The Hedgehog (Hh) pathway is essential for normal embryonic development and tissue repair. The role of Hh signaling in hematopoiesis has been studied primarily by modulating the activity of Patched and Smoothened, but results have been conflicting. Some studies demonstrate a requirement for pathway activity in hematopoiesis, whereas others report that it is dispensable. Hh activity converges on the Gli transcription factors, but the specific role of these downstream effectors in hematopoiesis has not been reported. We have analyzed hematopoietic stem cell (HSC) and progenitor function in mice with a homozygous deletion of Gli1 (Gli1null). Gli1null mice have more long-term HSCs that are more quiescent and show increased engraftment after transplantation. In contrast, myeloid development is adversely affected with decreased in vitro colony formation, decreased in vivo response to granulocyte colony-stimulating factor (G-CSF), and impaired leukocyte recovery after chemotherapy. Levels of the proto-oncogene Cyclin D1 are reduced in Gli1null mice and may explain the loss of proliferation seen in HSCs and progenitor cells. These data demonstrate that Gli1 regulates normal and stress hematopoiesis. Moreover, they suggest that Gli1 and Smoothened may not be functionally redundant, and direct GLI1 inhibitors may be needed to effectively block HH/GLI1 activity in human disease.
Introduction
The proliferation of hematopoietic stem cells (HSCs) and progenitors is tightly regulated during normal homeostasis. HSCs are normally quiescent in the adult mouse but they can be induced to proliferate in response to stress or cytokine stimulation. In contrast, progenitors are highly proliferative to maintain a constant supply of infection-fighting white blood cells. Precisely how HSC and progenitor proliferation are regulated is not completely understood, but recent data have implicated a role for developmental signaling pathways such as Wnt and Notch in the regulation of stem cell proliferation, self-renewal, and differentiation.1,,–4
The Hedgehog (Hh) signaling pathway in mammals consists of 3 closely related ligands, Sonic Hh (Shh), Indian Hh (Ihh), and Desert Hh (Dhh), that can each bind to the transmembrane protein Patched (Ptch). Upon ligand binding, Ptch inhibition of the positive effector Smoothened (Smo) is released and signaling is transduced. Three zinc finger transcription factors, Gli1, Gli2, and Gli3, lie downstream of Smo and mediate Hh's effects. Gli1 is a positive effector of signaling, Gli3 is predominantly a transcriptional inhibitor, and Gli2 can function in both roles.5 The precise role of Hh signaling in normal hematopoiesis, however, is not known and the literature is contradictory. One group has reported that loss of Smo activity leads to a severe defect in HSC function,6 whereas others have reported a more modest phenotype,7 or none at all.8,9 All of these studies have focused primarily on the upstream modulators of pathway activity, Ptch and Smo. To better understand the role of Hh signaling in normal hematopoiesis, we have focused on the common downstream positive effector Gli1. Using a Gli1null mouse model, we have analyzed the function of Gli1 in adult HSCs and myeloid progenitors. Our results demonstrate that this transcription factor is a key regulator of HSC and myeloid cell proliferation and differentiation, and that loss of Gli1 impairs stress hematopoiesis.
Methods
Analysis of bone marrow fractions and cell sorting
Gli1lacZ mice were originally obtained from Alexander Joyner (Memorial Sloan-Kettering Cancer Center), and the colony was bred and maintained at Johns Hopkins University. Genotyping was performed by polymerase chain reaction (PCR) as previously described.10 Mice were housed in accordance with Johns Hopkins University Institutional Animal Care and Use Committee policy and fed mouse chow and water ad libitum. Mice used for analysis were between 8 and 12 weeks old. Bone marrow was flushed from femurs and tibia with staining media (phosphate-buffered saline supplemented with 2% fetal bovine serum) and labeled at 108 cells/mL in staining media with the following antibodies as indicated: CD34-allophycocyanin–, FcRγ–phycoerythrin (PE)– or Flt3-PE–, c-Kit–allophycocyanin–Alexa Fluor 750–, Sca1–PE–cyanin 7–, and biotin streptavidin–peridinin-chlorophyll-protein complex–cyanin 5.5–labeled lineage cocktail (CD3e, Gr1, B220, Ter119) (eBioscience). Fractions were analyzed and sorted on a 2-laser FACSAria (BD Biosciences).
Quantitative PCR
Cell populations were isolated by fluorescence-activated cell sorting (FACS) as described in “Analysis of bone marrow fractions and cell sorting.” RNA was isolated using RNAqueous (Ambion), and cDNA was made using Superscript II (Invitrogen) primed with random hexamers per the manufacturer's protocol. Quantitative reverse-transcriptase PCR for β-galactosidase was performed using SYBR-green Master Mix (Applied Biosystems) with the following primers: (forward) 5′-TAATGTTGATGAAAGCTGGCT-3′ and (reverse) 5′-ATGCGCTCAGGTCAAATTCAG-3′ as described by Fleming et al and normalized to beta-actin (4367659; Applied Biosystems).11 Predesigned Taqman real-time primer/probes sets for Cyclin D1 (Mm00432360_m1; Applied Biosystems), Patched1 (Mm00_436031m1; Applied Biosystems), and 18sRNA (4319413E) were obtained and PCR was performed using TaqMan Universal Master Mix (Applied Biosystems) on a I-Cycler Real-Time PCR machine (Bio-Rad). Expression levels were normalized to 18sRNA and compared with the ΔΔ threshold cycle method.
Bone marrow transplantation
Five hundred c-Kit+Sca1+Linneg (KSL) sorted cells from Gli1wt and Gli1null male mice were cotransplanted with 250 000 whole bone marrow cells from female mice and transplanted into lethally irradiated female recipients. DNA was extracted from peripheral blood using the DNeasy Kit (QIAGEN), and engraftment was estimated by percentage of male DNA by PCR for sex-determining region Y (SRY).12
Analysis of in vivo proliferation by BrdU incorporation
Mice were treated with 2 injections of bromodeoxyuridine (5-bromo-2-deoxyuridine [BrdU]; BD Biosciences) 50 μg/g by intraperitoneal injection 8 and 2 hours before analysis. Marrow was obtained and labeled with antibodies as described in “Analysis of bone marrow fractions and cell sorting,” and then fixed and labeled with a fluorescein isothiocyanate–conjugated anti-BrdU antibody using the BrdU Flow Kit (BD Biosciences), according to the manufacturer's protocol.
5-Fluorouracil treatment
Mice were treated with 150 mg/kg 5-flourouracil (5-FU; American Pharmaceutical Partners Inc) by intraperitoneal injection. Peripheral blood was obtained by retrorbital puncture in heparinized capillaries, and complete blood counts were measured on a Hemavet hematology analyzer (Drew Scientific).
G-CSF treatment
Mice were treated with 10 mg/kg recombinant granulocyte colony-stimulating factor (G-CSF, filgrastim; Amgen) by daily intraperitoneal injection.
Colony-forming assays
Bone marrow was plated in methylcellulose media supplemented with cytokines (Methocult M3434; StemCell Technologies) according to the manufacturer's directions, and colonies were scored at 12 to 14 days. Images of colonies were obtained using a Nikon TE200 inverted microscope (Nikon Instruments Inc) equipped with a Nikon DMX1200 digital color camera and ACT software using 4× and 10× objectives and 10× eyepiece objectives. Images were imported into Adobe Photoshop.
Statistics
Statistical significance was calculated using a single-sided Student t test, and P values less than .05 were considered significant.
Results
Gli1 deficiency skews HSC and progenitor compartments
Gli1LacZ/LacZ mice, which are homozygous for a LacZ insertion in the first exon of Gli1 (hereafter referred to as Gli1null), are born in the expected Mendelian ratios and develop normally.10 We examined the peripheral blood of adult wild-type and mutant mice and found no difference in the levels of hemoglobin, platelets, leukocytes, or neutrophils (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). We examined the expression of Gli1 and the Hh target gene Ptch within the bone marrow cells of wild-type mice by quantitative reverse-transcription–PCR (q-PCR). Although we did not detect Gli1 mRNA within the c-Kit+Sca1+Linneg (KSL) compartment similar to other reports,8 we did find that expression of the Gli1 target Ptch was highest within the CD34negKSL long-term HSCs (LT-HSCs) and expressed in all fractions of the bone marrow (Figure 1A). It has recently been reported that Gli1 transcripts have a short half-life and are highly protein bound, which may lead to difficulties in detection of the transcript by reverse-transcription–PCR.13 Gli1Lacz mice have the LacZ transgene inserted in the first exon of Gli1, and Gli1LacZ/wt heterozygotes have been used to analyze patterns of Gli1 expression during development.10 Therefore, to quantify Gli1 expression levels in HSC and progenitor fractions, we performed q-PCR for the Gli1 reporter gene β-Galactosidase on bone marrow subsets FACS sorted from Gli1LacZ/wt mice. Using this reporter, we observed increased expression of Gli1 in LT-HSCs, CD34+KSL short-term HSCs (ST-HSCs), FcRγlowCD34+c-Kit+Sca1negLinneg common myeloid progenitors (CMPs), and FcRγhighCD34+c-Kit+Sca1negLinneg granulocyte-monocyte progenitors (GMPs) compared with Lin+ cells. This pattern was similar to Ptch expression in the same subsets, validating that β-Galactosidase levels corresponded to bona fide Gli1 activity (Figure 1A).
Gli1null KSL cells have increased engraftment after transplantation due to increased numbers of LT-HSCs. (A) Gli1 (β-Galactosidase) and Ptch expression levels by q-PCR in LT-HSCs (CD34negcKit+Sca1+Linneg), ST-HSCs (CD34+cKit+Sca1+Linneg), CMPs (FcRγlowCD34+cKit+Sca1negLinneg), and GMPs (FcRγhiCD34+cKit+Sca1negLinneg) from Gli1LacZ/wt mice. (B) Five hundred KSL cells from Gli1wt and Gli1null male mice with 250 000 female competitor bone marrow cells were transplanted into irradiated female hosts. Peripheral blood chimerism of individual mice at 16 weeks after transplantation is indicated by diamonds and the average chimerism, by a horizontal bar. *P < .05. (C) Whole bone marrow cells were labeled with fluorescent-labeled antibodies and analyzed by flow cytometry for the LT-HSC (CD34negFlt3negKSL), ST-HSC (CD34+Flt3negKSL), and MPP (CD34+Flt3+KSL) fractions within the KSL compartment. Representative results from one pair of mice are shown. Percentages are of the KSL fraction. (D) Results for KSL subsets are summarized. Results are mean percentage of total bone marrow mononuclear cells ± SEM from 3 individual mice. *P < .05.
Gli1null KSL cells have increased engraftment after transplantation due to increased numbers of LT-HSCs. (A) Gli1 (β-Galactosidase) and Ptch expression levels by q-PCR in LT-HSCs (CD34negcKit+Sca1+Linneg), ST-HSCs (CD34+cKit+Sca1+Linneg), CMPs (FcRγlowCD34+cKit+Sca1negLinneg), and GMPs (FcRγhiCD34+cKit+Sca1negLinneg) from Gli1LacZ/wt mice. (B) Five hundred KSL cells from Gli1wt and Gli1null male mice with 250 000 female competitor bone marrow cells were transplanted into irradiated female hosts. Peripheral blood chimerism of individual mice at 16 weeks after transplantation is indicated by diamonds and the average chimerism, by a horizontal bar. *P < .05. (C) Whole bone marrow cells were labeled with fluorescent-labeled antibodies and analyzed by flow cytometry for the LT-HSC (CD34negFlt3negKSL), ST-HSC (CD34+Flt3negKSL), and MPP (CD34+Flt3+KSL) fractions within the KSL compartment. Representative results from one pair of mice are shown. Percentages are of the KSL fraction. (D) Results for KSL subsets are summarized. Results are mean percentage of total bone marrow mononuclear cells ± SEM from 3 individual mice. *P < .05.
To robustly test the role of Gli1 in HSC function, we performed competitive transplantation experiments of KSL cells from Gli1wt or Gli1null mice into wild-type recipients and examined the peripheral blood for donor chimerism. Because the Gli1null mice are on an outbred Swiss-Webster background that precludes the use of the CD45.1/CD45.2 isogenic system typically used for monitoring peripheral blood chimerism, we transplanted HSCs from male mice into female recipients and measured chimerism by quantitative PCR for Y chromosome–specific DNA.12 We found that 500 KSL cells, cotransplanted with 250 000 wild-type female competitor marrow, gave an average peripheral blood chimerism of 6% for wild-type cells and 18% for Gli1null cells at 16 weeks after transplantation (P < .05, Figure 1B). To better understand the reason for increased engraftment seen with Gli1null HSCs, we quantified the LT-HSC (CD34negFlt3negKSL), ST-HSC (CD34+Flt3negKSL), and multipotent progenitor (MPP, CD34+Flt3+KSL) fractions within the KSL compartment (Figure 1C). LT-HSCs are the only cells that are capable of long-term engraftment beyond 16 weeks after transplantation.14 We observed a 2-fold increase in LT-HSCs in Gli1null mice that likely explains the increased engraftment in mice that received a transplant of Gli1null KSL cells (Figure 1D). These data show that Gli1 regulates HSC number in the most primitive stem cell compartment.
Gli1 loss impairs myeloid differentiation
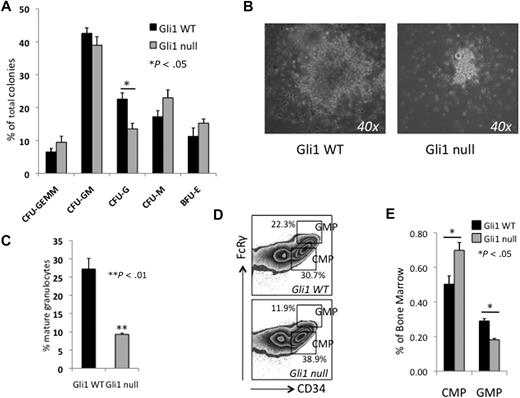
To examine the effect of Gli1 deficiency on myeloid development, we performed in vitro colony-forming assays in semisolid media supplemented with cytokines that allow for quantification of lineage-committed progenitors in mouse bone marrow. Plates were scored at 2 weeks for the presence of myeloid, erythroid, and mix-lineage colonies. We observed an almost 2-fold reduction in number and a marked decrease in the size of granulocyte colony-forming units (CFU-Gs) derived from Gli1null bone marrow (Figure 2A-B). Mixed granulocyte-macrophage colonies (CFU-GMs) were also smaller and had fewer granulocytes. We quantified total granulocyte formation by harvesting cells from the methylcellulose after 12 days of culture and measuring Gr1+/SSChigh mature granulocytes by flow cytometry. We observed a 3-fold (27% to 9%) reduction in the formation of granulocytes from Gli1null bone marrow (Figure 2C).
Loss of Gli1 impairs in vitro granulopoiesis and differentiation of myeloid progenitors in vivo. (A) Whole bone marrow cells from Gli1WT and Gli1null mice were plated in methylcellulose supplemented with cytokines (Methocult M3434). Twenty thousand cells were plated per well, in duplicate. Results are mean ± SEM from 4 separate experiments. Colonies were scored at 12 to 14 days. *P < .05 (B) Photomicrograph of CFU-Gs grown in Methocult M3434. Original magnification ×40. (C) In vitro granulocyte formation measured by harvesting cells grown in Methocult M3434, labeling with antibody against Gr1, and analyzing by flow cytometry. Mature granulocytes were Gr1+ with high side-scatter properties. **P < .01 (D) Whole bone marrow was labeled with fluorescent-labeled antibodies and analyzed by flow cytometry for common myeloid progenitors (CMPs, FcRγlowCD34+cKit+Sca1negLinneg) and granulocyte macrophage progenitors (GMPs, FcRγhiCD34+cKit+Sca1negLinneg). Representative results from one pair of mice are shown. Percentages are of the c-Kit+Sca1negLinneg fraction. (E) Results for the myeloid progenitor subsets are summarized. Results are the mean percentage of total bone marrow cells ± SEM. *P < .05.
Loss of Gli1 impairs in vitro granulopoiesis and differentiation of myeloid progenitors in vivo. (A) Whole bone marrow cells from Gli1WT and Gli1null mice were plated in methylcellulose supplemented with cytokines (Methocult M3434). Twenty thousand cells were plated per well, in duplicate. Results are mean ± SEM from 4 separate experiments. Colonies were scored at 12 to 14 days. *P < .05 (B) Photomicrograph of CFU-Gs grown in Methocult M3434. Original magnification ×40. (C) In vitro granulocyte formation measured by harvesting cells grown in Methocult M3434, labeling with antibody against Gr1, and analyzing by flow cytometry. Mature granulocytes were Gr1+ with high side-scatter properties. **P < .01 (D) Whole bone marrow was labeled with fluorescent-labeled antibodies and analyzed by flow cytometry for common myeloid progenitors (CMPs, FcRγlowCD34+cKit+Sca1negLinneg) and granulocyte macrophage progenitors (GMPs, FcRγhiCD34+cKit+Sca1negLinneg). Representative results from one pair of mice are shown. Percentages are of the c-Kit+Sca1negLinneg fraction. (E) Results for the myeloid progenitor subsets are summarized. Results are the mean percentage of total bone marrow cells ± SEM. *P < .05.
Analysis of the c-Kit+Sca1negLinneg (KL) myeloid progenitor compartment revealed an increase in FcRγlowCD34+KL common myeloid progenitors (CMPs) and decrease in the FcRγhighCD34+KL granulocyte-monocyte progenitors (GMPs; P < .05, Figure 2D-E).15 The GMP compartment gives rise to CFU-Gs in colony-forming assays,15 and the reduction in this compartment is consistent with the lower number of CFU-Gs observed (Figure 2A). In the hematopoietic developmental hierarchy, cells in the CMP compartment give rise to cells in the GMP,15 therefore, the accumulation of cells in the more primitive CMP and loss of cells in the more differentiated GMP compartments suggest that the reduced granulocyte development in Gli1null marrow may be a result of impaired progenitor differentiation.
Gli1null HSCs and progenitors are less proliferative and have lower levels of Cyclin D1
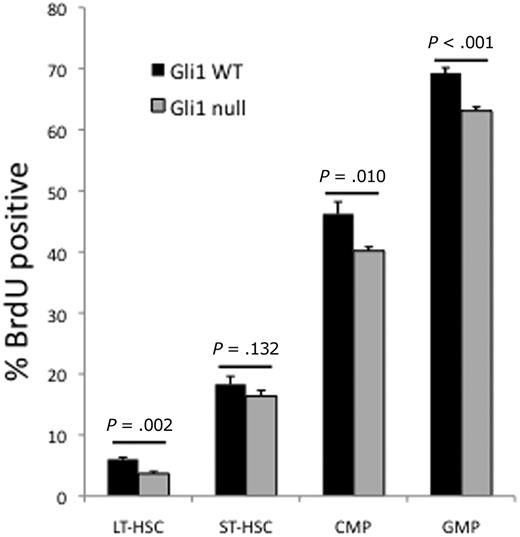
We speculated that the differences observed in the HSC and myeloid progenitor compartments may be due to impaired proliferation because both ST-HSCs and GMPs are more proliferative than LT-HSCs and CMPs, respectively. We analyzed these compartments for proliferation by in vivo BrdU incorporation and found fewer BrdU-positive cells, indicating decreased proliferation, in the LT-HSC, CMP, and GMP compartments of Gli1null mice (Figure 3). Proliferation and differentiation are closely linked processes in hematopoiesis. Therefore, the defect in proliferation results in reduced differentiation from LT-HSCs to ST-HSCs and from CMPs to GMPs and explains the skewing seen in these compartments.
HSCs and myeloid progenitors are less proliferative in Gli1null mice. Gli1wt and Gli1null mice were injected with BrdU, to mark actively cycling cells, and bone marrow was harvested at 8 hours, labeled with fluorescent antibodies, permeabilized, and labeled with anti-BrdU antibody. Cells were analyzed by flow cytometry. Data are the mean percentage of BrdU-positive cells for each bone marrow fraction in each genotype. A total of 4 mice in each group were analyzed. P values for each group are indicated.
HSCs and myeloid progenitors are less proliferative in Gli1null mice. Gli1wt and Gli1null mice were injected with BrdU, to mark actively cycling cells, and bone marrow was harvested at 8 hours, labeled with fluorescent antibodies, permeabilized, and labeled with anti-BrdU antibody. Cells were analyzed by flow cytometry. Data are the mean percentage of BrdU-positive cells for each bone marrow fraction in each genotype. A total of 4 mice in each group were analyzed. P values for each group are indicated.
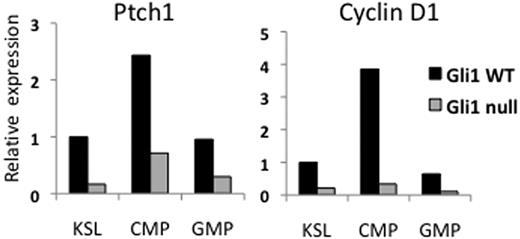
Hh signaling is known to be mitogenic in many tissues and Cyclin D1, broadly expressed throughout mitosis, has been shown to be a target of Gli1 activity.16 Cyclin D1 has been shown to be particularly important for the proliferation and differentiation of myeloid progenitors.17 We therefore measured expression levels of Cyclin D1 in Gli1wt and Gli1null marrow to determine whether this could explain the differences in proliferation between these 2 groups. Quantitative PCR of these compartments revealed a 4- to 10-fold decrease in Cyclin D1 levels in the KSL, CMP, and GMP compartments of Gli1null mice, suggesting that lower Cyclin D1 levels may account for the lower rates of proliferation in Gli1null cells (Figure 4).
Gli1null cells have lower RNA expression levels of the Hedgehog target genes Patched1 and Cyclin D1. Bone marrow cells were labeled with fluorescent antibodies and sorted by FACS. RNA was extracted from sorted populations, used to generate cDNA, and then used for quantitative PCR of the genes Patched1 (Ptch1) and the cell cycle regulator Cyclin D1. Relative expression is plotted on the vertical axis.
Gli1null cells have lower RNA expression levels of the Hedgehog target genes Patched1 and Cyclin D1. Bone marrow cells were labeled with fluorescent antibodies and sorted by FACS. RNA was extracted from sorted populations, used to generate cDNA, and then used for quantitative PCR of the genes Patched1 (Ptch1) and the cell cycle regulator Cyclin D1. Relative expression is plotted on the vertical axis.
Gli1null mice show impaired stress hematopoiesis
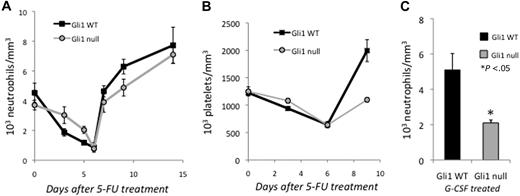
Although Gli1 appears to be dispensable for steady-state hematopoiesis, we hypothesized that it may be required for the rapid proliferation of progenitors needed during stress hematopoiesis. Hh signaling is well understood in neural development where it regulates the proliferation of neural stem cells, particularly after the depletion of neural progenitors with chemotherapy.18 To assess the role of Gli1 under stress conditions, we treated Gli1wt and Gli1null mice with 5-FU and measured the absolute neutrophil count (ANC) in the peripheral blood. Treatment with 5-FU selectively depletes the bone marrow of cycling cells and forces quiescent stem cells to proliferate and repopulate the marrow. As expected, the lower proliferation rate of Gli1null bone marrow progenitors led to a relative sparing of these cells after 5-FU treatment and resulted in a more gradual nadir of ANC in mutant mice after 5-FU treatment (Figure 5A). On recovery, however, the ANC increased more slowly in Gli1null mice, likely because of the reduced proliferation and differentiation of myeloid progenitors (Figures 2–3). Platelet counts showed a similar pattern of delayed recovery after 5-FU treatment in Gli1null mice, demonstrating that multiple blood lineages were affected by the loss of Gli1 (Figure 5B).
Loss of Gli1 impairs stress hematopoiesis. (A) Two cohorts of mice (total of 10 mice/group) were treated with 5-FU (150 mg/kg) and neutrophil counts measured. Each time point represents the mean ± SEM from 4 to 5 mice. (B) Mice were treated with 5-FU (150 mg/kg) and platelet counts measured. Each time point is the mean ± SEM from 5 mice. (C) Mice were treated with G-CSF (10 mg/kg) intraperitoneally daily for 3 days, and neutrophil counts measured after 3 days. Results are mean ± SEM from 5 mice. *P < .05.
Loss of Gli1 impairs stress hematopoiesis. (A) Two cohorts of mice (total of 10 mice/group) were treated with 5-FU (150 mg/kg) and neutrophil counts measured. Each time point represents the mean ± SEM from 4 to 5 mice. (B) Mice were treated with 5-FU (150 mg/kg) and platelet counts measured. Each time point is the mean ± SEM from 5 mice. (C) Mice were treated with G-CSF (10 mg/kg) intraperitoneally daily for 3 days, and neutrophil counts measured after 3 days. Results are mean ± SEM from 5 mice. *P < .05.
To investigate whether Gli1null mice also demonstrated an impaired neutrophil response to the cytokine G-CSF, mutant and wild-type mice were treated with daily G-CSF and their peripheral blood was monitored. Three days after G-CSF treatment, the ANC in Gli1null was approximately half of wild-type counterparts, demonstrating a defect in stress granulopoiesis in mutant mice (Figure 5C). These data clearly demonstrate that although Gli1 is not absolutely required for granulopoiesis, its loss has significant effects on myeloid progenitor function and impairs their ability to recover after cytotoxic injury or respond to stimulatory cytokines.
Discussion
Prior studies examining the role of Hedgehog signaling in normal hematopoiesis have reported conflicting results. An initial study reported that the induction of pathway activity by SHH results in the expansion of human HSCs in vitro.19 Furthermore, Patched heterozygote mice that display increased cellular Hh pathway activity demonstrate increased HSC proliferation and expansion.20 Taken together, these reports suggest that Hh pathway activity drives proliferation of HSCs and are consistent with our observation that loss of the downstream effector Gli1 leads to reduced proliferation.
Further evidence of a role for Hh signaling in normal blood development has been suggested by loss-of-function studies in zebrafish in which both Hh and Smo expression is required for definitive hematopoiesis.21 More recently, selective loss of Smo function in the in utero hematopoietic compartment of transgenic mice has been found to profoundly inhibit the engraftment capacity of HSCs.6 Because of the functional redundancy between Gli1 and Gli2, Gli1 is not absolutely necessary for Hh signal transduction, therefore, it is possible that upstream loss of Smo activity could lead to defects not seen with Gli1 deficiency alone.10 Indeed, Gli2-deficient mice show profound developmental defects and embryonic lethality, whereas Gli1-deficient mice show no gross developmental defects, and definitive hematopoiesis and long-term HSC function appear to be unaffected by the loss of Gli1.10,22 Our data demonstrate that Gli1 deficiency reduces the proliferation of HSCs and progenitors and has important consequences for normal hematopoiesis. At steady state, the loss of Gli1 in the HSC compartment does not lead to a loss of function, but rather, an increased proportion of long-term HSCs and improved engraftment after transplantation. In the highly proliferative progenitor compartment, however, the loss of proliferation leads to impairment in myeloid differentiation and defective stress hematopoiesis. These results are consistent with Gli1's role in signal amplification and positive feedback within the Hh signaling loop.
In contrast to the studies that support a role for Hh signaling in hematopoiesis, 2 recent reports have demonstrated that the conditional loss of Smo within adult HSCs has no discernable impact on hematopoiesis.8,9 Of note, Gao et al8 did examine the bone marrow of Gli1null mice and found no differences in the numbers of KSL progenitors, B cells, or T cells, but did not perform the subset analysis or functional studies we report here. Using a different model, Dierks et al reported that mice with a germline deletion of Smo showed no functional defect in HSCs isolated from the fetal liver, however they did observe a transient impairment in peripheral blood count recovery after 5-FU treatment in mice reconstituted with Smonull HSCs, similar to our findings in Gli1null mice.7 It is possible that the discrepancies between these findings and those reported by Zhao et al6 are because of differential requirements for Hh pathway activity within embryonic or adult hematopoiesis as some of these studies used conditional systems that impaired Hh signaling in adult animals, whereas in other studies Smo is absent in utero. Our data do not further elucidate the requirement for Smo at any stage of hematopoiesis because Gli1 is downstream of Smo. Our results do suggest that non–Smo-mediated activation of Gli1 may be operating in the hematopoietic system. Alternatively, the phenotype we observed may be the result of selective imbalance between the positive regulatory function of Gli1 and the negative regulatory function of the suppressor forms of Gli2 and Gli3. Furthermore, recent studies in animal models of pancreatic cancer have suggested that Hh signaling may regulate the tumor stroma.23 Thus, it is possible that some of the effects of Hh signaling in normal hematopoiesis may be in the bone marrow microenvironment, which may or may not be affected depending on the transgenic model used.
Our data demonstrate that Gli1null HSCs have decreased expression of the cell cycle regulator Cyclin D1, a known target gene of the Hh signaling pathway that is critical for the normal proliferation and differentiation of bone marrow cells.17 Cyclin D1, a proto-oncogene, is dysregulated in many types of cancer and abnormal Hh activity may be exerting its oncogenic effects though Cyclin D1, although further investigations will be needed to clarify this issue. Furthermore, these results suggest that abnormal Gli1 activity in myeloid leukemia may be important for driving the uncontrolled proliferation of myeloid cancer cells.6,7,24
Our findings may also have implications for the development of Hh pathway inhibitors as novel anticancer agents. The majority of agents in development target SMO, similar to the naturally occurring inhibitor cyclopamine. These agents are therefore highly effective at blocking Hh signal transduction in a normal developmental context. Our data, however, suggest a role for Smo-independent activation of Gli1 in normal hematopoiesis. Furthermore, emerging data from several types of cancer have revealed aberrancies in canonical signal transduction and demonstrated SMO-independent activation of GLI1 via other pathways such as TGF-β, K-Ras, Wnt, or the EWS-FLI fusion oncogene.13,25,–27 Previously we reported that up to one third of primary leukemia samples have evidence of GLI1 expression in the absence of SMO expression.28 Therefore, it is possible that SMO inhibition may not lead to complete inhibition of GLI1 activity and therefore may not be effective against all tumors with aberrant Hh pathway activity. Our findings support the suggestion of some investigators that direct GLI1 inhibitors may be needed in addition to SMO antagonists to effectively block the Hh pathway in cancer.29
In summary, we believe that Hh signaling is dispensable for the development of HSCs and the maintenance of steady-state hematopoiesis. This is likely due to a combination of non–Smo-mediated activation of Gli transcription factors, cross-talk between other developmental signaling pathways, and functional redundancies among the individual Gli factors. This study, however, clearly demonstrates that Hh/Gli1 signaling plays a critical role in the complex regulation of hematopoiesis during stress and injury.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Alexander Joyner of Memorial Sloan-Kettering Institute for the kind gift of Gli1LacZ mice and for helpful discussions of these results.
This work is supported by the American Society of Hematology Scholar Award, American Society for Clinical Oncology Young Investigator Award, American Association for Cancer Research-Astellas USA Fellowship for Basic Cancer Research (A.M.), the National Institutes of Health R01CA127574 and K23CA107040, the Gabrielle's Angel Foundation for Cancer Research, the Sidney Kimmel Foundation for Cancer Research, and the Goodwin Foundation (W.M.).
National Institutes of Health
Authorship
Contribution: A.M. designed and performed experiments, analyzed data, and prepared the paper; G.J., Q.W., and S.B. performed experiments; and W.M. designed experiments and prepared the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William Matsui, The Sidney Kimmel Comprehensive Cancer Center, Department of Oncology, Johns Hopkins University School of Medicine, CRB245, 1650 Orleans St, Baltimore, MD 21287; e-mail: matsuwi@jhmi.edu.