Abstract
Mice deficient in c-fms–like tyrosine kinase 3 (FLT3) signaling have reductions in early multipotent and lymphoid progenitors, whereas no evident myeloid phenotype has been reported. However, activating mutations of Flt3 are among the most common genetic events in acute myeloid leukemia and mice harboring internal tandem duplications within Flt3 (Flt3-ITD) develop myeloproliferative disease, with characteristic expansion of granulocyte-monocyte (GM) progenitors (GMP), possibly compatible with FLT3-ITD promoting a myeloid fate of multipotent progenitors. Alternatively, FLT3 might be expressed at the earliest stages of GM development. Herein, we investigated the expression, function, and role of FLT3 in recently identified early GMPs. Flt3-cre fate-mapping established that most progenitors and mature progeny of the GM lineage are derived from Flt3-expressing progenitors. A higher expression of FLT3 was found in preGMP compared with GMP, and preGMPs were more responsive to stimulation with FLT3 ligand (FL). Whereas preGMPs and GMPs were reduced in Fl−/− mice, megakaryocyte-erythroid progenitors were unaffected and lacked FLT3 expression. Notably, mice deficient in both thrombopoietin (THPO) and FL had a more pronounced GMP phenotype than Thpo−/− mice, establishing a role of FL in THPO-dependent and -independent regulation of GMPs, of likely significance for myeloid malignancies with Flt3-ITD mutations.
Introduction
Activating mutations in the c-fms–like tyrosine kinase 3 (Flt3) receptor, are one of the most common mutations observed in acute myeloid leukemia (AML). Internal tandem duplications (ITD) of the juxtamembrane domain are found in almost 1 of 4 patients and are associated with poor prognosis.1,2 The Flt3-ITD mutation leads to constitutive activation of the FLT3 receptor,3 and mice harboring an ITD in the Flt3 locus develop a myeloproliferative disease with increased numbers of granulocyte-monocyte (GM) progenitors (GMPs) and mature myeloid cells.4-6 The FLT3 receptor is also known as Flk2 and belongs to the family of class III receptor tyrosine kinases, which also include monocyte-colony stimulating factor receptor (M-CSFR) and the stem cell factor receptor (KIT). Studies of FLT3 ligand (Fl−/−) and FLT3 receptor (Flk2−/−) deficient mice have demonstrated a key role of FLT3 signaling in early lymphopoiesis, with reduced numbers of progenitors of multiple lymphoid lineages, including lymphoid-primed multipotent progenitors (LMPP),7-10 a progenitor common for the GM and lymphoid, but not megakaryocyte-erythroid (MkE) lineages. However, a role of FLT3 in normal myelopoiesis has yet not been established, as no evident myeloid phenotype has been reported in FLT3-deficient mice. Because activating Flt3 mutations are frequently found in myeloid malignancies, the mutations might preferentially target multipotent progenitors, such as LMPPs,11 promoting primarily a myeloid phenotype; or, a role of FLT3 at distinct stages of myelopoiesis has yet to be uncovered.
Previous studies have demonstrated that only a small fraction of GMPs have detectable cell-surface FLT3 expression,12,13 and myeloid progenitors in Flt3 ligand (FL)–deficient mice were not significantly affected,8,9 beyond an overall reduction in cellularity. This translated into reduced numbers of all progenitors, including those of the MkE lineage, on which the FLT3 receptor is not expressed, neither in mice nor in humans.8,12,14 In agreement with this, Flk2−/− mice lacked a myeloid phenotype.7 However, a recent study has identified earlier and more distinct stages of GM and MkE progenitors, including the GMP and preGMPs,15 and the role of FLT3 at these earliest stages of GM development have yet to be investigated. The low expression of FLT3 in GMPs could be compatible with higher and more functionally relevant expression levels in the preGMP population because it precedes the GMP developmental stage. Thus, we hypothesized that lack of FLT3 signaling might affect the preGMP population. As it is possible that such a myeloid phenotype might be subtle due to redundancy of other cytokines, the role of FL in myelopoiesis might be best uncovered in the absence of other nonredundant myeloid growth factors. One such candidate is the cytokine thrombopoietin (THPO), initially identified as a primary regulator of megakaryocytes and platelets16,17 and later shown to be a critical regulator of hematopoietic stem cells (HSCs) and myeloid progenitors.18-20 Of particular relevance, THPO and FL have been shown to act in potent synergy on early hematopoietic progenitors to promote myeloid expansion and survival.21
Herein, we demonstrate for the first time the importance of FL in THPO-dependent as well as THPO-independent maintenance of preGMPs as well as GMPs, establishing a role of FLT3 in normal myelopoiesis, of likely relevance for a better understanding of its impact in myeloid malignancies.
Methods
Animals
Congenic CD45.1 and CD45.2 wild-type (WT) C57BL/6J (BomBmsd) mice were used as controls and recipients in transplantation experiments. Fl−/− (Flt3 ligand knockout) and Thpo−/− (Thpo ligand knockout) mice on a pure C57Bl/6 background have previously been described.8,9,17,19 Fl−/− × Thpo−/− mice were generated by crossing of Thpo−/− and Fl−/− mice. In the analysis of Fl−/− mice, WT C57Bl/6 as well as WT littermate mice were used as controls, with similar results. Flt3-cre mice were mated with Rosa 26 (R26R)-eYFP mice and have previously been described.22,23 Mice were analyzed at the age of 7 to 13 weeks. Animal experiments were approved by the local animal ethics committees at Lund University and University of Oxford.
Fluorescence-activated cell sorting
Nucleated peripheral blood (PB) cells were stained with anti-CD3-FITC (145-2C11), anti-Gr-1-phycoerythrin (PE; RB6-8C5), anti-CD19-PECy7 (1D3), anti-Mac-1-allophycocyanin (APC; M1/70; all from BD Biosciences), anti-NK1.1 Pacific Blue (PK136; BioLegend), anti-CD45-Alexa Fluor 700 (30F11; eBioscience). Dead cells were excluded using propidium iodide (Invitrogen) or 7-Aminoactinomycin D (7-AAD; Sigma-Aldrich). Spleens were homogenized to single-cell suspension, incubated with anti-CD16/CD32 (2.4G2) purified antibodies, and stained as PB.
Bone marrow (BM) samples were obtained by isolating tibias and/or femurs/crista and crushing them with a mortar and pestle. To analyze myeloid progenitors, cells were incubated with antibodies against lineage markers of mature blood cells (purified rat anti-B220 [RA3-6B2], anti-CD5 [53-7.3], anti-CD8α [53-6.7], anti-Gr-1 [RB6-8C5], anti-Ter-119 [TER-119], anti-CD4 [H129.19], and anti-Mac-1 [M1/70]; all from BD Biosciences), stained with goat anti–rat-QDot605 (Invitrogen). For cell sorting, cells were enriched using anti-CD117 immunomagnetic beads (Miltenyi Biotec). Samples were further stained with anti-CD41-fluorescein isothiocyanate (FITC; MW/reg30; BD Biosciences), anti-CD135-PE (A2F10.1; BD Biosciences), anti-CD150-PECy7 (TC15-12F12.2; BioLegend), anti-CD16/32-APC (93; eBioscience), anti-c-Kit-APC-Alexa Fluor 750 (2B8; eBioscience), anti-Sca-1-Pacific Blue (D7; BioLegend), and anti-CD105-biotin (MJ7/18; eBioscience) antibodies. Biotin was visualized with streptavidin-QDot655 (Invitrogen). Dead cells were excluded using propidium iodide. Myeloid progenitors in Flt3-cre/R26R-eYFP mice were lineage stained with purified rat anti-B220, anti-CD8α (eBioscience), anti–GR-1, anti-CD4, and anti–Mac-1 (eBioscience) and further stained with anti-CD16/32-PE (2.4G2; BD Biosciences), anti-Ter119-PECy5.5 (Ter119; eBioscience), anti-CD41-PECy7 (MWReg30; eBioscience), anti-CD150-APC (TC15-12F12.2; BioLegend), anti-c-Kit-APCeF780 (2B8; eBioscience), anti-CD45.2-AF700 (104; BioLegend), and anti-Sca-1-Pacific Blue (E13-161.7; BioLegend) antibodies. Dead cells were excluded with 7-AAD. For lineage analysis of mature myeloid cells in Flt3-cre/R26R-eYFP BM, cells were stained with anti-CD41-PE (MWReg30; BD Biosciences), anti-Ter119-PE (Ter119; BD Biosciences), anti-Gr1-APC (RB6-8C5; BD Biosciences), anti-Mac1-AF700 (M1/70; eBioscience), anti-CD4-APCeF780 (RM4-5; eBioscience), anti-CD8-APCeF780 (53-6.7; eBioscience), anti-CD19-PECy7 (1D3; eBioscience), and anti-CD45.2-Pacific Blue (104; BioLegend) antibodies.
For fluorescence-activated cell sorting (FACS) analysis and sorting of the lineage-negative SCA-1+, KIT+ (LSK) population, cells were stained with lineage antibodies and enriched with anti-CD117 immunomagnetic beads, and incubated with anti-CD16/CD32 (2.4G2) purified antibodies. Polyclonal goat anti–rat Tricolor antibody (Invitrogen) was used to visualize lineage antibodies. The following antibodies were used: anti-Sca-1-FITC (E13-161.7; BD Biosciences) or Sca-1-PECy5.5 or Pacific Blue (D7; BioLegend), anti-c-Kit-APC or APC-AF750 (2B8; BD Biosciences and eBioscience), anti-CD34-FITC (RAM34; eBioscience), anti-CD135-PE (A2F10.1; BD Biosciences), anti-THPOR-biotin (AMM2; Mpl; generously provided by Kirin Brewery Co Ltd). Streptavidin-PECy7 (BD Biosciences) was used to visualize biotinylated anti-THPOR. Dead cells were excluded using 7-AAD. Cellularities were counted with Sysmex (KX-21N).
Clonogenic myeloid progenitor assays
Colony-forming-units-GM (CFU-GM) colonies were quantified by plating 10 000 unfractionated BM cells in duplicates in methylcellulose (MethoCult 3134; StemCell Technologies) supplemented with Iscove modified Dulbecco medium (PAA), 20% fetal calf serum (FCS; Sigma-Aldrich), 1% penicillin/streptomycin (PAA), 1% l-glutamine (2mM; PAA), 1% 2-Mercaptoethanol (0.1mM; Sigma-Aldrich), and the following cytokines: 10 ng/mL human granulocyte–colony stimulating factor (G-CSF; Amgen), 10 ng/mL human FL (Immunex), 2 ng/mL mouse interleukin-3 (IL-3; PeproTech), 5 ng/mL mouse GM–colony stimulating factor (GM-CSF; PeproTech). Colonies (> 40 cells) were scored after 7 to 8 days.24 For cytokine-response studies, single cells of indicated populations were either manually plated or sorted directly into Terasaki plates with X-vivo 15 (BioWhittaker) supplemented with 1% penicillin/streptomycin, 1% l-glutamine, 1% 2-Mercaptoethanol, 0.5% to 1% detoxified bovine serum albumin (BSA; Stem Cell Technologies), and cytokines; 50 ng/mL FL, 50 ng/mL human THPO (PeproTech), 50 ng/mL G-CSF, 50 ng/mL GM-CSF, and 50 ng/mL human monocyte–colony stimulating factor (M-CSF; Cetus Corp). For GM conditions, the following cytokines were used: 25 ng/mL mouse stem cell factor (SCF; PeproTech), 10 ng/mL IL-3, 25 ng/mL FL, 25 mg/mL G-CSF, 25 ng/mL GM-CSF, and 25 ng/mL THPO. Colonies were scored after 7 to 8 days. All cultures were incubated at 37°C with 5% CO2 in air and more than 95% humidity.
Transplantation assays
Lethally irradiated (900 cGy) C57Bl/6 CD45.2 recipient mice were transplanted with 1 × 106 WT, Fl−/−, Thpo−/−, or Fl−/− × Thpo−/− unfractionated BM CD45.1 cells, respectively (3 pooled mice per genotype) together with 1 × 106 WT unfractionated BM CD45.2 support cells. To determine multilineage reconstitution,25 nucleated PB cells were stained with anti–CD45.2-FITC (104; BD Biosciences) and anti–CD45.1-PE (A20; BD Biosciences) to determine total donor reconstitution; anti–CD4-PECy5 (H129.9; BD Biosciences) and anti–CD-8a-PECy5 (Ly2/53-67; BD Biosciences) to identify T cells; anti–CD19-PECy7 (1D3; BD Biosciences) for B cells; and anti-Mac-1-APC (M1/70; BD Biosciences) together with anti-NK1.1 PB (PK136; BioLegend) antibodies to determine myeloid cells, defined as NK1.1−Mac-1+. The detection level for reconstitution was as previously26 set at 0.1% of total test cells (of total mononuclear cells) and 0.02% (of total mononuclear cells) of each of the B, T, and myeloid test cell contribution.
Gene expression analysis of single cells by multiplex RT-PCR
Multiplex single-cell reverse transcription–polymerase chain reaction (RT-PCR) analysis was preformed as previously described,11,27 with the addition of the following primers for the Csf2r (Gm-csfr): Csf2r-1 TTGCGAACGACTTGTCACTGCT, Csf2r-2 AGTGAGGCCTACTTGGTCGTGA, Csf2r-3 AAGAGTTACAGGACGCGGTGAC, Csf2r-4 CCACTGGACC-TCAAACTGGAAG.
Statistics
Statistical significances were determined using the 2-tailed, unpaired Mann-Whitney test, or the paired Mann-Whitney test (only Figure 3F). Group comparison in Figure 5A was done with the Kruskal-Wallis test and with the unpaired Mann-Whitney test when significant (P < .05) data were further evaluated. P values were corrected for multiple comparisons using Bonferroni-Holm. The significance level was set at P less than .05. Data are shown as means plus or minus SD.
Results
Flt3 fate mapping demonstrates that steady-state GM differentiation derives through Flt3-expressing progenitors
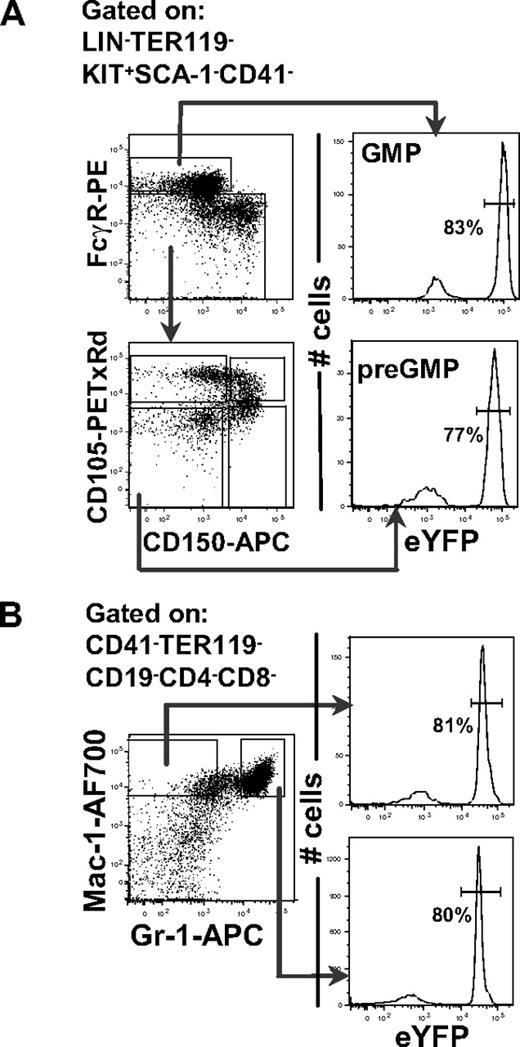
The LMPP—with combined GM and lymphoid potential—express high levels of FLT3,11 whereas the recently identified primitive common myeloid progenitor—with considerable GM potential—as well as the majority of GMPs, are regarded as negative for FLT3 expression.12,13,28 It is therefore currently unclear whether myeloid cells are normally generated through FLT3-positive or -negative progenitors. To establish to what degree GMPs and mature myeloid cells pass through Flt3-expressing progenitors, we pursued a cre-loxP fate-mapping approach, in which mice expressing the cre recombinase under control of the Flt3 promoter22 were crossed with mice having a loxP-flanked transcriptional termination sequence preceding the eYFP gene, under control of the ubiquitous Rosa26 promoter.23 When intercrossed, cells expressing Flt3 as well as all their progeny will express eYFP. Notably, using a recently developed staging strategy in which GMPs can be separated into preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low) and GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+),15 we found that as many as 77% plus or minus 7% of preGMPs and 83% plus or minus 6% of GMPs in the BM expressed eYFP (Figure 1A). Furthermore, as many as 80% plus or minus 6% of Mac-1+Gr-1hi (granulocytes)29 and 81% plus or minus 6% Mac-1+Gr-1lo (monocytes)29 in the BM also expressed eYFP (Figure 1B). Thus, genetic Flt3 fate-mapping establishes that most cells of the GM lineage are derived from Flt3-expressing progenitors.
Flt3 fate-mapping reveals that the majority of GMPs and their mature progeny are derived from Flt3-expressing progenitors. (A) FACS analysis of eYFP expression in preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low) and GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+) in BM of Flt3::Cre//R26R-eYFP mice. Histogram shows percentage of preGMPs and GMPs expressing eYFP. Percentages are the means of 3 mice, analyzed at 10 weeks of age. (B) Representative FACS profiles of mature (Mac-1+Gr-1neg-lo and Mac-1+Gr-1hi) myeloid cells in the BM of Flt3::Cre//R26R-eYFP mice. Histogram shows percentage of Mac-1+Gr-1neg-lo and Mac-1+Gr-1hi cells expressing eYFP. Percentages are the means of 3 mice, analyzed at 10 weeks of age.
Flt3 fate-mapping reveals that the majority of GMPs and their mature progeny are derived from Flt3-expressing progenitors. (A) FACS analysis of eYFP expression in preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low) and GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+) in BM of Flt3::Cre//R26R-eYFP mice. Histogram shows percentage of preGMPs and GMPs expressing eYFP. Percentages are the means of 3 mice, analyzed at 10 weeks of age. (B) Representative FACS profiles of mature (Mac-1+Gr-1neg-lo and Mac-1+Gr-1hi) myeloid cells in the BM of Flt3::Cre//R26R-eYFP mice. Histogram shows percentage of Mac-1+Gr-1neg-lo and Mac-1+Gr-1hi cells expressing eYFP. Percentages are the means of 3 mice, analyzed at 10 weeks of age.
GMPs express functionally relevant levels of cell-surface FLT3
FLT3 has been shown to be expressed only in a small fraction of GMPs, as previously defined.12,13,30 In agreement with this, we found that 13% plus or minus 4% of GMPs expressed FLT3 receptor at low levels as determined by FACS (Figure 2A). However, the expression was much more distinct on preGMPs, of which as many as 38% plus or minus 8% were FLT3-positive and, in addition, the expression levels were higher (Figure 2A). In contrast to GMPs and preGMPs, megakaryocyte progenitors (MkP; Lin−Kit+Sca-1−CD41+), pre–megakaryocyte erythroid progenitors (pMegE; Lin−Kit+Sca-1−CD41−CD150+FcγR−CD105low), and CFU-erythroid (CFU-E; Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105high) and preCFU-E (Lin−Kit+Sca-1−CD41−CD150+FcγR−CD105high)15 populations lacked detectable FLT3 expression (Figure 2A, and data not shown), supporting a specific expression pattern within the GM and not the MkE progenitors, and in particular within the fraction of early GMPs. Investigating Flt3 mRNA expression at the single-cell level, 66% plus or minus 8% of preGMPs were found to express Flt3, compared with 22% plus or minus 9% of GMPs (Figure 2B).
PreGMPs express functionally relevant levels of FLT3. (A) Representative FACS profiles showing FLT3 expression on different myeloid progenitors. Lineage-negative cells were first gated as Kit+Sca-1− cells as indicated. Further gating strategies are indicated with arrows. Histograms show FLT3 expression within indicated populations; preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low), GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+), MkP (Lin−Kit+Sca-1−CD41+), pMegE (Lin−Kit+Sca-1−CD41−CD150+FcγR−CD105low), and CFU-E (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105high). Gray areas indicate isotype control. Percentages in panels represent percentages of FLT3-positive cells in the investigated cell populations, and represent mean values from 8 experiments with pooled mice at 8 to 12 weeks of age. (B) Single-cell RT-PCR analysis of transcriptional expression of Flt3, Csf1r (M-csfr), Csf2r (Gm-csfr), and Csf3r (G-csfr) in preGMPs and GMPs, respectively. Only cells expressing Kit were analyzed further (> 93% of cells analyzed). ■ indicate percentage of cells coexpressing Flt3. Data are means (SD) from 2 experiments, in which 88 cells were analyzed per experiment. (C) Single preGMP and GMP cells were sorted directly into media with indicated cytokines. Clones were scored after 7 to 8 days. Data represent mean (SD) number of clones from 2 to 3 experiments, with 120 cells analyzed per experiment. (D) Single preGMP FLT3-positive and -negative cells were sorted directly into media with GM-promoting cytokines (see “Clonogenic myeloid progenitor assays”). Clones were scored after 7 to 8 days. Data represents mean (SD) number of clones from 3 experiments with 60 to 120 cells analyzed per experiment.
PreGMPs express functionally relevant levels of FLT3. (A) Representative FACS profiles showing FLT3 expression on different myeloid progenitors. Lineage-negative cells were first gated as Kit+Sca-1− cells as indicated. Further gating strategies are indicated with arrows. Histograms show FLT3 expression within indicated populations; preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low), GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+), MkP (Lin−Kit+Sca-1−CD41+), pMegE (Lin−Kit+Sca-1−CD41−CD150+FcγR−CD105low), and CFU-E (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105high). Gray areas indicate isotype control. Percentages in panels represent percentages of FLT3-positive cells in the investigated cell populations, and represent mean values from 8 experiments with pooled mice at 8 to 12 weeks of age. (B) Single-cell RT-PCR analysis of transcriptional expression of Flt3, Csf1r (M-csfr), Csf2r (Gm-csfr), and Csf3r (G-csfr) in preGMPs and GMPs, respectively. Only cells expressing Kit were analyzed further (> 93% of cells analyzed). ■ indicate percentage of cells coexpressing Flt3. Data are means (SD) from 2 experiments, in which 88 cells were analyzed per experiment. (C) Single preGMP and GMP cells were sorted directly into media with indicated cytokines. Clones were scored after 7 to 8 days. Data represent mean (SD) number of clones from 2 to 3 experiments, with 120 cells analyzed per experiment. (D) Single preGMP FLT3-positive and -negative cells were sorted directly into media with GM-promoting cytokines (see “Clonogenic myeloid progenitor assays”). Clones were scored after 7 to 8 days. Data represents mean (SD) number of clones from 3 experiments with 60 to 120 cells analyzed per experiment.
To explore whether the known synergy between other myeloid growth factors and FL,21,31 might in fact occur directly on preGMPs and GMPs, we also investigated transcriptional coexpression patterns for the receptors of important myeloid cytokines. Interestingly, more than 50% of the Csf2r (Gm-csfr)– or Csf3r (G-csfr)–positive cells in preGMPs coexpressed Flt3, whereas all Csf1r (M-csfr)–positive cells were Flt3-positive, although only 6% of preGMPs expressed Csf1r (Figure 2B). While at the GMP level increased frequency of cells expressed Csf1r, Csf2r and Csf3r, a smaller fraction of these were positive for Flt3 compared with preGMPs (Figure 2B).
To establish whether the expression of FLT3 observed in preGMPs and GMPs was physiologically relevant; we next examined the cytokine responsiveness of purified preGMPs and GMPs at the single-cell level. Consistent with the expression data, preGMPs and, to a more limited extent, GMPs proliferated in response to FL alone. Most notably, whereas both GMP subsets proliferated in the presence of GM-CSF, G-CSF, and M-CSF, the reported synergy between these cytokines and FL31-34 was only observed at the preGMP level (Figure 2C).
The preGMP population was further subfractionated based on FLT3 expression, and in the FLT3 receptor–positive population up to 70% of the cells formed large clones, compared with only 23% in the FLT3 receptor–negative population, in response to a cocktail of GM-promoting cytokines (Figure 2D). These data demonstrate that FLT3 is expressed at functionally relevant levels on primitive preGMPs and to a less extent on GMPs and that the synergistic interaction between FL and other myeloid growth factors is restricted to preGMPs.
Reduction of preGMPs and GMPs in FL-deficient mice
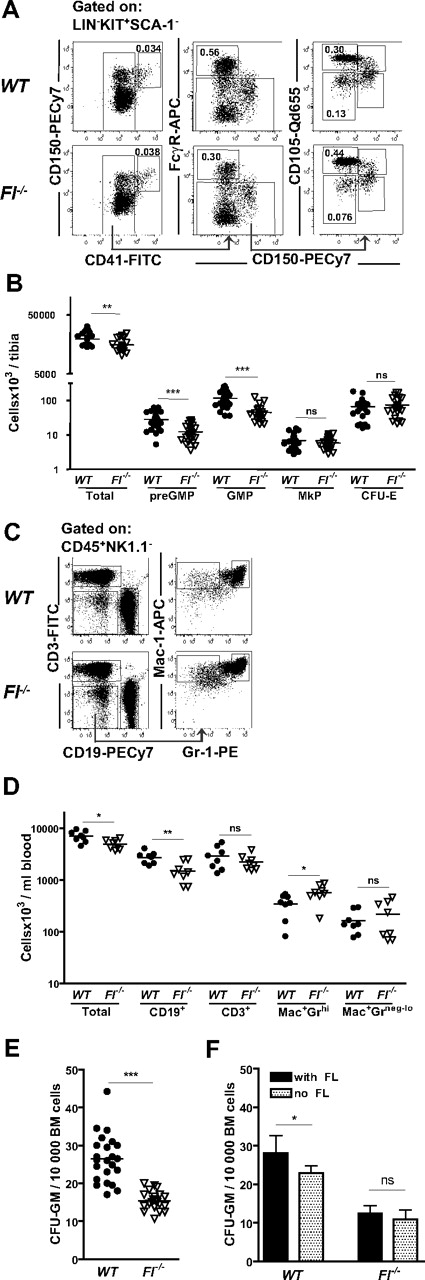
To investigate whether FLT3 has a nonredundant role in myelopoiesis, mice deficient in FL were next investigated. In BM, a slight but significant reduction in cellularity in Fl−/− mice was observed as reported previously8 (Figure 3B). The frequency of different subsets of MkE progenitors was not significantly reduced in Fl−/− compared with WT mice (Figure 3A-B), in agreement with the observed lack of FLT3 receptor expression on MkE progenitors. In striking contrast, we found a 43% reduction in the frequency of preGMPs in Fl−/− mice compared with WT mice (P < .001; Figure 3A) translating into an absolute reduction of 56%, whereas GMPs had an absolute reduction of 61% compared with WT (Figure 3A-B). There was, however, no significant reduction in FLT3 receptor expression on preGMPs in Fl−/− (39% ± 12%) compared with WT mice (43% ± 12%). While we confirmed the previously reported reduction in peripheral blood B cells, numbers of Mac-1+Gr-1hi (granulocytes)29 and Mac+Gr-1neg-low(monocytes)29 were not reduced in the blood of Fl−/− mice. However, a minor decrease in numbers of Mac+Gr-1neg-low was observed in the spleen (Figure 3C-D and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The GMPs were also quantified functionally using in vitro semisolid cultures. In contrast to previous studies of FLT3 ligand and receptor-deficient mice,7,8 we here included FL in the CFU-GM assay, as a potent factor for GMP growth.31-34 Notably, when investigating CFU-GM numbers under these conditions, we could demonstrate that the BM cells from Fl−/− mice have significantly reduced capacity to form CFU-GM compared with WT mice, reflected in reduced frequencies as well as absolute numbers of CFU-GM (Figure 3E), in agreement with the phenotypic analysis. We also evaluated CFU-GM growth without adding FL showing significantly higher cloning efficiency in WT BM cells in the presence than absence of FL, whereas in Fl−/− mice there were no significant differences, compatible with a selective reduction of FL-responsive progenitors in Fl−/− mice (Figure 3F). Taken together, these results show that FL is a nonredundant regulator of GM, but not MkE progenitors.
Selective deficiencies in GMPs and preGMPs in FL-deficient mice. (A) Representative FACS profiles and gating strategies for myeloid progenitors in WT and Fl−/− mice. preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low), GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+), MkP (Lin−Kit+Sca-1−CD41+), and CFU-E (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105high). Numbers represent frequencies of gated populations relative to total BM cells, and are mean values from more than 3 experiments. (B) Absolute number of total BM cells, preGMPs, GMPs, MkPs, and CFU-Es per tibia in WT and Fl−/− mice. Each dot represents an individual mouse. More than 3 litters were analyzed for each genotype and progenitor subset. ns indicates not significant; **P < .01, ***P < .001. (C) Representative FACS profiles of peripheral blood analysis of WT and Fl−/− mice. Cells were first gated as CD45+NK1.1−. For myeloid lineage analysis cells were as indicated first gated as CD19−CD3− cells and then analyzed for Gr-1 and Mac-1 expression. (D) Peripheral blood cell analysis in WT and Fl−/− mice based on same gating strategy as shown in panel C. Data shown are total number of cells per milliliter of blood, B cells (CD19+), T cells (CD3+), granulocytes (Mac-1+Gr-1hi), and monocytes (Mac-1+Gr-1neg-lo). Each dot represents an individual mouse. Data are from 3 experiments. ns indicates not significant; *P < .05, **P < .01. (E) CFU-GM cultures: 10 000 unfractionated BM cells from WT or Fl−/− mice were cultured in methylcellulose supplemented with FL, IL-3, GM-CSF, and G-CSF. Clones were scored after 7 to 8 days. Each dot represents data from 1 mouse (means of 2 replicates). Horizontal bars indicate mean values of all mice in each genotype. Data are from at least 3 experiments. ***P < .001. (F) Effect of FL on CFU-GM cultures; 10 000 unfractionated BM cells from WT or Fl−/− mice were added to methylcellulose supplemented with IL-3, GM-CSF, and G-CSF with or without FL. Clones were scored after 7 to 8 days. Data represent mean (SD) values from 6 mice/genotype, 2 experiments. Statistical significance was tested with the paired Mann-Whitney, between WT, with and without FL, and Fl−/− with and without FL, respectively. *P < .05.
Selective deficiencies in GMPs and preGMPs in FL-deficient mice. (A) Representative FACS profiles and gating strategies for myeloid progenitors in WT and Fl−/− mice. preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low), GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+), MkP (Lin−Kit+Sca-1−CD41+), and CFU-E (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105high). Numbers represent frequencies of gated populations relative to total BM cells, and are mean values from more than 3 experiments. (B) Absolute number of total BM cells, preGMPs, GMPs, MkPs, and CFU-Es per tibia in WT and Fl−/− mice. Each dot represents an individual mouse. More than 3 litters were analyzed for each genotype and progenitor subset. ns indicates not significant; **P < .01, ***P < .001. (C) Representative FACS profiles of peripheral blood analysis of WT and Fl−/− mice. Cells were first gated as CD45+NK1.1−. For myeloid lineage analysis cells were as indicated first gated as CD19−CD3− cells and then analyzed for Gr-1 and Mac-1 expression. (D) Peripheral blood cell analysis in WT and Fl−/− mice based on same gating strategy as shown in panel C. Data shown are total number of cells per milliliter of blood, B cells (CD19+), T cells (CD3+), granulocytes (Mac-1+Gr-1hi), and monocytes (Mac-1+Gr-1neg-lo). Each dot represents an individual mouse. Data are from 3 experiments. ns indicates not significant; *P < .05, **P < .01. (E) CFU-GM cultures: 10 000 unfractionated BM cells from WT or Fl−/− mice were cultured in methylcellulose supplemented with FL, IL-3, GM-CSF, and G-CSF. Clones were scored after 7 to 8 days. Each dot represents data from 1 mouse (means of 2 replicates). Horizontal bars indicate mean values of all mice in each genotype. Data are from at least 3 experiments. ***P < .001. (F) Effect of FL on CFU-GM cultures; 10 000 unfractionated BM cells from WT or Fl−/− mice were added to methylcellulose supplemented with IL-3, GM-CSF, and G-CSF with or without FL. Clones were scored after 7 to 8 days. Data represent mean (SD) values from 6 mice/genotype, 2 experiments. Statistical significance was tested with the paired Mann-Whitney, between WT, with and without FL, and Fl−/− with and without FL, respectively. *P < .05.
Mice double deficient in FL and THPO demonstrate a role of FL in THPO-independent regulation of GM but not MkE progenitors or HSCs
We next asked whether FL might be critically involved in sustaining the reduced pool of myeloid progenitors in the absence of THPO, another cytokine implicated in the regulation of multipotent myeloid progenitors and HSCs.35 Initially, we examined whether these 2 cytokines might interact at the level of GM and multipotent progenitors. Within the LIN−SCA+KIT+ (LSK) population, a fraction (63%; Figure 4A) of the FLT3 highest-expressing cells, the LMPPs, coexpressed cell-surface FLT3 and THPO receptor, (THPOR, also called c-Mpl), as previously demonstrated.36 Notably, although THPORhi LMPPs, in agreement with previous studies, failed to respond to THPO alone,26 a potent synergy was observed between FL and THPO in promoting the clonal growth and proliferative burst of THPORhi LMPPs but not of THPORneg LMPPs (Figure 4A-B). This interaction between FL and THPO was also seen in preGMPs, but to a smaller degree (Figure 4C). In agreement with these results, 29% plus or minus 11% of preGMPs expressed Thpor mRNA at the single-cell level and the majority of these cells coexpressed Flt3 (Figure 4D). The functional relevance of this coexpression and interaction was revealed in mice double deficient in FL and THPO, in which a 60%, 38%, and 34% further reductions in frequencies of preGMPs, GMPs, and CFU-GMs, respectively, were found compared with single Thpo−/− mice, whereas the already reduced MkE progenitors in Thpo−/− mice were not further affected in the double-deficient Fl−/− × Thpo−/− mice (Figure 4E-G). As in Fl−/− mice, a decrease in numbers of Mac-1+Gr-1neg-low cells was observed in the spleen of Fl−/− × Thpo−/− compared with Thpo−/−-deficient mice (supplemental Figure 1).
Role of FL in THPO-independent maintenance of GM but not MkE progenitors. (A) Representative FACS profile showing sorting gates for THPORhi and THPOR− LMPPs cells (top panels) and purity reanalysis (bottom panels). LSK cells were gated on the 25% highest FLT3 expressing cells to define LMPPs and further based on THPOR-high and -negative subpopulations. (B) THPORhi and THPOR− LMPPs were sorted and cultured at 1 cell per well (manually plated) in Terasaki plates with indicated cytokines and clones were scored after 7 to 8 days. Data represent mean (SD) values from 2 to 3 experiments, with 120 cells being evaluated per population and experiment. (C) The preGMPs were sorted and cultured at 1 cell per well (directly deposited from the sorter) in Terasaki plates with indicated cytokines and clones were scored after 7 to 8 days. Data represent mean (SD) values from 2 experiments, with 120 cells being evaluated per population and experiment. (D) Single-cell RT-PCR analysis of transcriptional expression of Flt3, Thpor, and coexpression of Flt3 and Thpor in preGMPs. Only cells expressing Kit were included for further analysis (> 98% of cells analyzed). Data are means (SD) from 2 experiments, in which 88 cells were analyzed per experiment. (E) Representative FACS profiles in WT and Thpo−/− and Fl−/− × Thpo−/− mice, showing frequencies of preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low) and GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+). Numbers represent frequencies of gated populations relative to total BM cells, and are mean values from more than 3 experiments and 10 to 13 mice per genotype. (F) Number of total BM cells, preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low), GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+), MkP (Lin−Kit+Sca-1−CD41+), and CFU-Es (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105high) in WT, Thpo−/−, and Fl−/− × Thpo−/− mice. Each dot represents an individual mouse. Data are from 3 experiments. Statistical significance was tested between Thpo−/− and Fl−/− × Thpo−/− mice. ns indicates not significant; **P < .01, ***P < .001. (G) CFU-GM cultures; 10 000 unfractionated BM cells from WT and Thpo−/− and Fl−/− × Thpo−/− mice, respectively, were added to methylcellulose supplemented with FL, IL-3, GM-CSF, and G-CSF. Clones were scored after 7 to 8 days. Each dot represents data from 1 mouse (means of 2 replicates). Data are from 3 experiments. Statistical significance was tested between Thpo−/− and Fl−/− × Thpo−/− mice. ***P < 0.001.
Role of FL in THPO-independent maintenance of GM but not MkE progenitors. (A) Representative FACS profile showing sorting gates for THPORhi and THPOR− LMPPs cells (top panels) and purity reanalysis (bottom panels). LSK cells were gated on the 25% highest FLT3 expressing cells to define LMPPs and further based on THPOR-high and -negative subpopulations. (B) THPORhi and THPOR− LMPPs were sorted and cultured at 1 cell per well (manually plated) in Terasaki plates with indicated cytokines and clones were scored after 7 to 8 days. Data represent mean (SD) values from 2 to 3 experiments, with 120 cells being evaluated per population and experiment. (C) The preGMPs were sorted and cultured at 1 cell per well (directly deposited from the sorter) in Terasaki plates with indicated cytokines and clones were scored after 7 to 8 days. Data represent mean (SD) values from 2 experiments, with 120 cells being evaluated per population and experiment. (D) Single-cell RT-PCR analysis of transcriptional expression of Flt3, Thpor, and coexpression of Flt3 and Thpor in preGMPs. Only cells expressing Kit were included for further analysis (> 98% of cells analyzed). Data are means (SD) from 2 experiments, in which 88 cells were analyzed per experiment. (E) Representative FACS profiles in WT and Thpo−/− and Fl−/− × Thpo−/− mice, showing frequencies of preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low) and GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+). Numbers represent frequencies of gated populations relative to total BM cells, and are mean values from more than 3 experiments and 10 to 13 mice per genotype. (F) Number of total BM cells, preGMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105low), GMPs (Lin−Kit+Sca-1−CD41−CD150−FcγR+), MkP (Lin−Kit+Sca-1−CD41+), and CFU-Es (Lin−Kit+Sca-1−CD41−CD150−FcγR−CD105high) in WT, Thpo−/−, and Fl−/− × Thpo−/− mice. Each dot represents an individual mouse. Data are from 3 experiments. Statistical significance was tested between Thpo−/− and Fl−/− × Thpo−/− mice. ns indicates not significant; **P < .01, ***P < .001. (G) CFU-GM cultures; 10 000 unfractionated BM cells from WT and Thpo−/− and Fl−/− × Thpo−/− mice, respectively, were added to methylcellulose supplemented with FL, IL-3, GM-CSF, and G-CSF. Clones were scored after 7 to 8 days. Each dot represents data from 1 mouse (means of 2 replicates). Data are from 3 experiments. Statistical significance was tested between Thpo−/− and Fl−/− × Thpo−/− mice. ***P < 0.001.
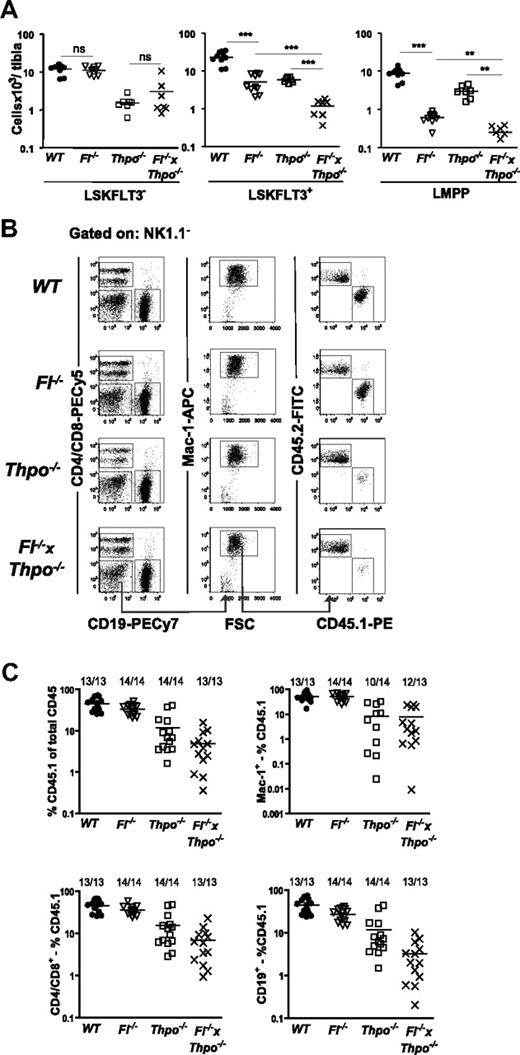
The role of FLT3 in regulation of HSCs in mice has been controversial, but recent studies do not support a role of the FLT3 receptor and ligand in HSC maintenance.7,9,37 In agreement with this, no further reduction in the LSKFLT3− HSC compartment was observed in Fl−/− × Thpo−/− mice compared with Thpo−/− mice (Figure 5A), suggesting that FL is not involved in THPO-independent regulation of HSCs, a conclusion further substantiated through competitive transplantation experiments (Figure 5B-C). However, in agreement with the potent synergistic interaction between FL and THPO found in LMPPs, LSKFLT3+ cells were markedly further reduced in Fl−/− × Thpo−/− mice, as much as 95% relative to WT mice (Figure 5A). Also when investigating the FLT3 highest-expressing cells, the LMPPs, we could demonstrate a further substantial reduction in Fl−/− × Thpo−/− mice compared with Fl−/− and Thpo−/− mice (Figure 5A), respectively. Thus, FL plays a role in THPO-independent and THPO-dependent regulation of GM and LMPP progenitors.
THPO-independent role of FL in regulation of LMPPs but not HSCs. (A) Number of LSKFLT3−, LSKFLT3+, and LMPPs (LSKFLT3hi) in WT, Fl−/−, Thpo−/−, and Fl−/− × Thpo−/− per tibia. Each dot represents an individual mouse. Data are from 3 to 4 experiments. Statistical significance was tested between WT and Fl−/−; Fl−/− and Fl−/− × Thpo−/− (it was not tested in LSKFlt3−); and Thpo−/− and Fl−/− × Thpo−/−. Due to multiple comparisons, Bonferroni-Holms correction of P values was performed (see “Statistics”). ns indicates not significant; **P < .01, ***P < .001. (B) Competitive transplantation assay: 1 million donor BM cells (CD45.1) of indicated genotypes were transplanted together with 1 million WT BM competitor (CD45.2) cells. Representative FACS profile from reconstitution analysis of peripheral blood from the different genotypes at 4 months posttransplantation. Cells were first gated as negative for NK1.1, further gating to identify the myeloid lineage and relative contribution of CD45.1 and CD45.2 is indicated by the gates and arrows. (C) Competitive transplantation assay: peripheral blood analysis at 4 months posttransplantation. Gating strategy is shown in panel B. (Top left panel) The percentage of CD45.1+ cells of total CD45 (CD45.1 and CD45.2) cells; (top right panel) the percentage of CD45.1+ myeloid cells (of total myeloid cells). (Bottom left panel) The percentage of CD45.1+ T cells (of total T cells); (bottom right panel) the percentage of CD45.1+ B cells (of total B cells). Myeloid cells were defined as NK1.1−Mac-1+, B cells as CD19+ and T cells as CD4/CD8+. Each dot represents an individual mouse. Numbers indicate frequencies of reconstituted mice. Mice were defined as reconstituted when at least 0.1% donor (CD45.1) cells were detected among total CD45+ cells, and lineage (myeloid, T, and B cell) reconstituted if a minimum of 0.02% of all CD45+ cells were CD45.1+ and coexpressing the markers for the specific lineage investigated. Data are from 2 experiments with 6 to 7 mice analyzed per genotype and experiment.
THPO-independent role of FL in regulation of LMPPs but not HSCs. (A) Number of LSKFLT3−, LSKFLT3+, and LMPPs (LSKFLT3hi) in WT, Fl−/−, Thpo−/−, and Fl−/− × Thpo−/− per tibia. Each dot represents an individual mouse. Data are from 3 to 4 experiments. Statistical significance was tested between WT and Fl−/−; Fl−/− and Fl−/− × Thpo−/− (it was not tested in LSKFlt3−); and Thpo−/− and Fl−/− × Thpo−/−. Due to multiple comparisons, Bonferroni-Holms correction of P values was performed (see “Statistics”). ns indicates not significant; **P < .01, ***P < .001. (B) Competitive transplantation assay: 1 million donor BM cells (CD45.1) of indicated genotypes were transplanted together with 1 million WT BM competitor (CD45.2) cells. Representative FACS profile from reconstitution analysis of peripheral blood from the different genotypes at 4 months posttransplantation. Cells were first gated as negative for NK1.1, further gating to identify the myeloid lineage and relative contribution of CD45.1 and CD45.2 is indicated by the gates and arrows. (C) Competitive transplantation assay: peripheral blood analysis at 4 months posttransplantation. Gating strategy is shown in panel B. (Top left panel) The percentage of CD45.1+ cells of total CD45 (CD45.1 and CD45.2) cells; (top right panel) the percentage of CD45.1+ myeloid cells (of total myeloid cells). (Bottom left panel) The percentage of CD45.1+ T cells (of total T cells); (bottom right panel) the percentage of CD45.1+ B cells (of total B cells). Myeloid cells were defined as NK1.1−Mac-1+, B cells as CD19+ and T cells as CD4/CD8+. Each dot represents an individual mouse. Numbers indicate frequencies of reconstituted mice. Mice were defined as reconstituted when at least 0.1% donor (CD45.1) cells were detected among total CD45+ cells, and lineage (myeloid, T, and B cell) reconstituted if a minimum of 0.02% of all CD45+ cells were CD45.1+ and coexpressing the markers for the specific lineage investigated. Data are from 2 experiments with 6 to 7 mice analyzed per genotype and experiment.
Discussion
We here demonstrate that the lack of FLT3 signaling confers hematopoietic deficiencies not only in early progenitors of multiple lymphoid lineages,7-9 but also in the earliest stages of GMPs. In contrast, the MkE progenitors are unaffected, and lack FLT3 receptor expression in agreement with previous studies.12,13 The restricted myeloid expression and function of FLT3 in the GM (and not MkE) lineages are of interest and relevant in regard to findings demonstrating that up-regulation of FLT3 expression on LMPPs is accompanied by loss of MkE potential, but sustained GM and lymphoid potentials,11,27 in support of a closer link between the lymphoid and GM lineages than lymphoid and MkE lineages. Furthermore, studies of common myeloid progenitors with combined GM and MkE potential have also been found to lack FLT3 expression.28
Although we found that Fl−/− mice have reduced GMPs as well as preGMPs, there are several lines of evidence suggesting that the primary role of FLT3 is at the preGMP stage and that reductions in GMPs might be secondary to decreased numbers of preGMPs. First, only a small fraction of GMPs expressed FLT3 at the cell surface, in contrast to preGMPs, in which almost half of the cells express FLT3. Second, the potent synergistic interaction between FL and other myeloid growth factors is almost exclusively restricted to the preGMP stage. It is also notable that the expression and function of FLT3 in both the GM, B-, and T-cell lineages appear to be restricted to the earliest progenitors.38,39
Although several cytokines have been demonstrated to potently promote growth of GMPs, many of these have been found to be redundant, as mice deficient in expression of GM-CSF and IL-3 do not exhibit a myeloid phenotype.40,41 A notable exception is THPO,35 and we here show that most preGMPs coexpress Thpor and Flt3. The functional significance of this is supported by synergy between FL and THPO at the preGMP level, and a pronounced reduction of preGMPs is observed in Fl−/− × Thpo−/− mice. Intriguingly, whereas we confirmed that LMPPs coexpressing the THPOR fail to respond to THPO alone,26 we observed a potent synergy between FL and THPO on LMPPs, and most strikingly a dramatic reduction in LMPPs in Fl−/− × Thpo−/− mice.
Despite of the reduction in preGMPs and GMPs in Fl−/− and Fl−/− × Thpo−/− mice, we observed little or no changes in mature myeloid cells in steady-state hematopoiesis. This is reminiscent of the B-cell (and T-cell) phenotype of Fl−/− mice, in which early B-cell progenitors are drastically reduced but mature B cells only slightly affected.9 The explanation for this is most likely that the functional expression of FLT3 is restricted to the earliest stages of the progenitors of these lineages, and therefore there is ample opportunity for compensatory expansion to occur at subsequent FL-independent stages. This, however, does not rule out a more crucial role of FL in the GM lineage. In the case of B lymphopoiesis, FL has been shown to have a critical role in B-cell generation after BM transplantation,42 becomes increasingly important with age, and—most notably IL-7–independent B lymphopoiesis—is strictly FL dependent.10 Thus, further studies might better reveal conditions in which FL is critically involved in generation and regeneration of cells of the GM lineage.
Previous studies have failed to support a role of FL in regulation of mouse HSCs,9,37 and this conclusion was further substantiated here as the HSC deficiency in Thpo−/− mice was not further accelerated in Fl−/− × Thpo−/− mice, in contrast to the preGMP and LMPP phenotypes.
The present study, demonstrating a distinct expression and a nonredundant role of FLT3 at the preGMP stage, is likely to have considerable implications for a better understanding of how FLT3-ITD drives myeloid malignancies, as well as for evaluating the therapeutic potential of FLT3 inhibitors.1,43 Although it remains possible that FLT3-ITD exerts its predominant myeloproliferative effect at a multipotent progenitor level, it is clear that preGMPs will also be directly targeted by activated FLT3-ITD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Monika Posaric Bauden and Zhi Ma for expert technical support, Dr Thomas Boehm for providing the Flt3-Cre mice, Dr Shankar Srinivas for the R26R-eYFP mice, and Fredrik Nilsson for helpful statistical advice.
This work was supported by grants from The Swedish Cancer Society, ALF (Government Public Health Grant) Region Skåne, The Göran Gustafsson's Foundation, Hemato-Linne (Swedish Research Council), Torsten o Ragnar Söderbergs Foundation, The Crafoord Foundation, The Swedish Society of Medicine, and the Medical Research Council UK (MRC reference: G0801073; Grant ID: 87511). A.H. was supported by The Swedish Cancer Society and S.E.W.J. through a strategic appointment from the Medical Research Council UK.
Authorship
Contribution: C.B. and S.E.W.J. designed and conceptualized the overall research, analyzed the data, and wrote the manuscript; C.B. performed the transplantation and synergy experiments and the phenotypic characterization of the different knockout mice; N.B.-V. designed and performed the analysis of the Flt3-cre-YFP mice and also contributed to analysis of data in the manuscript; C.B. and C.T.J. performed the LMPP response experiments; C.J.H.P. provided expertise in FACS analysis of myeloid progenitors; S.K. interpreted data; L.W. provided technical expertise in the animal work; E.S. contributed to the design and execution of experiments; and A.H. designed and performed the gene expression experiments and contributed to the design and execution of experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sten Eirik W. Jacobsen Haematopoietic Stem Cell Laboratory, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Headington, Oxford OX3 9DS, United, Kingdom; e-mail: sten.jacobsen@imm.ox.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal