Abstract
Interaction of chronic lymphocytic leukemia (CLL) B cells with tissue microenvironment has been suggested to favor disease progression by promoting malignant B-cell growth. Previous work has shown expression in peripheral blood (PB) of CLL B cells of activation-induced cytidine deaminase (AID) among CLL patients with an unmutated (UM) profile of immunoglobulin genes and with ongoing class switch recombination (CSR) process. Because AID expression results from interaction with activated tissue microenvironment, we speculated whether the small subset with ongoing CSR is responsible for high levels of AID expression and could be derived from this particular microenvironment. In this work, we quantified AID expression and ongoing CSR in PB of 50 CLL patients and characterized the expression of different molecules related to microenvironment interaction. Our results show that among UM patients (1) high AID expression is restricted to the subpopulation of tumoral cells ongoing CSR; (2) this small subset expresses high levels of proliferation, antiapoptotic and progression markers (Ki-67, c-myc, Bcl-2, CD49d, and CCL3/4 chemokines). Overall, this work outlines the importance of a cellular subset in PB of UM CLL patients with a poor clinical outcome, high AID levels, and ongoing CSR, whose presence might be a hallmark of a recent contact with the microenvironment.
Introduction
Chronic lymphocytic leukemia (CLL) is a heterogeneous disease following a variable course with survival ranging from months to decades. One-third of patients never require treatment and have a long survival; in another third, an initial indolent phase is followed by disease progression and treatment requirement; the remaining third exhibits an aggressive disease at the onset and needs immediate treatment.1 The Rai2 and Binet3 staging systems provided a foundation for the prognosis and design of therapeutic strategies. An unmutated (UM) profile of immunoglobulin (Ig) VH genes,4,5 as well as the presence of genetic lesions at chromosome 17p13, or at 11q23,6 constitute, to date, the poor prognosis indicators.7 In addition, in the last years, CD38 and ζ-associated protein 70 (ZAP-70) expressions have shown important prognostic information.8 However, neither Rai/Binet staging systems nor the molecular markers described to date are able to completely predict the progression of disease and/or explain the heterogeneous progression profile in the clinical course of CLL. One of the long-term goals of the hematologic community is to provide a molecular explanation for this marked clinical heterogeneity of CLL highlighted by the differential mutational profile.9
The traditional view has been that CLL is a disease deriving from an inherent defect in apoptosis, or programmed cell death in which, slowly proliferating B lymphocytes accumulate because of this diminished cell death. Increased expression of antiapoptotic Bcl-2 protein and blockade of tumoral CLL B cells in a G0-G1 phase support this observation. However, recent studies showed that CLL is a dynamic process which results from cells that proliferate and die, often at appreciable levels.10 These observations have turned the attention toward the occurrence of different subpopulations inside the tumoral clone in which a homeostatic balance exists in patients with stable lymphocyte counts and good clinical course or an imbalance in patients with rising lymphocyte counts and poor outcome. It is clear that most, if not all, proliferative events occur in the tissues where leukemic cells are able to exploit microenvironment interactions to avoid apoptosis and to acquire tumoral growing conditions.10
Activation-induced cytidine deaminase (AID), a B cell–restricted enzyme, is principally induced through the contact of T and B cells via CD40-CD40 ligand (CD40L) interactions and is required for somatic hypermutation (HMS) and class switch recombination (CSR) process.11 The mutational activity of AID identifies this enzyme as the first genome mutator in humans with oncogenic potential.12 Supporting this view, different works report that constitutive AID expression is associated with a loss in the target specificity and with lymphoproliferative disorders.12,13 Interestingly, in CLL disease we have described that AID is expressed in a percentage of patients with CLL with UM VH genes and with active CSR.14 Despite expression of a functional AID as assessed by an active CSR and mutations induced in the preswitch μ region, CLL B cells in these patients did not succeed to achieve the process of somatic hypermutation.14 Although clonal CSR has been described in CLL B cells long ago15,16 and different works have shown that this process occurs principally in patients with UM disease,14,17 the origin and the biologic implications of this subpopulation in the physiopathology of CLL remains elusive.
Inhibition of apoptosis may occur in vivo in pseudofollicles observed in the lymph nodes, and in the cell clusters described in the bone marrow. These pseudofollicles include in close contact with proliferating B cells increased numbers of CD4 T cells expressing CD40L, which is necessary for AID expression. These activated CD4 T cells could be recruited by tumor B cells through the expression of T cell–attracting chemokines such as CCL17 and CCL2218 and/or CCL3 and CCL4.19 Besides this, the CD38 and CD49d proteins appear to be important additional players interacting with nurselike, stromal, and endothelial cells to complete the activation pathway within the proliferative centers.20 Overall, these observations favor the view that certain cellular subsets in CLL could receive survival signals in the specific microenvironments, increasing their proliferative potential and consequently associated with a more aggressive disease.
Because AID expression in CLL is associated with ongoing CSR in patients with UM disease, we investigated the relation of AID expression, CSR process, and microenvironment activation in peripheral blood (PB) of patients with CLL with different clinical profiles. In this work we examined whether the small subset of tumoral cells with ongoing CSR is responsible for AID expression and whether this subpopulation could have an increased activated and proliferative potential related to the progression of the disease. Our results show that high expression of AID is almost exclusively restricted to the subpopulation of tumoral B cells having an active CSR process. This subset expresses higher levels of proliferation and antiapoptotic molecules such as Ki-67, c-myc, and Bcl-2. In addition, present are high levels of CD49d and CCL3/CCL4 chemokines, as well as a decreased expression of cell cycle inhibitor p27−kip1 compared with their quiescent counterpart IgM B cells. Finally, the presence of this subpopulation in patients with UM CLL is closely related to an aggressive course of the disease.
Methods
Patient samples
PB was obtained from 50 patients with a typical diagnosis of B-cell CLL (B-CLL), displaying, respectively an UM profile in 25 and a mutated (MUT) profile in 25 (Table 1). Among the patients with MUT disease with a median follow-up of 36 months (range, 6-240 months), 21 corresponded to stage A and 3 to stage C; 3 required treatment, and 2 among them died. With a median follow-up of 48 months (range, 3-108 months) there were 13 patients in stage A, 8 in stage B, and 4 in stage C in the UM group; 16 required treatment and 5 died. All patients were followed at the Hospital Maciel from Montevideo and provided an informed consent in accordance with the ethical regulations from Uruguay and the Declaration of Helsinki. The study was approved by the Institutional Review Board of Institut Pasteur Montivideo. The diagnosis of B-CLL relied on cytologic features of mature lymphocytes and a characteristic phenotype (CD5+, CD23+, low expression of CD79b, and of surface immunoglobulin).
Phenotypic and functional studies of B cells
Blood collection was carried out in a period close to the diagnostic time for each patient as indicated in Table 1. The peripheral blood mononuclear cells (PBMCs) were isolated by centrifugation on Ficoll-Hypaque (Pharmacia Fine Chemicals) and immediately cryopreserved in liquid nitrogen. Phenotypic analysis of leukemic cells was performed with anti-CD19 phycoerythrin (PE), anti-CD5 fluorescein-isothiocyanate (FITC), anti–human μ chains F(ab′)2 conjugated with PE, and anti–human γ chains F(ab′)2 conjugated with FITC. Negative controls were performed by incubating the cells with irrelevant F-(ab′)2 antibodies conjugated to PE or FITC. Forward and side scatters were used to gate out contaminating debris and the cells killed during the staining procedure. All antibodies were from Dako SA. Data were acquired, and analysis was performed with the use of a CyAn Flow Cytometer (Beckman Coulter).
Analysis of progression and proliferation markers by flow cytometry
PBMCs (1 × 106) obtained from patients with CLL were incubated for 30 minutes at 4°C with the antibody in phosphate-buffered saline buffer supplemented with 0.5% bovine serum albumin, washed twice, and analyzed by flow cytometry. Anti-CD49d conjugated with allophycocyanin antibody was obtained from BioLegend, and anti-CD38 conjugated with PE–Texas Red was obtained from Invitrogen. For intracellular detection of Bcl-2 and Ki-67 proteins, 1 × 106 PBMCs were fixed in phosphate-buffered saline 4% paraformaldehyde and permeabilized in the same buffer containing 0.5% saponin and 5% fetal bovine serum before the addition of the specific antibody (10 μg/mL). After 45 minutes of incubation at 4°C, with anti–human Ki-67 Alexa Fluor 647 or anti–human Bcl-2 Alexa Fluor 647 the cells were washed twice and analyzed by flow cytometry. Negative isotype controls were performed by incubating the cells with irrelevant antibody in the same experimental conditions. Anti–Ki-67, anti–Bcl-2, and negative isotype control conjugated to Alexa Fluor 647 were obtained from Santa Cruz Biotechnology Inc. In all cases, expression of these CLL progression markers (CD49d and CD38) as well the proliferation and antiapoptotic markers (Ki-67 and Bcl-2) were analyzed in the different CLL B subsets (IgM+, IgG+, and IgM+/IgG+) with the use of Summit v4.3 from Dako.
Cell-sorting studies
Sorting experiments of B-CLL cells were performed with the use of the MoFlo cell sorter (Beckman Coulter) with the same antibodies described earlier to isolate the following 3 different populations of CLL B cells: (1) IgM+ subset expressing CD19+, CD5+, and IgM surface markers; (2) IgG+ subset expressing CD19+, CD5+, and IgG membrane proteins; and (3) IgM+/IgG+ subset corresponding to cells CD19+, CD5+ markers and expressing simultaneously IgM and IgG. In all cases, purity of isolated subpopulations was shown to be greater than 98% before flow cytometric evaluation.
Extraction and analysis of RNA transcripts by reverse transcription–PCR
RNA from total CLL B cells was isolated from 1 to 5 × 106 cells, and cDNA synthesis was performed as described.21 When RNA extraction was performed from isolated IgM+, IgG+, and IgM+/IgG+ subpopulations before cell sorter assays, the mirVana isolation kit (Applied Biosystems), rRNAsin, RNase inhibitor (Promega), and Superscript II reverse transcriptase (Invitrogen) were used to achieve a maximal performance in the mRNA extraction. Despite this care, the mRNA from the IgM+/IgG+ subset was only successful performed in CLL 01, whereas this could not be successfully achieved in the other CLL cases, given the low amounts of cells showing the double marking. Amplifications of circular transcripts (CTs), AID, c-myc, p27−Kip1, and CCL3 and CCL4 chemokines were performed with an initial denaturation step at 95°C for 4 minutes, followed by 30 cycles of 1 minute at 95°C, 1 minute at 62°C, and 1 minute at 72°C and final elongation step at 72°C for 5 minutes. For gene expression analyses the used primers were as follows: for CTs (forward, 5′-GGC CCT TCC AGA TCT TTG AG-3′, and reverse, 5′-CTC TCA GGA CTG ATG GGA AGC CCC G-3′), for AID (forward, 5′-GAG GCA AGA AGA CAC TCT GG-3′, and reverse, 5′-CTA CTT CTG TGA GGA CCG C-3′), for c-myc (forward, 5′-CTT TGT GTG CCC CGC TCC AG-3′, and reverse, 5′-GCG CTC AGA TCC TGC AGG TA-3′), for p27−Kip1 (forward, 5′-AGG TGC GAG TGT CTA ACG GG-3′, and reverse, 5′-GCG CAT TGC TCC GCT AAC CC-3′), for CCL3 (forward, 5′-GAC ACT CGA GCC CAC ATT CCG-3′, and reverse, 5′-CCCCTCAGGCACTCAGCTCC-3′), and for CCL4 (forward, 5′-CCA CCA ATA CCA TGA AGC TCT G-3′, and reverse, 5′-CCT AAT ACA ATA ACA CGG CAC ATA A-3′). In the case of CTs the fragments obtained in polymerase chain reaction (PCR) amplification were transferred and hybridized with specific probes labeled for Cμ and glyceraldehyde phosphate dehydrogenase (GAPDH) α-[32P] deoxycytidine-5′-triphosphate to increase the signal response.
Quantitative real-time PCR
For gene expression analyses of AID, we used Corbette Rotor Gene 6000 Real-Time PCR and the SYBR Green I dye. Primers used in this study were the same that for the reverse transcription (RT)–PCR analysis. Total RNA (1 μg) was isolated from 5 × 106 B cells and retro-transcribed as described.21 One microliter from a 20-μL cDNA reaction was used for AID and GAPDH amplification in a PCR reaction including 40 cycles of amplification (95°C for 20 seconds, 60°C for 30 seconds, 72°C for 30 seconds). Positive calibrator values were obtained from tonsil samples and from CD40L and interleukin-4 (IL-4)–activated CLL B cells. For this, kinetic studies of AID expression were made to evaluate the higher expression of mRNA before activation. Stimulation of CLL B cells was carried out in vitro by monolayer culturing of 1 × 106/mL fibroblast expressing recombinant soluble CD40L and IL-4 (1000 U/mL; PharMingen).
Statistical analyses
Expression of AID mRNA and Ki-67, Bcl-2, and CD49d proteins were compared between IgM+, IgM+/IgG+, and IgG+ subsets with the Mann-Whitney test. On finding significant differences across groups, a Bonferroni-like adjusted pairwise comparison was made to determine which groups differed from each another. The Spearman rank correlation coefficient was calculated to determine the strength of association between AID expression, percentage of clonal-related CSR, and progression-free survival (PFS). Variables with P values less than .05 were considered to be significant. Overall survival (OS) and PFS were calculated from Table 1 and used to perform the Kaplan-Meier method. All analyses were done with GraphPad Prism, Version 4.0 (GraphPad Software Inc).
Results
Analysis of AID expression in CLL B cells
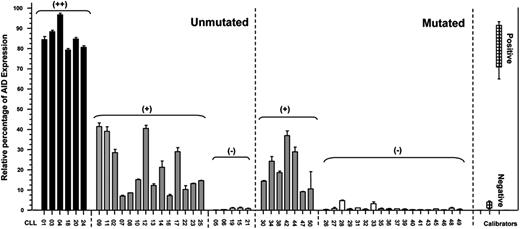
To obtain a positive calibrator for the quantitative PCR we compared levels of AID expression between tonsil samples, Daudi cell line, and B-CLL stimulated through CD40L and IL-4. Taking into account results from Guikema et al22 and our observations, we assumed that mRNA AID expression levels after CD40L/IL-4 activation are comparable to those of B cells in an activated microenvironment. In addition, values from PBMCs from 8 healthy donors were used as negative controls. In this context, we considered as having high AID expression (AID+/+) those patients with CLL whose levels ranged between 80% and 100% of relative units of AID/GAPDH mRNA transcripts (Figure 1). Patients with an AID expression ranging between 40% and 80% and less than 40% of the positive control were considered as having low AID expression (AID+) or negative (AIDneg), respectively (Figure 1). According to these criteria, we analyzed the mRNA AID expression in 50 patients with CLL (UM = 25; MUT = 25). Results show that 6 of the 25 patients with UM CLL corresponded to group AID+/+, 14 to AID+, and 5 to AIDneg. Interestingly, no AID+/+ expression could be detected among patients with a MUT profile, 7 patients displayed lower expression of AID (AID+), whereas no AID transcripts were found in the remaining 18 MUT CLLs. In summary, in our series, 80% of patients with UM disease express AID transcripts, whereas in MUT CLLs this only occurs in 28%. These results are close to those previously reported by Heintel et al.23
AID mRNA expression levels in patients with mutated and unmutated CLL. Quantitative PCR for AID and GAPDH as the endogenous control was performed on PBMCs from patients with MUT and UM CLL. CLL B cells stimulated with CD40L/IL-4, tonsil samples, and PBMCs from 8 healthy donors were used as positive and negative calibrators, respectively. By subtraction of the mean threshold cycle (Ct) triplicate AID measurements with the mean Ct from triplicate GAPDH measurements, the mean ΔCt was calculated. The ΔΔCt values were calculated with the mean ΔCt of the 8 healthy donors and the 3 independently experiments of CLL B cells activated with CD40L/IL-4 as calibrators. AID negativity was defined by the absence of AID expression in duplicate analysis. The expression factor difference and range were calculated by the following formulas: 2−ΔΔCt (mean factor difference); 2−(ΔΔCt − ΔCt SD) and 2−(ΔΔCt + ΔCt SD) (error bars indicate range factor difference). The factor difference conversion of the ΔΔCt is depicted in the graph in relative percentages of AID expression.
AID mRNA expression levels in patients with mutated and unmutated CLL. Quantitative PCR for AID and GAPDH as the endogenous control was performed on PBMCs from patients with MUT and UM CLL. CLL B cells stimulated with CD40L/IL-4, tonsil samples, and PBMCs from 8 healthy donors were used as positive and negative calibrators, respectively. By subtraction of the mean threshold cycle (Ct) triplicate AID measurements with the mean Ct from triplicate GAPDH measurements, the mean ΔCt was calculated. The ΔΔCt values were calculated with the mean ΔCt of the 8 healthy donors and the 3 independently experiments of CLL B cells activated with CD40L/IL-4 as calibrators. AID negativity was defined by the absence of AID expression in duplicate analysis. The expression factor difference and range were calculated by the following formulas: 2−ΔΔCt (mean factor difference); 2−(ΔΔCt − ΔCt SD) and 2−(ΔΔCt + ΔCt SD) (error bars indicate range factor difference). The factor difference conversion of the ΔΔCt is depicted in the graph in relative percentages of AID expression.
Analysis of CSR fraction cells in CLL
The in vivo plasticity of CLL is further underscored by reports showing that IgM+ leukemic cells can give rise to clonally related IgG+ or IgA+ elements, possibly by ongoing in vivo CSR process in the proliferating centers.21,24 However, the implication of this activation and of this tumoral subpopulation in the physiopathology of CLL remains elusive as yet.
To better characterize the tumoral subset with ongoing CSR we studied CLL B cells with specific human antibodies (anti-μ, anti-γ, anti-CD5, and anti-CD19) from 50 patients and analyzed the clonal identity for cells with ongoing CSR. Results show the presence of IgG+ and IgG+/IgM+ subpopulations ranged from 0.5% to 8% in 44 of the 50 patents with CLL by cytometric assays (Table 1). In addition, we performed RT-PCR assays with specific familial VH genes and Cγ primers and sequenced the obtained products to evaluate the correspondence with the tumoral clone. Amplifications were obtained for 36 of the 44 patients with CLL (72% of the 50 patients with CLL). However, an identical tumoral VH gene sequence was found in only 16 of these 36 CLLs, representing 32% of the total population (6 from the UM AID+/+ CLL B cells, 7 from UM AID+ CLL B cells, and 3 from MUT AID+ CLL B cells; Table 1). Thus, CSR related to the tumoral clone ranged between 2.5% and 8.5% of the tumoral population among patients with UM disease. In contrast, unrelated tumoral CSR never exceeded 2% and is associated with low or no expression of AID mRNA (Table 1).
Clinical and molecular characterization of patients with CLL
| CLL no. . | Progression status . | Binet stage . | Lymphocyte count, ×103/μL . | CD38* . | LPL . | Diagnostic and blood collection . | TFT, mo . | A/D† . | AID (Q-PCR) . | Percentage of CSR‡ . | VDJ clonal identity . | Mutational status§ . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1¶ | Progressor | C | 128.0 | + | + | 2000 | 9 | D | +/+ | 8.2 | Yes | UM |
| 2‖ | Progressor | B | 80.0 | + | + | 1999 | 18 | D | + | 4.1 | Yes | UM |
| 3¶ | Progressor | C | 31.00 | + | + | 1998 | 6 | D | +/+ | 5.4 | Yes | UM |
| 4¶ | Progressor | B | 105.00 | + | + | 2000 | 8 | D | +/+ | 6.4 | Yes | UM |
| 5** | Indolent | A | 20.00 | + | + | 1999 | N/T | A | − | 1.3 | N/A | UM |
| 6** | Progressor | B | 32.50 | − | + | 2001 | N/T | A | − | 0.3 | N/A | UM |
| 7‖ | Indolent | B | 17.00 | + | − | 2000 | N/T | A | + | 2.7 | Yes | UM |
| 8‖ | Progressor | A | 70.00 | − | − | 2002 | 42 | A | + | 1.6 | N/A | UM |
| 9‖ | Progressor | B | 60.00 | + | + | 2005 | 12 | A | + | 4 | Yes | UM |
| 10‖ | Indolent | A | 10.00 | + | + | 1999 | N/T | A | + | 0.7 | N/A | UM |
| 11‖ | Progressor | C | 88.00 | + | + | 2001 | 15 | A | + | 3.8 | Yes | UM |
| 12‖ | Progressor | B | 54.00 | + | + | 2001 | 6 | A | + | 4.5 | Yes | UM |
| 13‖ | Indolent | A | 20.00 | − | + | 2005 | 54 | A | + | 1.3 | No | UM |
| 14‖ | Indolent | A | 7.00 | + | + | 2006 | 36 | A | + | 3.3 | Yes | UM |
| 15** | Indolent | A | 20.40 | + | + | 2004 | N/T | A | − | 0.3 | No | UM |
| 16‖ | Progressor | B | 148.00 | + | + | 2005 | 3 | A | + | 1.4 | No | UM |
| 17‖ | Progressor | A | 54.50 | − | + | 2007 | 17 | A | + | 2.5 | No | UM |
| 18†† | Indolent | A | 15.00 | + | + | 2006 | 12 | A | +/+ | 5.9 | Yes | UM |
| 19** | Indolent | A | 11.00 | + | + | 2005 | N/T | A | − | 0.6 | No | UM |
| 20†† | Progressor | C | 220.00 | + | + | 2007 | 3 | D | +/+ | 7.4 | Yes | UM |
| 21** | Indolent | A | 8.90 | + | − | 2008 | N/T | A | − | 1.1 | No | UM |
| 22‖ | Progressor | B | 17.80 | + | + | 2008 | 11 | A | + | 3.5 | Yes | UM |
| 23‖ | Indolent | A | 28.00 | − | + | 2008 | N/T | A | + | 4.1 | No | UM |
| 24†† | Progressor | A | 80.00 | + | + | 2009 | 10 | A | +/+ | 5.5 | Yes | UM |
| 25‖ | Progressor | A | 10.50 | + | + | 2005 | N/T | A | + | 1 | N/A | UM |
| 26** | Progressor | A | 17.20 | − | − | 1999 | N/T | A | − | 0.9 | N/A | Mut |
| 27** | Progressor | A | 14.00 | − | − | 2000 | N/T | A | − | 0.4 | N/A | Mut |
| 28** | Progressor | A | 5.00 | + | − | 1989 | N/T | A | − | 0.6 | No | Mut |
| 29** | Progressor | A | 11.60 | + | − | 2001 | N/T | A | − | 0.1 | N/A | Mut |
| 30‖ | Progressor | A | 18.00 | − | − | 1999 | N/T | A | + | 1.8 | Yes | Mut |
| 31** | Progressor | B | 38.00 | + | − | 2000 | N/T | D | − | 0.5 | N/A | Mut |
| 32** | Indolent | A | 47.60 | − | − | 2002 | N/T | A | − | 2.5 | No | Mut |
| 33** | Indolent | A | 15.40 | − | − | 2006 | N/T | A | − | 0.8 | No | Mut |
| 34‖ | Indolent | A | 15.00 | − | + | 2007 | N/T | A | + | 0.5 | N/A | Mut |
| 35** | Indolent | A | 10.20 | − | + | 2006 | 25 | A | − | 1.2 | No | Mut |
| 36** | Indolent | A | 27.60 | − | + | 2007 | N/T | A | − | 0.4 | N/A | Mut |
| 37** | Indolent | A | 15.00 | − | − | 2006 | N/T | A | − | 0.3 | No | Mut |
| 38‖ | Indolent | A | 12.80 | − | + | 2004 | N/T | A | + | 2.1 | No | Mut |
| 39** | Indolent | A | 15.60 | − | + | 2006 | N/T | A | − | 0.5 | No | Mut |
| 40** | Indolent | A | 12.00 | − | + | 1997 | N/T | A | − | 0.1 | N/A | Mut |
| 41** | Progressor | C | 18.00 | − | + | 2007 | 6 | D | − | 1.0 | N/A | Mut |
| 42‖ | Indolent | A | 3.40 | − | − | 2008 | N/T | A | + | 1.7 | Yes | Mut |
| 43** | Indolent | A | 28.40 | − | + | 2007 | N/T | A | − | 2.0 | No | Mut |
| 44‖ | Indolent | A | 8.00 | − | − | 2006 | N/T | A | + | 2.2 | No | Mut |
| 45** | Progressor | B | 105.00 | − | + | 2007 | 12 | A | − | 2.0 | No | Mut |
| 46** | Indolent | A | 2.47 | − | − | 2008 | N/T | A | − | 0.1 | N/A | Mut |
| 47‖ | Indolent | C | 5.00 | − | − | 2009 | N/T | A | + | 1.8 | No | Mut |
| 48** | Indolent | A | 12.60 | − | − | 2008 | N/T | A | − | 2.1 | No | Mut |
| 49** | Indolent | A | 5.80 | − | − | 2008 | N/T | A | − | 1.1 | N/A | Mut |
| 50‖ | Indolent | A | 11.50 | − | − | 2008 | N/T | A | + | 2.1 | Yes | Mut |
| CLL no. . | Progression status . | Binet stage . | Lymphocyte count, ×103/μL . | CD38* . | LPL . | Diagnostic and blood collection . | TFT, mo . | A/D† . | AID (Q-PCR) . | Percentage of CSR‡ . | VDJ clonal identity . | Mutational status§ . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1¶ | Progressor | C | 128.0 | + | + | 2000 | 9 | D | +/+ | 8.2 | Yes | UM |
| 2‖ | Progressor | B | 80.0 | + | + | 1999 | 18 | D | + | 4.1 | Yes | UM |
| 3¶ | Progressor | C | 31.00 | + | + | 1998 | 6 | D | +/+ | 5.4 | Yes | UM |
| 4¶ | Progressor | B | 105.00 | + | + | 2000 | 8 | D | +/+ | 6.4 | Yes | UM |
| 5** | Indolent | A | 20.00 | + | + | 1999 | N/T | A | − | 1.3 | N/A | UM |
| 6** | Progressor | B | 32.50 | − | + | 2001 | N/T | A | − | 0.3 | N/A | UM |
| 7‖ | Indolent | B | 17.00 | + | − | 2000 | N/T | A | + | 2.7 | Yes | UM |
| 8‖ | Progressor | A | 70.00 | − | − | 2002 | 42 | A | + | 1.6 | N/A | UM |
| 9‖ | Progressor | B | 60.00 | + | + | 2005 | 12 | A | + | 4 | Yes | UM |
| 10‖ | Indolent | A | 10.00 | + | + | 1999 | N/T | A | + | 0.7 | N/A | UM |
| 11‖ | Progressor | C | 88.00 | + | + | 2001 | 15 | A | + | 3.8 | Yes | UM |
| 12‖ | Progressor | B | 54.00 | + | + | 2001 | 6 | A | + | 4.5 | Yes | UM |
| 13‖ | Indolent | A | 20.00 | − | + | 2005 | 54 | A | + | 1.3 | No | UM |
| 14‖ | Indolent | A | 7.00 | + | + | 2006 | 36 | A | + | 3.3 | Yes | UM |
| 15** | Indolent | A | 20.40 | + | + | 2004 | N/T | A | − | 0.3 | No | UM |
| 16‖ | Progressor | B | 148.00 | + | + | 2005 | 3 | A | + | 1.4 | No | UM |
| 17‖ | Progressor | A | 54.50 | − | + | 2007 | 17 | A | + | 2.5 | No | UM |
| 18†† | Indolent | A | 15.00 | + | + | 2006 | 12 | A | +/+ | 5.9 | Yes | UM |
| 19** | Indolent | A | 11.00 | + | + | 2005 | N/T | A | − | 0.6 | No | UM |
| 20†† | Progressor | C | 220.00 | + | + | 2007 | 3 | D | +/+ | 7.4 | Yes | UM |
| 21** | Indolent | A | 8.90 | + | − | 2008 | N/T | A | − | 1.1 | No | UM |
| 22‖ | Progressor | B | 17.80 | + | + | 2008 | 11 | A | + | 3.5 | Yes | UM |
| 23‖ | Indolent | A | 28.00 | − | + | 2008 | N/T | A | + | 4.1 | No | UM |
| 24†† | Progressor | A | 80.00 | + | + | 2009 | 10 | A | +/+ | 5.5 | Yes | UM |
| 25‖ | Progressor | A | 10.50 | + | + | 2005 | N/T | A | + | 1 | N/A | UM |
| 26** | Progressor | A | 17.20 | − | − | 1999 | N/T | A | − | 0.9 | N/A | Mut |
| 27** | Progressor | A | 14.00 | − | − | 2000 | N/T | A | − | 0.4 | N/A | Mut |
| 28** | Progressor | A | 5.00 | + | − | 1989 | N/T | A | − | 0.6 | No | Mut |
| 29** | Progressor | A | 11.60 | + | − | 2001 | N/T | A | − | 0.1 | N/A | Mut |
| 30‖ | Progressor | A | 18.00 | − | − | 1999 | N/T | A | + | 1.8 | Yes | Mut |
| 31** | Progressor | B | 38.00 | + | − | 2000 | N/T | D | − | 0.5 | N/A | Mut |
| 32** | Indolent | A | 47.60 | − | − | 2002 | N/T | A | − | 2.5 | No | Mut |
| 33** | Indolent | A | 15.40 | − | − | 2006 | N/T | A | − | 0.8 | No | Mut |
| 34‖ | Indolent | A | 15.00 | − | + | 2007 | N/T | A | + | 0.5 | N/A | Mut |
| 35** | Indolent | A | 10.20 | − | + | 2006 | 25 | A | − | 1.2 | No | Mut |
| 36** | Indolent | A | 27.60 | − | + | 2007 | N/T | A | − | 0.4 | N/A | Mut |
| 37** | Indolent | A | 15.00 | − | − | 2006 | N/T | A | − | 0.3 | No | Mut |
| 38‖ | Indolent | A | 12.80 | − | + | 2004 | N/T | A | + | 2.1 | No | Mut |
| 39** | Indolent | A | 15.60 | − | + | 2006 | N/T | A | − | 0.5 | No | Mut |
| 40** | Indolent | A | 12.00 | − | + | 1997 | N/T | A | − | 0.1 | N/A | Mut |
| 41** | Progressor | C | 18.00 | − | + | 2007 | 6 | D | − | 1.0 | N/A | Mut |
| 42‖ | Indolent | A | 3.40 | − | − | 2008 | N/T | A | + | 1.7 | Yes | Mut |
| 43** | Indolent | A | 28.40 | − | + | 2007 | N/T | A | − | 2.0 | No | Mut |
| 44‖ | Indolent | A | 8.00 | − | − | 2006 | N/T | A | + | 2.2 | No | Mut |
| 45** | Progressor | B | 105.00 | − | + | 2007 | 12 | A | − | 2.0 | No | Mut |
| 46** | Indolent | A | 2.47 | − | − | 2008 | N/T | A | − | 0.1 | N/A | Mut |
| 47‖ | Indolent | C | 5.00 | − | − | 2009 | N/T | A | + | 1.8 | No | Mut |
| 48** | Indolent | A | 12.60 | − | − | 2008 | N/T | A | − | 2.1 | No | Mut |
| 49** | Indolent | A | 5.80 | − | − | 2008 | N/T | A | − | 1.1 | N/A | Mut |
| 50‖ | Indolent | A | 11.50 | − | − | 2008 | N/T | A | + | 2.1 | Yes | Mut |
LPL indicates lipoprotein lipase; TFT, time from initial diagnosis to first treatment for clinical progression; A/D, alive/dead; CSR, class switch recombination; UM, unmutated; Mut, mutated; N/T, no treatment; and N/A, no amplification.
Obtained results in cytometric assays, using a CD38 cutoff of 30%.
Related to decease.
Percentage of IgG+ and IgM+/IgG+ CLL B cells by cytometric studies.
Less than or equal to 2% difference from germline gene defined patients with UM disease, ≥ 2% difference defines patients with MUT.
High AID expression and CSR ≥ 5%.
Low AID expression and CSR ≤ 5%.
No AID expression and CSR ≤ 2.5%.
AID expression is mainly restricted to the subset of tumoral cells displaying an ongoing CSR
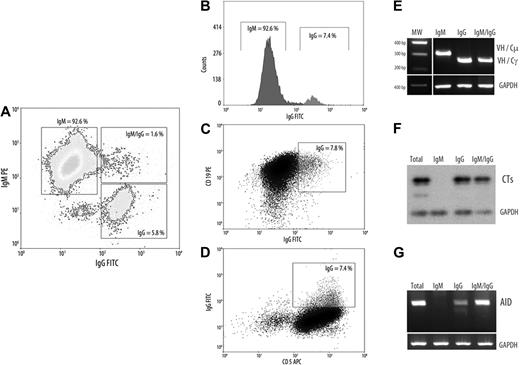
As reported by Cerutti et al24 B cells with ongoing CSR are absent in normal PB and segregates within founder germinal center (GC) and GC B cells. By contrast, ongoing CSR has been described in the PB of patients with UM disease expressing AID constitutively.14 Because AID expression in CLL B cells is confined to a small subset25 and results from signals received through interaction with microenvironment and CD4 T cells, we examined whether its expression is restricted to tumoral cells with ongoing CSR. To substantiate this possibility, we isolated by cell sorting cytometry the subsets expressing the different immunoglobulin isotypes within the tumoral clone in the 3 AID+/+ UM CLLs. Tumoral B cells expressing clonal membrane IgG (IgG+) or intracellular IgM and IgG (B cells in transition state, IgM+/IgG+) and the typical surface IgM (IgM+) were isolated with a greater than 98% purity as assessed by cytometric assays. A representative patient with CLL is shown in Figure 2A. Afterward, the mRNA was extracted, and RT-PCR was performed to determine CSR transcripts (Figure 2E), CTs, and AID expression in these different subsets. Because CTs are rapidly degraded by nucleases, it constitutes the specific molecular markers of ongoing CSR in B cells. Results from semiquantitative RT-PCR indicate that both subpopulations IgG+ and IgG+/IgM+ express CTs, confirming that ongoing CSR occurs in both subpopulations (Figure 2F). Finally, we found that AID expression is mainly restricted to the subpopulations IgG+ and IgG+/IgM+ (Figure 2G).
Characterization of CLL B cells with ongoing IgG CSR. (A-D) Representative flow cytometric profile from a patient with UM AID++ CLL. The 3 CLL B-cell subsets as well as the surface expression of CD5 and CD19 markers are depicted (IgM+, IgM+/IgG+, and IgG+). (E-G) Semiquantitative RT-PCR from the 3 cell sorter–isolated subpopulations. (E) Clonal isotype switch transcripts with tumor-related VH and Cμ or Cγ primers. (F) Amplification of CTs and subsequently hybridization with Cμ probe encompassing 1-180 nt of the first Cμ exon. (D) AID amplification from isolated CLL subsets. For all PCR analysis putative B-cell contamination between isolated subsets, moreover to cytometric purity analysis, was estimated by amplifying tumor-related VH-Cμ inside the IgG+ subset and with VH-Cγ inside the IgM+ subset. GAPDH was amplified in all cases as internal control.
Characterization of CLL B cells with ongoing IgG CSR. (A-D) Representative flow cytometric profile from a patient with UM AID++ CLL. The 3 CLL B-cell subsets as well as the surface expression of CD5 and CD19 markers are depicted (IgM+, IgM+/IgG+, and IgG+). (E-G) Semiquantitative RT-PCR from the 3 cell sorter–isolated subpopulations. (E) Clonal isotype switch transcripts with tumor-related VH and Cμ or Cγ primers. (F) Amplification of CTs and subsequently hybridization with Cμ probe encompassing 1-180 nt of the first Cμ exon. (D) AID amplification from isolated CLL subsets. For all PCR analysis putative B-cell contamination between isolated subsets, moreover to cytometric purity analysis, was estimated by amplifying tumor-related VH-Cμ inside the IgG+ subset and with VH-Cγ inside the IgM+ subset. GAPDH was amplified in all cases as internal control.
The subpopulation with high levels of AID and ongoing CSR displays an increased proliferative potential
Because AID is up-regulated after antigen activation and is the principal enzyme implicated in CSR, which occurs in GCs, we determined the relationship between AID expression with different proliferation, antiapoptotic, and activation markers such as Ki-67, Bcl-2, and c-myc, respectively, in the 3 tumoral subpopulations (IgM+, IgG+, and IgM+/IgG+).
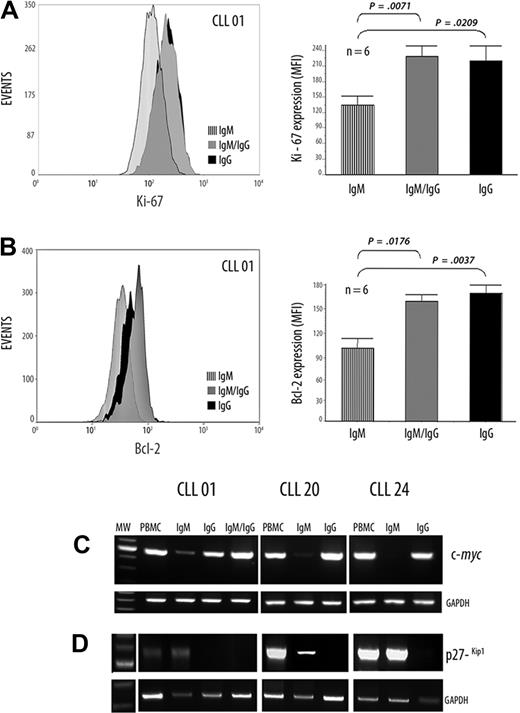
The cytometric assays from one representative patient for Ki-67 expression are depicted in Figure 3A. In addition, in the 6 patients studied, results show significantly increased expression of Ki-67 in the IgG+ and IgM+/IgG+ subsets compared with the IgM+ subpopulation (comparing IgM+ [mean = 92.25] with IgM+/IgG+ [mean = 150.8]; P < .01, Mann-Whitney test, and comparing IgM+ [mean = 92.25] with IgG+ [mean = 145.25]; P ≤ .01, Mann-Whitney test; Figure 3A).
Differential expression of Ki-67, Bcl-2, c-myc, and p27−kip1 in CLL B cells with ongoing IgG CSR and in their IgM counterpart. (A-B) Flow cytometric analysis, in a representative patient with UM CLL, showing the Ki-67 and Bcl-2 protein expressions, respectively. Values are mean ± SE of mean fluorescence intensity (MFI) of, respectively, Ki-67 and Bcl-2 expression from 6 patients with UM CLL with high AID expression levels. (C-D) Semiquantitative RT-PCR from the cell sorter–isolated subpopulations with c-myc–specific primers (C) and with p27−kip1-specific primers (D) are depicted. GAPDH was amplified in all cases as internal semiquantitative control.
Differential expression of Ki-67, Bcl-2, c-myc, and p27−kip1 in CLL B cells with ongoing IgG CSR and in their IgM counterpart. (A-B) Flow cytometric analysis, in a representative patient with UM CLL, showing the Ki-67 and Bcl-2 protein expressions, respectively. Values are mean ± SE of mean fluorescence intensity (MFI) of, respectively, Ki-67 and Bcl-2 expression from 6 patients with UM CLL with high AID expression levels. (C-D) Semiquantitative RT-PCR from the cell sorter–isolated subpopulations with c-myc–specific primers (C) and with p27−kip1-specific primers (D) are depicted. GAPDH was amplified in all cases as internal semiquantitative control.
Expression of Bcl-2 protein is shown in Figure 3B in the same representative patient among the 6 studied. Significantly higher expressions were found comparing IgM+ (mean = 31.3) with IgM+/IgG+ (mean = 47.62) (P ≤ .01, Mann-Whitney test). A more significant difference was found in the case of IgM+ with IgG+ subpopulations (IgM+: mean = 31.3; IgG+: mean = 49.75; P < .01, Mann-Whitney test), suggesting a more increased antiapoptotic profile for both IgG+ and IgM+/IgG+ than in the typically CLL IgM B cell (Figure 3B).
Because c-myc protein was observed in the highest amount at the proliferative B-lymphoblast stage and undetectable in plasma cells,26 we evaluated at the RNA level c-myc expression in the 3 subpopulations. Results show higher expression of c-myc RNA in the IgG+ and IgM+/IgG+ subsets compared with the IgM+ tumoral counterpart (Figure 3C).
The cyclin-dependent kinase inhibitor p27−kip1 promotes exit to cell cycle and is overexpressed in CLL B cells.27 Therefore, we examined its expression at the mRNA level in the 3 isolated subpopulations. Semiquantitative RT-PCR results show a high expression of p27−Kip1 in the IgM+ isolated subset; whereas no, or very weak, expression was found in both IgG+ and IgM+/IgG+ isolated subpopulations (Figure 3D). Taken together, these results outline the idea that IgG+ and IgM+/IgG+ subsets exhibit an activated phenotypic profile containing CLL B cells with increased proliferative and antiapoptotic potential.
AID+/+ subpopulation with ongoing CSR cells displays expression molecules resulting from proliferation center contact
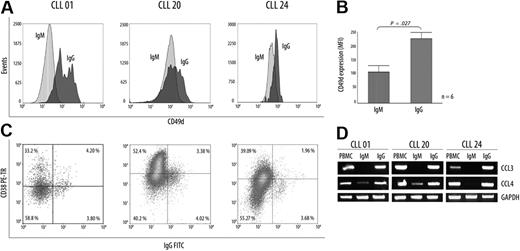
Expression of CD49d/α4-integrin has been associated with high levels of CD38 glycoprotein and expression of CCL3/CCL4 chemokines in progressive disease.20 Because all these molecules appear to be important players in the survival of CLL B cells, we compared in the IgM+ and the IgG+ subpopulations the levels of protein expression for CD49d and CD38 markers. Results show that CLL B-cell IgG+ express increased membrane CD49d levels compared with its IgM+ counterpart (IgM+: mean = 115.8; IgG+: mean = 239.2; P < .01, Mann-Whitney test; Figure 4A). Interestingly, when the IgG+ subset was studied for CD38 expression in the 3 representative patients, only a fraction (< 50%) of this subpopulation was competent to express CD38 (Figure 4C). Finally, we evaluated CCL3 and CCL4 chemokine mRNA expression profiles in both subpopulations in the same 3 patients. Results show that transcripts for CCL3 are exclusively observed within the IgG+ subset, whereas higher levels of CCL4 transcripts were observed in this same subpopulation compared with its IgM+ counterpart. Overall, these results favor the view that the tumoral subset with ongoing CSR and high AID expression constitutes an activated subset expressing molecules associated with the progressive disease and that may result from a recent contact with proliferative centers.
Differential expression of CD49d, CCL3, and CCL4 chemokines in CLL B cells with ongoing IgG CSR and in their IgM counterpart. (A) Flow cytometric analysis from CD49d protein expression in 3 representative patients. Although increased levels of CD49d are found in the UM CLLs, expression values for this protein were always higher in the IgG+ subset that in their counterpart IgM+ subset. (B) Values are mean (± SE) for MFI of CD49d expression in the 6 patients with CLL with high AID expression levels. (C) Flow cytometric profiles of CD38 expression inside the IgG subset from 3 representative patients with CLL. Numbers in quadrants indicate percentages of cells. (D) CCL3 and CCL4 mRNA profile expressions in the different isolated subsets (IgM+ and IgG+) from the 3 representative patients with UM CLL.
Differential expression of CD49d, CCL3, and CCL4 chemokines in CLL B cells with ongoing IgG CSR and in their IgM counterpart. (A) Flow cytometric analysis from CD49d protein expression in 3 representative patients. Although increased levels of CD49d are found in the UM CLLs, expression values for this protein were always higher in the IgG+ subset that in their counterpart IgM+ subset. (B) Values are mean (± SE) for MFI of CD49d expression in the 6 patients with CLL with high AID expression levels. (C) Flow cytometric profiles of CD38 expression inside the IgG subset from 3 representative patients with CLL. Numbers in quadrants indicate percentages of cells. (D) CCL3 and CCL4 mRNA profile expressions in the different isolated subsets (IgM+ and IgG+) from the 3 representative patients with UM CLL.
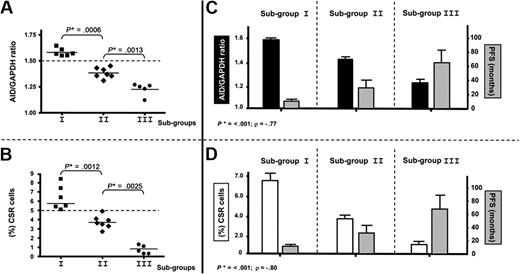
High expression of AID and high percentage of clonal CSR in PBMCs delineate a subgroup with poor prognosis among patients with UM CLL
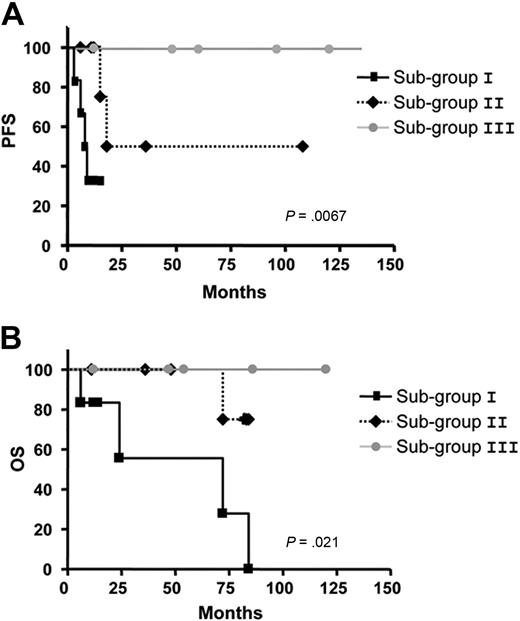
Significant differences concerning AID expression in the 3 subgroups (AID+/+: mean = 1.58; AID+: mean = 1.40; AIDneg: mean = 1.22) were observed in patients with UM CLL after quantitative PCR analysis (P < .001, Mann-Whitney test; Figure 5A). As for the clonally related CSR process, the same 3 subgroups were also identified displaying significant differences between high clonally related CSR (mean = 6.36), low clonally related CSR (mean = 3.75), and not clonally related CSR (mean 0.72,) processes (P < .001, Mann-Whitney test; Figure 5B). Because our results suggest that CLL B cells with high expression of AID and high percentage of ongoing CSR correspond to an activated and proliferative subpopulation (see Figures 3-4), we speculated whether this subset could be associated with the disease progression. Thus, we segregated patients with UM CLL into 3 subgroups: subgroup I, AID+/+ with higher percentage of clonally related CSR (≥ 5%); subgroup II, AID+ and lower percentage of clonally related CSR; and subgroup III, AIDneg without clonally related CSR. Results show that among the 6 patients from subgroup I, all required treatment at 3, 6, 8, 9, 10, and 12 months. Four of these 6 patients died of causes related to disease at 6, 24, 72, and 84 months after diagnosis. Interestingly, all these patients displayed high values of AID expression and tumoral-related CSR with values higher than 5% (Table 1; Figure 5A-B). Subgroup II contained 7 patients. Six of the 7 patients received treatment at 6, 11, 12, 15, 18, and 36 months, and 1 of them died at 70 months from disease-related causes. In subgroup III, the 5 patients failing to express AID, no tumoral CSR was found, and none of them received treatment and remain alive to date. To link a clinical poor outcome with high AID expression and high clonally related CSR, a Spearman rank correlation test was carried out. A significant negative correlation comparing the 3 subgroups was found between AID expression and PFS (P < .001; Spearman rank coefficient ρ = −0.77), as well as between CSR percentage and PFS (P < .001; Spearman rank coefficient ρ = −0.80; Figure 5C and 5D, respectively). Despite the low number of patients with CLL with clonally related CSR in the UM group, PFS and OS analyses were performed. Results showed significant differences for PFS analysis (P = .007) and low but still significant difference for OS analysis (P = .021; Figure 6A and 6B, respectively).
AID expression levels and high CSR segregate patients with UM CLL into 3 subgroups with different clinical progression. (A-B) Significant differences in P values in mRNA AID expression (A) and percentage of CLL B cells with clonally related CSR (B) allow us to segregate patients with UM disease into 3 different groups: subgroup I (6 patients) identified as AID+/+ and high clonal CSR (≥ 5%), subgroup II (7 patients) identified as AID+ and low clonal CSR, and subgroup III (5 patients) identified as AIDneg without clonal CSR. Significant P values are shown (*) (P ≤ .001, Mann-Whitney test). (C-D) Correlations of AID expression and PFS (C) and clonal CSR and PFS (D) for the 3 subgroups are plotted. Both analyses indicate a significant negative correlation; P ≤ .001 considered significant (*) by Spearman rank test. P < .001 (Spearman rank coefficient ρ = −0.77) for AID expression correlated to PFS and P < .001 (Spearman rank coefficient ρ = −0.80) for clonal CSR linked to PFS.
AID expression levels and high CSR segregate patients with UM CLL into 3 subgroups with different clinical progression. (A-B) Significant differences in P values in mRNA AID expression (A) and percentage of CLL B cells with clonally related CSR (B) allow us to segregate patients with UM disease into 3 different groups: subgroup I (6 patients) identified as AID+/+ and high clonal CSR (≥ 5%), subgroup II (7 patients) identified as AID+ and low clonal CSR, and subgroup III (5 patients) identified as AIDneg without clonal CSR. Significant P values are shown (*) (P ≤ .001, Mann-Whitney test). (C-D) Correlations of AID expression and PFS (C) and clonal CSR and PFS (D) for the 3 subgroups are plotted. Both analyses indicate a significant negative correlation; P ≤ .001 considered significant (*) by Spearman rank test. P < .001 (Spearman rank coefficient ρ = −0.77) for AID expression correlated to PFS and P < .001 (Spearman rank coefficient ρ = −0.80) for clonal CSR linked to PFS.
Kaplan-Meier curves based on AID and CSR expression in the 3 subgroups in patients with UM CLL comparing the PFS and OS. The Kaplan-Meier method was used to construct survival curves for PFS (A) and OS (B), and results were compared with the log-rank test Spearman. P values refer to the log-rank test. Subgroup I corresponds to UM CLL AID+/+ and CSR ≥ 5%; subgroup II to UM CLLs AID+ and low clonally related CSR, and subgroup III to UM CLL AIDneg and CSR not related to the tumoral clone.
Kaplan-Meier curves based on AID and CSR expression in the 3 subgroups in patients with UM CLL comparing the PFS and OS. The Kaplan-Meier method was used to construct survival curves for PFS (A) and OS (B), and results were compared with the log-rank test Spearman. P values refer to the log-rank test. Subgroup I corresponds to UM CLL AID+/+ and CSR ≥ 5%; subgroup II to UM CLLs AID+ and low clonally related CSR, and subgroup III to UM CLL AIDneg and CSR not related to the tumoral clone.
Among the patients with MUT disease, none of the 6 patients expressing low levels of AID required treatment, and all patients are alive with a mean time of follow-up of 36 months. Among the 19 patients with MUT disease failing to express AID transcripts, 3 required treatment at 6, 12, and 25 months, and 1 died of causes unrelated to CLL at 120 months without receiving previous treatment. Overall, these results outline the importance of the subset with ongoing high clonally related CSR and expressing high levels of AID among patients with UM disease and suggest that the analysis of this subpopulation could be important to identify a more aggressive form in CLL disease.
Discussion
Important progress resulting in high levels of clinical and even molecular remissions has been recently achieved in CLL treatment. However, CLL remains an incurable disease. Recently, compelling evidence suggests that crosstalk with accessory cells in specialized tissue microenvironments, such as the bone marrow and secondary lymphoid organs, favors disease progression by promoting malignant B-cell growth and drug resistance. Therefore, understanding the crosstalk between malignant B cells and their milieu could give us new keys into the cellular and molecular biology of CLL that can finally lead to novel strategies in the treatment of this disease. Nevertheless, we need a more proper knowledge about the signals received and/or transmitted by CLL B lymphocytes, interacting with T lymphocytes, and/or with stromal, endothelial, dendritic, and nurselike cells in the particular CLL microenvironment. The simple observation that CLL B cells progressively accumulate in vivo, but undergo apoptosis when cultured in vitro, draws attention to the microenvironment and its ability to deliver signals that may ensure the survival of malignant cells. Further, it is self-evident that the accumulated CLL B cells in the PB are constantly nourished by an upstream proliferation cell compartment. It is reasonable to assume that the balance between the 2 compartments may be at the basis of the highly variable clinical course of CLL, which may behave as a stable and indolent monoclonal lymphocytosis or as an aggressive disease.
In this study we attempted to provide additional evidence related to the importance of the microenvironment signals in the clinical course of CLL. For this, we analyzed a particular tumoral subpopulation displaying ongoing CSR17,28 and high AID expression in the PB of patients with UM CLL. Ongoing CSR process24 and high expression of AID29 are physiologically confined to the GC, a highly specialized and activated microenvironment originated during an immune response. However, in previous work, we demonstrated expression of AID in the PB of patients with UM CLL, which additionally displayed a subpopulation of tumoral cells with ongoing CSR.14 Because a subsequent work has shown that AID expression is restricted to a small proportion of the tumoral CLL clone,30 the patients with UM disease with this subpopulation and high expression of a potential mutator such as AID are interesting to study. Given this, we hypothesized that the subset with ongoing CSR could be responsible for AID expression and might correspond to a CLL in which an activated tumor microenvironment is present, being that this is responsible for a poorer clinical outcome. We speculated that this continuous activation might lead to a highly proliferative disease whose hallmark is the presence of this particular subset in the PB of this UM CLL.
Our results show that almost all mRNA AID expression is restricted to the subpopulation of tumoral cells with ongoing CSR. Furthermore, this subpopulation is clonally related to the tumoral IgM counterpart but expresses higher levels of Ki-67, c-myc, Bcl-2, CD49d, and CCL3/4 chemokines as well as lower levels of the cell-cycle inhibitory protein p27−kip1 and, more importantly, is associated with disease progression. Active CSR process is currently measured by the presence of switch circle or their transcribed chimeric I-Cμ product referred as the CTs. Because they are rapidly degraded by nucleases, they constitute specific molecular markers of ongoing CSR. Our results show the presence of CTs in the tumoral subset IgM+/IgG+, which is a minor pool of CLL B cells in a transitional state of CSR. Interestingly, in the established IgG+ CLL B cells CTs were also identified, suggesting that at least some of them come out of an active CSR process. High expression of AID was observed in the IgM+/IgG+ and IgG+ subpopulations but not in their IgM+ counterpart. These results suggest that the subset with ongoing CSR is responsible for almost all AID expression and that this activation might have recently been triggered, in an activated CLL microenvironment. To clarify the importance of the microenvironment interactions with the different clinical progression of these patients with UM CLL, we asked, which is the stimulus for this constitutive expression and where does it occur? These interesting questions remain to be elucidated.
In this context, we have evaluated the expression of different key molecules implicated in cell-cycle regulation, proliferation, and tumoral progression. Higher expression of Ki-67, c-myc, Bcl-2, as well as the underexpression of the cell-cycle inhibitor p27−kip1, was found among the tumoral subsets with ongoing CSR and high AID expression, compared with their IgM counterpart. The Ki-67 is a nuclear protein up-regulated in the G1, S, G2, and M phases of the cell cycle but absent in resting cells (G0 phase).31 In addition, c-myc oncogene has been proposed to be expressed at a stage of differentiation in B cells. Studies of Larsson et al26 show that CLL B cells could be induced to proliferate and differentiate, after in vitro incubation with 12-O-tetradecanoyl-phorpol-13-acetate. In this case the CLL B cells change from G0 to G1, and this process is accompanied by a dramatic increase in the expression of c-myc.26 In our studies, both subpopulations (IgM+/IgG+ and IgG+) depicted significantly higher levels of Ki-67 and Bcl-2 protein and the c-myc mRNA, suggesting an activated state for these particular subsets of CLL B cells. Because some reports indicated that AID expression could vary during evolution of CLL disease,23,25 we have studied 2 patients (CLL 18 and 20) at 2 time points with a 2-year interval. Results showed no significant changes in AID, CSR, and Ki-67 expression, suggesting that the presence of IgM+/IgG+ and IgG+ subsets as well as AID expression are maintained during the course of the disease (data not shown).
Recent reports suggest that CD38 and CD49d molecules are negative prognostic factors in CLL and could be implicated in the molecular crosstalk between malignant B cells and their microenvironment.20,32 In addition,CCL/3 and CCL/4 chemokines have been identified to contribute to the recruitment of cells from the monocyte macrophage lineage in these important interactions with CLL B cells and stromal cells.19,20 Interestingly, our results in the IgG+ subsets show higher expression of CD49d protein and CCL3/4 mRNA than in the IgM+ tumoral subpopulation. These results are in agreement with previous reports showing that these molecules are important players in the activation of CLL B cells interacting with their microenvironment.
With the use of a nonradioactive, stable isotopic labeling method to measure CLL kinetics, Messmer et al33 demonstrated that B-CLL is not a static disease that results simply from accumulation of long-lived lymphocytes, but it is a dynamic process in which cells proliferate and die. Markers such as CD38 have been proposed as helping to identify the proliferative pool.34 However, the fact that CD38 is expressed in a high percentage of tumoral cells in patients with UM disease and recent results from Calissano et al35 fail to establish a strong correlation between the percentage of CD38+ proliferating cells in CLL clones and survival and disease progression indicate that CD38+ tumoral cells constitute a heterogeneous population, including a small fraction of cells with an increased proliferative potential. Results from Messmer et al33 indicating that proliferating rates of CLL cells range from 0.08% to 1.7% suggest that, as in the case of CD38+ subset, not all the AID+/+ subpopulation with ongoing CSR correspond to cells in a state of active proliferation. Our results showing that only a fraction of tumoral cells displaying ongoing CSR coexpress CD38 support this last view. To our knowledge only one work has correlated AID and CD38 expression in CLL B cells.36 Because a positive correlation was found, we can presume that this small CD38+ subset within the IgG+ B cells could be the same that express high levels of AID enzyme. Further studies isolating the tumoral IgG+ subset coexpressing CD38 from IgG+CD38− cells are warranted.
Intriguingly, it is difficult to determine the precise role of these highly proliferating activated tumoral B cells. Because the presence of this subset is clearly associated with poor prognosis, it might have an adjuvant role in the maintenance of the CLL proliferative pool. However, given their increased proliferative potential, they should normally outnumber the IgM+ cells, but this is not the case. Thus, we could assume that these cells should undergo apoptosis once leaving the pseudofollicles. A recent work suggesting a link between AID expression and B-cell apoptosis in the GC favors this view.37 In these conditions, the IgG+ subset could reflect the existence of an active microenvironment leading to permanent stimulation of the IgM+ pool, which would turn on the CSR machinery maintaining this IgG+ subset in the PB. If true, the hypothesis that in the UM subgroup stimulation of BCR takes place by an unknown autoantigen38-40 and that this is responsible for consecutive stimulations and is able to sustain survival/expansion signals in the tumoral clone results in an interesting issue highlighted by these results.
Finally, our results indicate that among UM CLLs exists a subgroup of patients identified by high levels of AID expression and ongoing clonally related CSR, which displays an even worse prognosis. These findings have been obtained in a retrospective series of patients with a short follow-up. Prospective studies in larger series of patients are needed to raise conclusive evidence to our results and to determine whether expression of the different CH isotypes in tumoral cells should be routinely included in flow cytometric studies to better assess prognosis among patients with UM CLL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Michele Goodhardt for critical comments, Dr Hugo Naya for technical assistance in statistical analysis, and Mrs Ivana Faccini for helpful secretarial assistance.
This work was supported by the Lady Tata Memorial Trust and Fondo Clemente Estable (grant FCE_365).
Authorship
Contribution: F.P. and P. Moreno performed experiments and wrote the paper; P. Morande, C.A., A.C., and V.P. performed experiments and collected CLL samples; A.I.L. and R.G. performed clinical activities and data collection of patients with CLL; M.G., G.D., and O.P. contributed to scientific design and revised the paper; and P.O. designed research, coordinated the study and data analysis, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Oppezzo Pablo, Institut Pasteur de Montevideo, Unit of Recombinant Protein, Mataojo 2020, Montevideo (11400), Uruguay; e-mail: poppezzo@pasteur.edu.uy.
References
Author notes
F.P. and P. Moreno contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal