARE), HOXA1, HOXB1, CYP26A1, p21-WAF1, FGF8, and CEBPϵ gene. (B) Gel-shift assays were performed using in vivo–expressed R1A-RARα protein incubated with radiolabeled DR5G (hot probe) without or with unlabeled DR5G (cold probe) in the amounts indicated. (C) Gel-shift assays were performed with α2RARE radiolabeled probe incubated with in vivo–expressed proteins R1A-RARα, R1A-RARα(ΔRIIa), and PML-RARα without or with RXRα as indicated. (D) Gel-shift assay analysis of the equivalent in vitro translation proteins R1A-RARα, R1A-RARα(ΔRIIa), R1A-RARα-M412R/T415R, and R1A-RARα(ΔRIIa)-M374R/T377R alone or plus RXRα, as indicated.
Binding of RAREs by R1A-RARα fusion protein homodimer. (A) Binding of R1A-RARα homodimer to a series of RAREs assayed by gel-shift assays. Equivalent amounts of in vivo–expressed R1A-RARα, PML-RARα, NPM-RARα, or RARα/RXRα proteins were incubated with the following radiolabeled RARE probes: the synthetic RARE sequences, DR1G to DR5G, and the RAREs from the natural enhancer/promoter regions of the human RARβ2 (β2RARE), RARα2 (α2RARE), HOXA1, HOXB1, CYP26A1, p21-WAF1, FGF8, and CEBPϵ gene. (B) Gel-shift assays were performed using in vivo–expressed R1A-RARα protein incubated with radiolabeled DR5G (hot probe) without or with unlabeled DR5G (cold probe) in the amounts indicated. (C) Gel-shift assays were performed with α2RARE radiolabeled probe incubated with in vivo–expressed proteins R1A-RARα, R1A-RARα(ΔRIIa), and PML-RARα without or with RXRα as indicated. (D) Gel-shift assay analysis of the equivalent in vitro translation proteins R1A-RARα, R1A-RARα(ΔRIIa), R1A-RARα-M412R/T415R, and R1A-RARα(ΔRIIa)-M374R/T377R alone or plus RXRα, as indicated.
The RIIa domain of R1A-RARα is responsible for its homodimerization
To confirm that the RIIa domain really is responsible for R1A-RARα homodimer formation, we performed gel-shift assays using R1A-RARα(ΔRIIa) (Figure 2C). As expected, deletion of this protein interaction domain from R1A-RARα prevented homodimeric/RARE complex formation but did not affect its ability to form heterodimers with RXRα to bind DNA (Figure 2C lanes 4-5), which was also confirmed by gel-shift assay using the in vitro–translated proteins (Figure 2D). This is reminiscent of our previous finding on STAT5b-RARα where deletion of the coiled-coil dimerization domain compromised its homodimeric DNA-binding property but not its transformation ability.14,15
The RIIa domain of R1A-RARα is also responsible for dimerization with wild-type R1A
To further determine and confirm that the R1A-RARα protein could form a complex with itself as well as with wild-type R1A, we performed coimmunoprecipitation assays using M2-tagged R1A-RARα. As shown in Figure 3A, M2-R1A-RARα was able to pull down Myc-tagged R1A-RARα itself and wild-type Myc-R1A but not Myc-R1A-RARα(ΔRIIa), Myc-PLZF-RARα, or Myc-PAX5, confirming our gel-shift assay results. Similar results were obtained using M2-R1A-RARα plus HA-tagged R1A-RARα, R1A or R1A-RARα(ΔRIIa) expression vectors (Figure 3B). Furthermore, when we coexpressed M2-tagged R1A plus Myc-R1A-RARα, Myc-R1A, or Myc-R1A-RARα(ΔRIIa) in 293T cells (Figure 3C), we found that M2-R1A was able to pull down Myc-R1A-RARα and Myc-R1A, but not Myc-R1A-RARα(ΔRIIa).
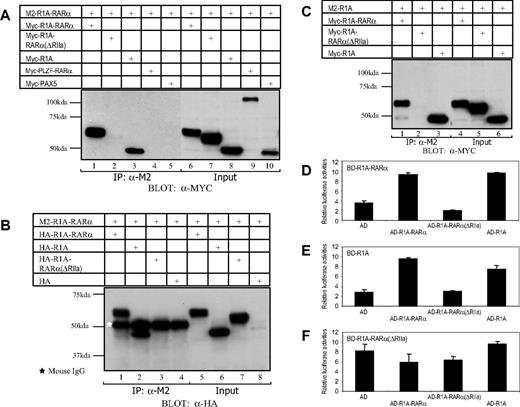
The RIIa domain-mediated protein interaction of R1A-RARα/R1A-RARα and R1A-RARα/R1A. The 293T cells were transiently transfected with the following combined expression vectors: (A) M2-R1A-RARα + Myc-R1A-RARα, M2-R1A-RARα + Myc-R1A-RARα(ΔRIIa), M2-R1A-RARα + Myc-R1A, M2-R1A-RARα + Myc-PLZF-RARα, or M2-R1A-RARα + Myc-PAX5; (B) M2-R1A-RARα + HA-R1A-RARα, M2-R1A-RARα + HA-R1A, M2-R1A-RARα + HA-R1A-RARα(ΔRIIa), or M2-R1A-RARα + HA-containing empty vector; (C) M2-R1A + Myc-R1A-RARα, M2-R1A + Myc-R1A-RARα(ΔRIIa), or M2-R1A + Myc-R1A, as indicated. The white star represents the position of mouse IgG. Cell lysates were separated by SDS-PAGE directly (Input) or after immunoprecipitation using antibody against the M2-FLAG epitope (immunoprecipitation, IP) and subsequently immunoblotted using antibody against Myc or HA epitope. The protein-protein interactions between R1A-RARα fusion protein and wild-type R1A were analyzed using mammalian 2-hybrid assay (D-F). The AD (activation domain) alone or its fusion with R1A-RARα, R1A-RARα(ΔRIIa), or R1A was expressed in HeLa cells together with a luciferase reporter containing 4 copies of Gal4-binding sites (4xUAS-TK luc) as well as Gal4 DBD-fused R1A-RARα (BD-R1A-RARα) (D), Gal4 DBD-fused R1A (BD-R1A) (E), or Gal4 DBD-fused R1A-RARα(ΔRIIa) (BD-R1A-RARα(ΔRIIa)) (F). The relative luciferase activities are averages of 3 independent transfection experiments normalized to β-galactosidase activity.
The RIIa domain-mediated protein interaction of R1A-RARα/R1A-RARα and R1A-RARα/R1A. The 293T cells were transiently transfected with the following combined expression vectors: (A) M2-R1A-RARα + Myc-R1A-RARα, M2-R1A-RARα + Myc-R1A-RARα(ΔRIIa), M2-R1A-RARα + Myc-R1A, M2-R1A-RARα + Myc-PLZF-RARα, or M2-R1A-RARα + Myc-PAX5; (B) M2-R1A-RARα + HA-R1A-RARα, M2-R1A-RARα + HA-R1A, M2-R1A-RARα + HA-R1A-RARα(ΔRIIa), or M2-R1A-RARα + HA-containing empty vector; (C) M2-R1A + Myc-R1A-RARα, M2-R1A + Myc-R1A-RARα(ΔRIIa), or M2-R1A + Myc-R1A, as indicated. The white star represents the position of mouse IgG. Cell lysates were separated by SDS-PAGE directly (Input) or after immunoprecipitation using antibody against the M2-FLAG epitope (immunoprecipitation, IP) and subsequently immunoblotted using antibody against Myc or HA epitope. The protein-protein interactions between R1A-RARα fusion protein and wild-type R1A were analyzed using mammalian 2-hybrid assay (D-F). The AD (activation domain) alone or its fusion with R1A-RARα, R1A-RARα(ΔRIIa), or R1A was expressed in HeLa cells together with a luciferase reporter containing 4 copies of Gal4-binding sites (4xUAS-TK luc) as well as Gal4 DBD-fused R1A-RARα (BD-R1A-RARα) (D), Gal4 DBD-fused R1A (BD-R1A) (E), or Gal4 DBD-fused R1A-RARα(ΔRIIa) (BD-R1A-RARα(ΔRIIa)) (F). The relative luciferase activities are averages of 3 independent transfection experiments normalized to β-galactosidase activity.
Next, we examined whether these proteins interact within the cell. To this end, a mammalian 2-hybrid assay using transfected HeLa cells was used. Transient transfection of a Gal4 DBD-R1A-RARα fusion (BD-R1A-RARα) plus AD-fused R1A (AD-R1A) (Figure 3D lane 4) or AD-R1A-RARα (Figure 3D lane 2) into HeLa cells resulted in strong stimulation of gene expression from a luciferase reporter containing 4 copies of Gal4-binding sites compared with the combination of one plus AD empty vector (Figure 3D lane 1). However, this gene activation effect was not observed with the combination of BD-R1A-RARα plus AD-R1A-RARα(ΔRIIa) in the same experimental setting (Figure 3D lane 3). Similar results were obtained when we coexpressed Gal4 DBD-R1A (BD-R1A) fusion plus AD-R1A, AD-R1A-RARα, or AD-R1A-RARα(ΔRIIa) (Figure 3E) in HeLa cells. In contrast, when Gal4 DBD-R1A-RARα(ΔRIIa) was used, no augmentation of luciferase activity was observed (Figure 3F). The results of these 2-hybrid studies together with the results of coimmunoprecipitation experiments provide strong evidence that the R1A-RARα fusion protein can form a complex with itself as well as wild-type R1A and that the RIIa domain of R1A-RARα is required to mediate these complex formations.
The next question we wanted to address is whether wild-type R1A, like RXRα, can be recruited to a chromatin array occupied by the R1A-RARα fusion protein. To address this question, we used the lac operator/lac repressor system used in our previous studies.13 Coexpression of CFP-LacR-R1A-RARα with YFP-R1A in A03_1 cells showed that, whereas a portion of the YFP-R1A remained within the cytoplasm (supplemental Figure 2 first row), a substantial fraction of YFP-R1A colocalized with CFP-LacR-R1A-RARα on the array, indicating that wild-type R1A can form a complex with R1A-RARα complex in the context of chromatin. In addition, coexpression of CFP-LacR-R1A-RARα with YFP-R1A-RARα in A03_1 cells revealed that most of the YFP-R1A-RARα colocalized with CFP-LacR-R1A-RARα on the array (supplemental Figure 2 second row), whereas coexpression of either CFP-LacR-R1A-RARα plus YFP-R1A-RARα(ΔRIIa) or CFP-LacR-R1A-RARα(ΔRIIa) plus YFP-R1A-RARα in A03_1 cells did not reveal recruitment of YFP-R1A-RARα(ΔRIIa) or YFP-R1A-RARα onto the lac operator array (supplemental Figure 2 third and fourth rows). These results confirmed, at the single-cell level and in the context of chromatin, the results obtained from gel-shift, coimmunoprecipitation, and mammalian 2-hybrid assays.
R1A-RARα has a similar nuclear localization as the wild-type RARα
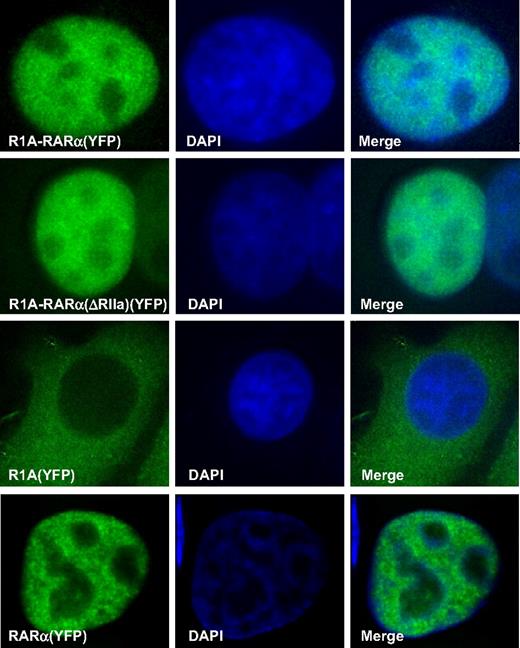
We have reported that X-RARα fusion proteins have a distinct cellular localization compared with wild-type RARα, which may be linked to its functional activities.16,19 To determine the distribution of R1A-RARα within cells, the contribution of the R1A domain to its distribution and to compare its distribution with that of each component of the fusion gene product, namely, wild-type RARα and wild-type R1A, we subcloned the coding sequences of R1A-RARα, R1A-RARα(ΔRIIa), and R1A into pEYFP-S5 to generate a YFP-tagged version of each. Fluorescent microscopic examination of HeLa cells was performed after transient expression of these YFP-tagged proteins. As shown in Figure 4, YFP-R1A-RARα and YFP-R1A-RARα(ΔRIIa) were localized predominantly in the nucleus and exhibited a diffuse granular pattern similar to wild-type YFP-RARα.16 However, YFP-R1A was located predominantly in the cytoplasm (Figure 4), confirming the data published by other investigators.8
Cellular localization of RARα-containing proteins. Deconvolution fluorescence microscopy was used to analyze HeLa cells transiently transfected with the indicated YFP-tagged proteins, R1A-RARα, R1A-RARα(ΔRIIa), wild-type R1A, or wild-type RARα. Nuclei were visualized by counterstaining with DAPI (blue), and representative cells are shown.
Cellular localization of RARα-containing proteins. Deconvolution fluorescence microscopy was used to analyze HeLa cells transiently transfected with the indicated YFP-tagged proteins, R1A-RARα, R1A-RARα(ΔRIIa), wild-type R1A, or wild-type RARα. Nuclei were visualized by counterstaining with DAPI (blue), and representative cells are shown.
R1A-RARα forms hetero-oligomeric complexes with RXRα to bind DNA
As shown in Figure 2C, several retarded bands were observed with α2RARE probe after the addition of RXRα to R1A-RARα in DNA-binding assays (Figure 2C lanes 2-3). The slowest migrating band most probably represents R1A-RARα homodimer plus 2 molecules of RXRα, whereas the faster migrating band represents the R1A-RARα homodimer itself. To determine whether R1A-RARα/RXRα heterodimer can bind to other RAREs, a series of RARE probes also were examined in gel-shift assays (Figure 5A). We demonstrated that in vivo–expressed R1A-RARα/RXRα (Figure 5A top panel) and R1A-RARα(ΔRIIa)/RXRα proteins (Figure 5A bottom panel) bound to all of the RAREs tested. However, different R1A-RARα/RXRα protein/DNA-binding complex species were observed within these RAREs. Using the same amounts of in vivo–expressed R1A-RARα and RXRα proteins, we found that R1A-RARα and RXRα preferred to form R1A-RARα homodimers plus a single complex with RXRα (type 2 R1A-RARα/RXRα heterodimer, Figure 5) when incubated with DR2G, DR3G, DR4G, HOXB1, and CYP26A1. In contrast, when incubated with DR1G, DR5G, β2RARE, α2RARE, HOXA1, p21, FGF8, and CEBPϵ, 3 types of DNA-binding complexes were formed: R1A-RARα homodimer, type 2 R1A-RARα/RXRα heterodimer, and a third complex that migrates between these 2 complexes referred to as type 1 R1A-RARα/RXRα heterodimer (Figure 5), in which both type 2 and type 1 R1A-RARα/RXRα could be supershifted by RARα or RXRα antibody (supplemental Figure 1).
Binding of RAREs by R1A-RARα/RXRα heterodimer. (A) Binding of R1A-RARα/RXRα heterodimer to a series of RAREs. The equivalent amounts of in vivo–expressed R1A-RARα/RXRα (top panel) or R1A-RARα(ΔRIIa)/RXRα (bottom panel) proteins were incubated with the radiolabeled RARE probes for DNA-binding assays. Gel-shift assays using α2RARE (B) or p21 RARE (C) hot probes were performed with in vivo–expressed R1A-RARα protein without or with the increasing amount of in vivo–expressed RXRα protein.
Binding of RAREs by R1A-RARα/RXRα heterodimer. (A) Binding of R1A-RARα/RXRα heterodimer to a series of RAREs. The equivalent amounts of in vivo–expressed R1A-RARα/RXRα (top panel) or R1A-RARα(ΔRIIa)/RXRα (bottom panel) proteins were incubated with the radiolabeled RARE probes for DNA-binding assays. Gel-shift assays using α2RARE (B) or p21 RARE (C) hot probes were performed with in vivo–expressed R1A-RARα protein without or with the increasing amount of in vivo–expressed RXRα protein.
To further establish the composition of the type 1 and type 2 R1A-RARα homodimer/RXRα complexes, α2RARE and p21 RARE probes were chosen for further study (Figure 5B-C). When incubated alone, R1A-RARα protein bound to both RAREs, forming a homodimer band (Figure 5B lane 1; Figure 5C lane 1). After addition of increasing amounts of in vivo–expressed RXRα protein to a constant amount of R1A-RARα protein, the R1A-RARα homodimer band became weaker and gradually disappeared. A decrease in the homodimer band was accompanied by the appearance of 2 different R1A-RARα homodimer/RXRα complexes, type 1 and type 2. The type 1 complex appeared with addition of the lowest amount of RXRα. As more RXRα protein is added, type 2 was the only complex detected in both RAREs (Figure 5B lane 4; Figure 5C lane 4). The type 2 complex appeared after addition of a lower amount of RXRα for α2RARE than for p21 RARE. These results suggested that different ratios of R1A-RARα to RXRα could give distinct R1A-RARα/RXRα/RARE complexes depending on the availability of R1A-RARα and RXRα and the RARE. Consistent with our previous report on STAT5-RARα,13 R1A-RARα preferably forms hetero-oligmeric complexes with RXRα, which also resurrects DNA-binding property to transformation-competent R1A-RARα(ΔRIIa) mutant.
Next, we applied the lac operator/lac repressor system to examine the interaction between R1A-RARα and RXRα in vivo at the single-cell level and in the context of chromatin.13 Coexpression of CFP-LacR-R1A-RARα or CFP-LacR-R1A-RARα(ΔRIIa) plus YFP-RXRα in A03_1 cells revealed that virtually all the YFP-RXRα colocalized onto the chromatin array bound by CFP-LacR-R1A-RARα (supplemental Figure 3 top panel) or CFP-LacR-R1A-RARα(ΔRIIa) (supplemental Figure 3 middle panel). In contrast, coexpression of CFP-LacR plus YFP-RXRα (data not shown) or CFP-LacR-R1A-RARα plus YFP-RARα (supplemental Figure 3 bottom panel) in cells revealed that YFP-RXRα or YFP-RARα distributed diffusely throughout the nucleus; no colocalization of YFP-RXRα with CFP-LacR or YFP-RARα with CFP-LacR-R1A-RARα on the lac operator array was observed, respectively. These findings are consistent with other data published by us and other investigators revealing that the RARα fusion protein specifically recruits RXRα but not wild-type RARα.13
RXRα is required for R1A-RARα fusion–mediated transformation
Recently, we and others have demonstrated that RXRα is an essential component of oncogenic X-RARα fusion protein-containing complexes.14,20,29 To determine whether RXRα was required for transformation of primary hematopoietic cells by R1A-RARα, we constructed several MSCV-based vectors, R1A-RARα-M412R/T415R, R1A-RARα(ΔRIIa)-M374R/T377R, PLZF-RARα-M775R/T778R, and PML-RARα-M872R/T875R, in which methionine (M) at the position corresponding to residue 379 of wild-type RARα and threonine (T) at the position corresponding to residue 382 of wild-type RARα were mutated to arginine (R) to eliminate the ability of X-RARα, including R1A-RARα to interact with nuclear receptor cofactor RXRα, as was confirmed by gel-shift assay (Figure 2D).20 Hematopoietic progenitor/stem cells transduced with R1A-RARα(ΔRIIa)-M374R/T377R failed to produce colonies, even in the second plating (Figure 1A). There were a few colonies after transduction with R1A-RARα-M412R/T415R after the third plating; however, the numbers of colonies were significantly reduced compared with wild-type R1A-RARα (Figure 1A), and the sizes of the residual colonies were smaller compared with R1A-RARα (data not shown). Similarly, PLZF-RARα-M775R/T778R and PML-RARα-M872R/T875R transduced bone marrow cells also displayed reduced colony formation as expected in the end of third-round plating.
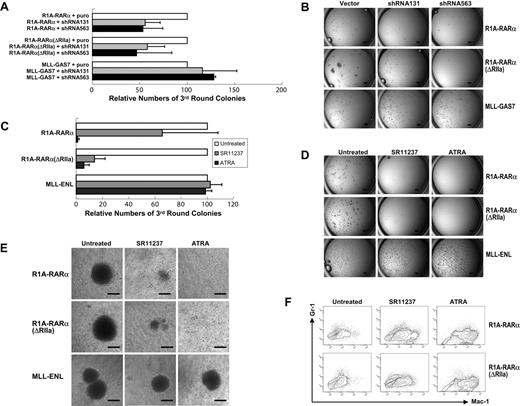
To confirm and extend these results, we used a shRNA approach in which 2 independent shRNAs (sh563 and sh131) targeting RXRα14 were used to knock down the expression of murine RXRα (mRXRα) in bone marrow progenitor/stem cells. Cells cotransduced with R1A-RARα or R1A-RARα(ΔRIIa) and vector control formed compact colonies in the third round of plating (Figure 6A). In contrast, third-round colony formation by cells cotransduced with R1A-RARα and either shRNA131 or shRNA563 were reduced by 55% and 53%, respectively. Similarly, third-round colony formation by cells cotransduced with R1A-RARα(ΔRIIa) and either shRNA131 or shRNA563 were reduced by 58% and 46%, respectively (Figure 6A-B). Importantly, neither of these mRXRα shRNAs affected MLL-GAS7-mediated transformation (Figure 6A-B), indicating that inhibition by these shRNA was specific for the R1A-RARα oncogenic pathway.
RXRα is required for R1A-RARα–mediated bone marrow cell transformation. (A) The bar chart represents the relative numbers of third-round colonies of primary hematopoietic cells cotransduced with R1A-RARα, R1A-RARα(ΔRIIa), or MLL-GAS7 plus empty vector, sh563, or sh131, respectively. Data are mean ± SD of 3 independent experiments. (B) Typical colony pictures of progenitor/stem cells cotransduced with retrovirus carrying the indicated leukemia fusion gene and empty vector, sh563, or sh131 after third-round plating in methylcellulose. Scale bars represent 1 mm. (C) The bar chart represents the relative numbers of third-round colonies of primary bone marrow cells transduced with the indicated constructs in the absent or presence of indicated drugs for 7 days. Data are mean ± SD of 3 independent experiments. (D) Typical third-round colony morphology in the absence or presence of the indicated drugs generated from transduced primary hematopoietic cells. Scale bars represent 1 mm. (E) Typical morphology of methylcellulose colonies generated from bone marrow cells transduced with retroviruses expressing R1A-RARα (top panel), R1A-RARα(ΔRIIa) (middle panel), or MLL-ENL (bottom panel), in the absence or presence of the indicated drugs. Scale bars represent 100 μm. (F) Flow cytograms are shown of Mac-1– and Gr-1–stained cells transduced by R1A-RARα (top panel) or R1A-RARα(ΔRIIa) (bottom panel) in the absence or presence of indicated drugs after third-round plating.
RXRα is required for R1A-RARα–mediated bone marrow cell transformation. (A) The bar chart represents the relative numbers of third-round colonies of primary hematopoietic cells cotransduced with R1A-RARα, R1A-RARα(ΔRIIa), or MLL-GAS7 plus empty vector, sh563, or sh131, respectively. Data are mean ± SD of 3 independent experiments. (B) Typical colony pictures of progenitor/stem cells cotransduced with retrovirus carrying the indicated leukemia fusion gene and empty vector, sh563, or sh131 after third-round plating in methylcellulose. Scale bars represent 1 mm. (C) The bar chart represents the relative numbers of third-round colonies of primary bone marrow cells transduced with the indicated constructs in the absent or presence of indicated drugs for 7 days. Data are mean ± SD of 3 independent experiments. (D) Typical third-round colony morphology in the absence or presence of the indicated drugs generated from transduced primary hematopoietic cells. Scale bars represent 1 mm. (E) Typical morphology of methylcellulose colonies generated from bone marrow cells transduced with retroviruses expressing R1A-RARα (top panel), R1A-RARα(ΔRIIa) (middle panel), or MLL-ENL (bottom panel), in the absence or presence of the indicated drugs. Scale bars represent 100 μm. (F) Flow cytograms are shown of Mac-1– and Gr-1–stained cells transduced by R1A-RARα (top panel) or R1A-RARα(ΔRIIa) (bottom panel) in the absence or presence of indicated drugs after third-round plating.
To further investigate that RXRα was required for R1A-RARα–mediated transformation of primary hematopoietic cells, we used a well-characterized pan-RXR agonist, SR11237.14 Cells that emerged from second-round plating after transduction with either R1A-RARα or R1A-RARα(ΔRIIa) were split and plated in the absence or presence of SR11237 or retinoic acid (RA), as a positive control. Treatment of R1A-RARα-transduced cells with SR11237 reduced colony formation by 35% compared with untreated controls (Figure 6C). Treatment of R1A-RARα(ΔRIIa)-transduced cells with SR11237 had an even more profound effect reducing colony number by 77% compared with untreated controls (Figure 6C-D). Like PML-RARα (supplemental Figure 4), treatment of both R1A-RARα- and R1A-RARα(ΔRIIa)-transduced cells with ATRA reduced colony formation by more than 90%, as expected. Inhibition of colony formation was not the result of a nonspecific toxic effect of the drugs because treatment of MLL-ENL-transformed cells with either SR11237 or ATRA did not affect colony number (Figure 6C-E). Cells treated with SR11237 or RA were analyzed for differentiation markers by FACS. ATRA treatment, as expected, increased expression of myeloid differentiation markers Mac-1 and Gr-1 (Figure 6F), but so did SR11237 treatment, although to a somewhat lesser extent (Figure 6F). In addition, May-Grunwald-Giemsa staining of cells treated with ATRA and, to a lesser extent SR11237, was smaller and exhibited multilobulated nuclei (supplemental Figure 5).
Discussion
In this study, we demonstrated that the newly described R1A-RARα chimeric APL protein is capable of transforming murine bone marrow hematopoietic progenitor/stem cells. We also showed that, although R1A-RARα bound RARE as either a homodimer or as a heterodimer with RXRα, its transformation activity critically depended on its ability to form complexes with RXRα.29 Unlike wild-type R1A, which was localized exclusively to the cytoplasm, R1A-RARα localized within the nucleus in a pattern that resembled wild-type RARα. These finding suggest that R1A-RARα contributes to APL oncogenesis either by binding to RARE as a R1A-RARα/RXRα hetero-oligomeric complex (thereby altering transcriptional activity at these sites) and/or by aberrantly sequestering key cAMP/PKA signaling intermediates within the nucleus (thereby interfering with PKA activity within the cytoplasm).
We also showed that homodimeric DNA binding by R1A-RARα is dispensable, as a mutant of R1A-RARα lacking the RIIa domain still could competently transform bone marrow cells. This result is reminiscent of STAT5b-RARα in which deletion of its coiled-coil domain required for dimerization did not abolish its transformation ability, which is dependent on its capability of forming high molecular weight hetero-oligomeric complexes with RXRα.14 However, it has been shown that the strength of self-association partly correlates with the transformation ability of the RARα fusions;11,13,22 thus, the robust transformation by R1A-RARα(ΔRIIa) is unexpected. It is notable that R1A is a key component in regulating cAMP-dependent PKA enzymatic activity and has a pivotal role in modifying signal transduction in a wide variety of intracellular pathways.9 R1A was shown in our studies to form a complex with R1A-RARα and to be partly recruited into the chromatin region by R1A-RARα through its RIIa-dimerization motif. These findings raise the possibility that R1A-RARα may sequester in the nucleus molecules essential for normal cAMP/PKA function in the cytoplasm. R1A germline or somatic mutations, which resulted in haploinsufficiency, are associated with increased PKA activity resulting from reduction of cytoplasmic R1A levels and have been implicated in oncogenesis.6 It was reported that a cAMP analog was able to induce differentiation of HL60 leukemia cells and to have a synergistic effect with arsenic in triggering maturation of APL cells. In addition, the addition of cAMP agonists in APL cells led to a decrease in the concentration of ATRA needed to induce leukemic cell differentiation, whereas a cAMP antagonist inhibited, in part, the differentiation by ATRA in APL.30-33 In R1A-RARα+ APL cells, loss of R1A on one chromosome allele combined with R1A sequestration in the nucleus by R1A-RARα chimeric protein would be expected to reduce cytoplasmic R1A levels, which may lead to the increased PKA kinase activity in R1A-RARα+ leukemic cell compared with other APL cells. This enhanced PKA kinase activity would lead to a favorable response to ATRA and arsenic treatment in R1A-RARα+ APL patients.5 However, with the deletion of RIIa dimerization domain in R1A-RARα chimeric protein, the recruited R1A by R1A-RARα would be released back into cytoplasm, resulting in decreased PKA activity, which might enhance the oncogenic effect of the fusion and may explain, in part, its robust transformation observed in our RTTA experiments (Figure 1A).
On the other hand, R1A-RARα–mediated transformation ability after deletion of protein-protein interaction domain of R1A moiety could also be the result of the loss of potential tumor suppressor activity. The RIIa dimerization domain may interact with an unknown protein, which possesses tumor suppressor activity. It has been reported that the RIIα subunit of protein kinase A interacted with ETO through the RIIa protein interaction domain.34 Therefore, R1A-RARα chimeric protein may interact with ETO as well as the related protein ETO-2/MTG16. ETO is a transcriptional repressor that partners with AML1 to form the AML1-ETO chimeric protein found in a subset of in AML. ETO-2 has been shown to be a putative tumor suppressor since introduction of ETO-2 gene into different breast tumor cell lines with low levels of ETO-2 gene expression significantly reduced the colony growth.35 Through the RIIa domain, R1A-RARα chimeric protein may recruit ETO or ETO-2 into its chromatin region, leading to potential tumor-suppressor activity by an unknown mechanism. The loss of interaction with ETO or ETO-2 after deletion of the RIIa domain would thus result in increased ability of R1A-RARα(ΔRIIa) to transform progenitor/stem cells compared with R1A-RARα. It remains to be seen whether ETO or ETO-2 can be recruited into the chromatin region of R1A-RARα.
APL fusion proteins have been previously shown to associate with RXRα to form high-molecular-weight hetero-oligomeric complexes, which can bind to RARE.1-3 We and others have demonstrated recently that RXRα recruitment is a critical determinant of the transforming potential of several RARα-fusion proteins, such as PML-RARα, STAT5b-RARα, and NuMA-RARα.14,20,29,36 In our present study, R1A-RARα was shown to bind RARE as a hetero-oligomer with RXRα, and both the ratio of R1A-RARα to RXRα and the specific RARE itself determined the pattern of R1A-RARα/RXRα hetero-oligomeric complexes formed. RXRα appears to be required for full execution of the transcriptional and transformation programs imposed by R1A-RARα on its target cell. These considerations raised the possibility of interventions in APL by targeting RXR, although its actual functional contribution for transformation is still not completely clear. R1A-RARα/RXRα may suppress and deregulate RARE-containing or non–RARE-containing gene transcriptional activities, which are important for leukemogenesis. Thus identification of the underlying mechanisms and key gene(s) dysregulated by the R1A-RARα/RXRα complex may be important not only for APL but also for other types of leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr M. Mavrakis (Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD) for kindly providing the GFP-PRKAR1A expression vector, which facilitated our making a series of R1A and R1A-RARα expression vectors; and Dr M. Busslinger from Research Institute of Molecular Pathology, Vienna, Austria, for the PAX5 expression vector.
This work was supported in part by the Albert and Margaret Alkek Foundation (J.J.Q., S.D.) and the Kay Kendall Leukemia Fund (B.B.Z., C.W.E.S.).
Authorship
Contribution: J.J.Q. designed and performed research and wrote the paper; X.L. performed research and wrote the paper; B.B.Z. performed research; Z.M., X.C., and S.C. analyzed data; H.G., D.J.T., and C.W.E.S. analyzed data and edited the paper; and S.D. supervised and designed research, interpreted results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shuo Dong, Department of Medicine, Baylor College of Medicine, One Baylor Plaza, BCM 286, Room N1317.03, Houston, TX 77030; e-mail: sdong@bcm.tmc.edu.
References
Author notes
J.J.Q. and X.L. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal