Abstract
Previous studies have shown that ADAMTS13 spacer domain is required for cleavage of von Willebrand factor (VWF). However, the exact amino acid residues within this domain critical for substrate recognition are not known. Epitope mapping of anti-ADAMTS13 immunoglobulin G from patients with thrombotic thrombocytopenic purpura and sequence alignment of the ADAMTS13 spacer domains of human, mouse, and zebrafish with these of human and murine ADAMTS1, a closely related member of ADAMTS family, have provided hints to investigate the role of the amino acid residues between Arg659 and Glu664 of the ADAMTS13 spacer domain in substrate recognition. A deletion of all these 6 amino acid residues (ie, Arg659-Glu664) from the ADAMTS13 spacer domain resulted in dramatically reduced proteolytic activity toward VWF73 peptides, guanidine-HCl denatured VWF, and native VWF under fluid shear stress, as well as ultralarge VWF on endothelial cells. Site-directed mutagenesis, kinetic analyses, and peptide inhibition assays have further identified a role for amino acid residues Arg659, Arg660, and Tyr661 in proteolytic cleavage of various substrates under static and fluid shear stress conditions. These findings may provide novel insight into the structural-function relationship of ADAMTS13 and help us to understand pathogenesis of thrombotic thrombocytopenic purpura and other arterial thromboses associated with compromised VWF proteolysis.
Introduction
ADAMTS13, a member of A Disintegrin And Metalloprotease with ThromboSpondin type 1 repeats (ADAMTS) family,1-5 cleaves von Willebrand factor (VWF) at the Tyr1605-Met1606 bond.6,7 This proteolytic cleavage appears to be necessary to eliminate ultralarge (UL)–VWF multimers in circulation, thereby preventing the formation of platelet-rich thrombi in small arterioles and capillaries.8-10 Several studies11-15 have demonstrated that the spacer domain of ADAMTS13 is not only required for efficient cleavage of VWF after being denatured with 1.5M urea or guanidine-HCl but also necessary for productive cleavage of VWF73, a fragment derived from the central A2 domain of VWF. The ADAMTS13 variants lacking the spacer domain are still able to cleave the small peptidyl substrates with much slower kinetics11-13 but have little or no proteolytic activity toward soluble multimeric VWF under denaturing conditions.11,16 Furthermore, we17 and others18 have recently shown that the Cys-rich and spacer domains of ADAMTS13 play a critical role in proteolytic cleavage of newly released UL-VWF on endothelial cells in the absence or presence of fluid shear stress. The spacer domain appears to directly interact with the distal portion (amino acid residues Glu1660-Arg1668) of the VWF73 peptide. The deletion of these C-terminal 9 amino acid residues from the VWF73 (ie, construct VWF64) results in dramatically reduced proteolytic cleavage of the shorter peptide by ADAMTS13 under static conditions.13,19
The importance of the ADATMS13 spacer domain is also highlighted by the fact that this region is frequently targeted by anti-ADAMTS13 autoantibodies in patients with acquired idiopathic thrombotic thrombocytopenic purpura (TTP),20-22 a potentially fatal syndrome characterized with disseminated thromboses in small arteries and capillaries.10,23 The epitope mapping study24 has suggested that the anti-ADAMTS13 immunoglobulin G (IgG) in some TTP patients specifically binds the amino acid residues between Tyr572 and Asn579 or between Val657and Gly666 on the ADAMTS13 spacer domain. Interestingly, most of the residues Val657-Gly666 are absent in the spacer domain of ADAMTS1, which does not bind and cleave VWF. Both the antibody mapping results and the sequence alignment findings provide hints to investigate the role of the amino acid residues between Arg659 and Glu664 in the ADAMTS13 spacer domain in substrate recognition. By using site-directed mutagenesis, recombinant proteins, and biochemical analyses, we are able to demonstrate that the amino acid residues Arg659, Arg660, and Tyr661 in the ADAMTS13 spacer domain play a critical role in recognition and cleavage of the peptidyl substrates, soluble multimeric VWF, and cell-bound UL-VWF polymers under static and flow conditions. Our findings may provide novel insight into the structure-function relationship of ADAMTS13 and shed some light on the mechanism of how ADAMTS13 mutations and anti-ADAMTS13 autoantibodies that specifically target at the spacer domain result in compromised ADAMTS13 activity, leading to microvascular thromboses in patients with hereditary and acquired TTP.
Methods
Constructs
The QuickChange site-directed mutagenesis was used to generate a deletion of 6 amino acid residues (Arg659-Glu664; del6aa) and other mutants (Arg659Ala, Arg660Ala, Tyr661Ala, Gly662Ala, Glu663Ala, and Glu664Ala; Stratagene). The primers used for constructing all mutants are listed in Table 1. pcDNA3.1-ADAMTS13-V5-His encoding human wild-type ADAMTS13 tagged at the C-terminus with V5-His epitope11,15 was used as a template. A fragment encoding the spacer domain (Ser556-Ala865) of ADATMS13 was generated by polymerase chain reaction and cloned into Champion pET151/D directional TOPO vector (Invitrogen) as previously described.11 The entire coding region of the plasmids was determined by sequencing at the Nucleic Acid Core Facility, The Children's Hospital of Philadelphia. No unintended mutation in the coding region was found in all constructs.
Primers used for generation of ADAMTS13 mutants
| Primer name . | Sequences . |
|---|---|
| 1. Del6aa | F: CAG GAA GAT GCT GAC ATC CAG GTT TAC- - -TAT GGC AAC CTC ACC CGC CCA GAC AT |
| R: GAT GTC TGG GCG GGT GAG GTT GCC ATA- - -GTA AAC CTG GAT GTC AGC ATC TTC CTG | |
| 2. R659A | F: GCT GAC ATC CAG GTT TAC GCG CGG TAT GGC GAG GAG TAT |
| R: ATA CTC CTC GCC ATA CCG CGC GTA AAC CTG GAT GTC AGC | |
| 3. R660A | F: GAC ATC CAG GTT TAC AGG GCG TAT GGC GAG GAG TAT GGC |
| R: GCC ATA CTC CTC GCC ATA CGC CCT GTA AAC CTG GAT GTC | |
| 4. Y661A | F: ATC CAG GTT TAC AGG CGG GCT GGC GAG GAG TAT GGC AAC |
| R: GTT GCC ATA CTC CTC GCC AGC CCG CCT GTA AAC CTG GAT | |
| 5. G662A | F: CAG GTT TAC AGG CGG TAT GCC GAG GAG TAT GGC AAC CTC |
| R: GAG GTT GCC ATA CTC CTC GGC ATA CCG CCT GTA AAC CTG | |
| 6. E663A | F: TAC AGG CGG TAT GGC GCG GAG TAT GGC AAC CTC |
| R: GAG GTT GCC ATA CTC CGC GCC ATA CCG CCT GTA | |
| 7. E664A | F: TAC AGG CGG TAT GGC GAG GCG TAT GGC AAC CTC ACC CGC |
| R: GCG GGT GAG GTT GCC ATA CGC CTC GCC ATA CCG CCT GTA |
| Primer name . | Sequences . |
|---|---|
| 1. Del6aa | F: CAG GAA GAT GCT GAC ATC CAG GTT TAC- - -TAT GGC AAC CTC ACC CGC CCA GAC AT |
| R: GAT GTC TGG GCG GGT GAG GTT GCC ATA- - -GTA AAC CTG GAT GTC AGC ATC TTC CTG | |
| 2. R659A | F: GCT GAC ATC CAG GTT TAC GCG CGG TAT GGC GAG GAG TAT |
| R: ATA CTC CTC GCC ATA CCG CGC GTA AAC CTG GAT GTC AGC | |
| 3. R660A | F: GAC ATC CAG GTT TAC AGG GCG TAT GGC GAG GAG TAT GGC |
| R: GCC ATA CTC CTC GCC ATA CGC CCT GTA AAC CTG GAT GTC | |
| 4. Y661A | F: ATC CAG GTT TAC AGG CGG GCT GGC GAG GAG TAT GGC AAC |
| R: GTT GCC ATA CTC CTC GCC AGC CCG CCT GTA AAC CTG GAT | |
| 5. G662A | F: CAG GTT TAC AGG CGG TAT GCC GAG GAG TAT GGC AAC CTC |
| R: GAG GTT GCC ATA CTC CTC GGC ATA CCG CCT GTA AAC CTG | |
| 6. E663A | F: TAC AGG CGG TAT GGC GCG GAG TAT GGC AAC CTC |
| R: GAG GTT GCC ATA CTC CGC GCC ATA CCG CCT GTA | |
| 7. E664A | F: TAC AGG CGG TAT GGC GAG GCG TAT GGC AAC CTC ACC CGC |
| R: GCG GGT GAG GTT GCC ATA CGC CTC GCC ATA CCG CCT GTA |
The dotted line between nucleotides indicates the portion being deleted; the nucleotides for a single amino acid substitution are underlined.
Preparation of fluorescein-labeled VWF73
Plasmid encoding VWF73 with substitutions by 2 cysteine residues at the amino residues Gln1599 and Pro1611 flanking the Tyr-Met bond was a kind gift from Dr John Owen (Department of Medicine, Wake Forest University School of Medicine). Recombinant VWF73 was expressed, purified, and labeled with fluorescein-5′-maleimide (Pierce) by use of the protocol described previously.25,26 The amount of purified fluorescein-labeled VWF73 designated rF-VWF73 was determined by the absorbance at 280 nm (1 optical density [OD] = 0.94 mg/mL). Because of the homoquenching effect resulting from close proximity of 2 labeled-fluorescent dyes, no fluorescence emits. When the Tyr-Met bond is cleaved by ADAMTS13, 2 fluorescent dyes are separated, and fluorescence emits at excitation wavelength of 485 nm, which can be detected at the emission wavelength of approximately 540 nm.
Preparation of recombinant ADAMTS13 and mutants
ADAMTS13 and mutants were purified and quantified with the method described previously.11,27 Alternatively, conditioned medium containing ADAMTS13 or mutants was concentrated with Centri-Prep30 (Millipore). The concentration of recombinant ADAMTS13 and mutants in the concentrated medium was quantified by Western blot with anti-V5 IgG (1:5000), followed by IR dye 800CW–labeled rabbit anti–mouse IgG (1:12 500; LI-COR Bioscience) in 20mM Tris-HCl, pH 7.5, 150mM NaCl, 0.005% Tween 20 (TBST) containing 1% casein. Purified wild-type ADAMTS13 was used for calibration.
Preparation of recombinant ADAMTS13 spacer domain
Recombinant ADAMTS13 spacer domain was expressed in Escherichia coli, purified, and refolded with the use of a protocol previously described.11 The refolded protein was dialyzed against 20mM Tris-HCl, pH 7.5; 150mM NaCl; and 5mM benzamidine-HCl. The protein purity was determined by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining. The concentration was determined by absorbance at 280 nm, corrected with light scattering at 320 nm (1AU = 0.86 mg/mL).
Preparation of synthetic peptides
Short peptides corresponding to amino acid residues (Arg659-Glu664) of the ADAMTS13 spacer domain were synthesized at Peptide 2.0. Three perfectly matched peptides, Arg-Arg-Tyr-Gly-Glu-Glu (RRYGEE), Arg-Arg-Tyr (RRY), and Gly-Glu-Glu (GEE), and 2 scrambled peptides with identical amino acid residues, Glu-Gly-Arg-Glu-Tyr-Arg (EGREYR) and Arg-Tyr-Arg (RYR), were synthesized. All peptides were purified by high-performance liquid chromatography system to greater than 92% purity. The mass spectrometry analysis showed that the masses [M +2H+]2+ for RRYGEE, EGREYR, RRY, RYR, and GEE were 406.2, 406.2, 248.5, 248.5, and 335.0, respectively. The peptides were lyophilized in 0.1% trifluoroacetate salt. For reconstitution, distilled water was added to concentration of 10 mg/mL. The stock peptides were stored in aliquots at −80°C.
Cleavage of glutathione S-transferase-VWF73 by ADAMTS13 and mutants
Purified glutathione S-transferase-VWF73 6xHis (GST-VWF73; 50nM) was incubated with the concentrated conditioned medium or purified ADAMTS13 or mutants (2.5nM) at 37°C for 1 and 3 hours. The proteolytic cleavage was determined by 4% to 20% SDS-PAGE (Bio-Rad) and Western blot. After being transferred onto polyvinylidene fluoride membrane (Millipore), the membrane was blocked with TBST containing 1% casein. The membrane was washed with TBST and then incubated with rabbit anti-GST IgG (1:800; Invitrogen) at 4°C overnight. After being washed 3 times with TBST, the bound antibody was detected by incubation with IRDye 800CW-labeled goat anti–rabbit IgG (1:12 500; LI-COR Biosciences) at 25°C for 1 hour. The fluorescent signal was determined by an Odyssey infrared imaging system (LI-COR Biosciences).
Kinetic cleavage of rF-VWF73 by ADAMTS13 and mutants
Purified recombinant Alexa-488–labeled VWF73 (rF-VWF73) at various concentrations (0-12μM) was incubated with wild-type ADAMTS13 or mutants (2.5nM) in 5mM Bis-Tris, pH 6.0, containing 25mM CaCl2 and 0.005% Tween 20 (total volume, 200 μL) in a 96-well black plate (Corning). Chymotrypsin that preferentially hydrolyzes peptide bonds after tyrosine was used for calibration. Chymotrypsin (20nM) converted all substrate to the products within 10 minutes. The rate of fluorescence generation was monitored at 37°C with a fluorescent microtiter plate reader (Molecular Devices; excitation [ex]/emission [em] 485/535 nm) every 2 minutes for 60 minutes. The maximal rate of fluorescence generation (Vmax) in cleaving rF-VWF73 by chymotrypsin was used to determine the relative rate of hydrolysis of rF-VWF73 by ADAMTS13 and mutants. The kinetic constants kcat and Km were determined by fitting the data into a Michaelis-Menten equation with the SigmaPlot software (Systat).
For the peptide inhibition assay, rF-VWF73 (1μM) was incubated with various concentrations (0-200μM) of synthetic peptides for 10 minutes. Recombinant ADAMTS13 (2.5nM) was added, and proteolytic cleavage of rF-VWF73 was monitored on the same fluorescent microtiter plate reader (ex 485 nm and em 543 nm) every minute for 20 minutes. The maximal rate of fluorescent generation per second (units/second) was plotted against the concentrations of synthetic peptides used. The experiments were repeated 3 times under the same conditions. The concentration achieving a half-maximal inhibition was determined by fitting the data into the following nonlinear equation:
Y = Bottom + (Top − Bottom)/[1 + 10̂((LogIC50 − X)*HillSlope)].
Here Top and Bottom are the highest and the lowest rates of fluorescent generation in the absence of peptide and in the presence of maximal concentration of the peptides.
Cleavage of multimeric VWF by ADAMTS13 and mutants under denaturing conditions
Purified plasma VWF (375 μg/mL) was incubated with 1.5M guanidine-HCl at 37°C for 1 hour. The denatured VWF was diluted to a final concentration of 37.5 μg/mL with 50mM Tris-HCl, pH 8.0; 50mM NaCl; 0.25% bovine serum albumin (BSA); and 5mM CaCl2. Purified ADAMTS13 or mutants (20nM) were added in a total volume of 50 μL. The reaction was incubated at 37°C for 8 hours. The digested material (10 μL) was denatured at 60°C for 20 minutes with sample buffer (70mM Tris-HCl, pH 6.5; 2.4% SDS; 0.67M urea; and 4mM ethylenediaminetetraacetic acid [EDTA]). The denatured proteins were fractionated in a 1% (wt/vol) SeaKem HGT agarose (Cambrex Corporation) gel in a Mini-protein III gel cassette (Bio-Rad) at 15 mA for 90 minutes. The proteins were then transferred onto a polyvinylidene fluoride membrane (Millipore) at 100 mA for 60 minutes. The VWF multimers were detected with rabbit anti-VWF IgG (1:5000), followed by IRDye 800CW-labeled goat anti-rabbit IgG (1:12 500; LI-COR Biosciences) in TBST containing 1% casein. The fluorescent signal was determined by the Odyssey infrared imaging system (LI-COR Biosciences). The 350-kDa band was quantified by ImageJ software (National Institutes of Health).
Cleavage of native VWF by ADAMTS13 and mutants under fluid shear stress
Purified VWF (37.5 μg/mL) was incubated at 37°C for 60 minutes with or without purified spacer domain or synthetic peptides at various concentrations in 50mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.5, containing 1 mg/mL BSA, 150mM NaCl, and 5mM CaCl2 (total volume, 20 μL). ADAMTS13 (50nM) with or without recombinant FVIII (10nM) and lyophilized platelets (210 × 103/μL) was added into the reaction. The reaction mixture was subjected to vortexing (2500 rpm, with an estimated fluid shear stress of ∼ 54 dyne/cm2) for 5 minutes with a mini-vortexer (Fisher Scientific). The reaction was stopped by heating at 60°C for 20 minutes with 2× sample buffer, and the proteolytic cleavage products were determined by 1% agarose gel as described previously.
Cleavage of cell-bound UL-VWF by ADAMTS13 and mutants in the presence and absence of fluid shear stress
Human umbilical vascular endothelial cells were grown on 6-well plates or gelatin-coated glass cover slides in EBM-2 medium containing 2% fetal bovine serum (Cambrex Corporation). The cells were treated at 37°C with 100μM histamine (0.5 mL/well) for 2 minutes and then gently washed 3 times with phosphate-buffered saline (PBS; Invitrogen). The histamine-stimulated and washed cells on the glass slides were assembled into a rectangular parallel flow chamber (flow width = 1.0 cm, thickness = 0.0127 cm; GlycoTech). The cells were then perfused with the perfusion buffer (20mM HEPES, pH 7.5; 150mM NaCl) alone or the buffer containing recombinant ADAMTS13 or mutants (10nM) for 5 minutes under flow shear stress (2.5 dyne/cm2). The perfusion buffers were collected, and EDTA (20mM) was added to inactivate the residual ADAMTS13. Alternatively, the histamine-stimulated and washed cells in the 6-well plates were treated with ADAMTS13 and mutants without flow. The UL-VWF released into the perfusion buffer or culture medium was concentrated (20-fold) by centri-con10 (Millipore). The VWF antigen and proteolytic cleavage products in the conditioned medium were determined by 5% SDS-PAGE and Western blot as described previously.17
Binding assays
A microtiter plate (Thermo Fisher Scientific) was coated without or with 100 μL of purified GST-VWF73 (2.0 μg/mL) or plasma VWF (12.5 μg/mL) for 1 hour. After being blocked with 2.5% BSA in 20mM Tris-HCl and 150mM NaCl, 100 μL of ADAMTS13 and various mutants at various concentrations in the presence of 10mM EDTA were added and incubated for 2 hours. The plates were washed 3 times with PBS and incubated with peroxidase-conjugated mouse anti-V5 IgG (Invitrogen; 1:1000) for 1 hour. After being washed 3 times with TBS, 0.05% Tween 20 and the bound antibody were quantified by 3,3′,5,5′-tetramethylbenzidine substrate (Thermo Fisher Scientific). After the addition of 50 μL of 1.5M H2SO4, the plate was read with the SpectroMax plate reader (Molecular Devices) at wavelength of 450 nm. The specific binding was obtained by subtracting the absorbance values in the nonligand controls from those in the ligand-coated wells. The data were then plotted as the bound against the concentrations of ADATMS13 and mutants. The data were fitted into a Michaelis-Menten equation with SigmaPlot software (Systat Software) to obtain the apparent dissociation constants (KD) and the maximal binding (Bmax).
Results
ADAMTS13 mutant constructs and purified proteins
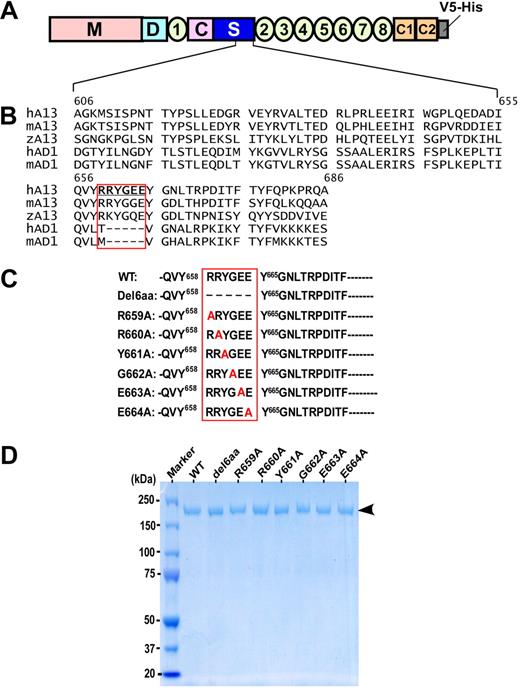
The authors of previous studies11-13,15,16 have shown that the ADAMTS13 spacer domain is required for recognition of VWF substrate. The amino acid residues between Val657and Gly666 in the spacer domain of ADAMTS13 are shown to be frequently targeted by anti-ADAMTS13 IgG autoantibodies isolated from patients with acquired TTP (Figure 1A).24 Interestingly, most of the amino acid residues (Arg659-Glu664) in this region were absent in the spacer domains of human and murine ADAMTS1, which cleaves a cartilage proteoglycan, aggrecan,28,29 but does not cleave VWF. These residues are, however, highly conserved in the spacer domains of zebrafish, murine, and human ADAMTS13 (Figure 1A). We therefore hypothesized that the amino acid residues (Arg659-Glu664) may participate in substrate recognition. To test this hypothesis, all 6 amino acid residues (Arg659-Glu664) in this region were either deleted (construct del6aa) or substituted with an alanine one by one (Figure 1B). The recombinant wild-type ADAMTS13 and mutants were stably expressed in HEK293 cells and purified to homogeneity. All purified recombinant proteins exhibited a single band of ∼195 kDa on 4% to 20% SDS-PAGE with Coomassie blue staining (Figure 1C arrowhead). The purified ADAMTS13 and mutants were exchanged by gel filtration to 20mM HEPES, pH 7.5; 150mM NaCl; and 5mM CaCl2. The concentrations of the purified ADAMTS13 and mutants were determined by absorbance (OD) at 280 nm, corrected with light scattering at 320 nm in a 1-cm length curvette. One corrected OD280 equals ∼0.68 mg/mL as previously described.27
ADAMTS13 domain organization, sequence alignment of the partial spacer domains, and purified recombinant ADAMTS13 mutants. (A) ADAMTS13 is consisted of a catalytic domain (M) and multiple noncatalytic domains, including the disintegrin domain (D), the first thrombospondin type 1 repeat (1), Cys-rich (C) domain, and spacer domain (S). The more distal C-terminus contains TSP1 2-8 repeats and 2 CUB domains (C1 and C2). (B) Sequence alignment of the partial ADAMTS13 (A13) spacer domains (amino acid residues between Ala606 and Ala686) of human (h), mouse (m), and zebrafish (z), as well as the ADAMTS1 spacer domains of human and mouse (AD1). The boxed area indicates the potential motif that may interact with VWF-A2 domain and autoantibodies against ADAMTS13 in some patients with acquired TTP. (C) Site-directed mutagenesis generates the full-length ADAMTS13 mutants with deletion or substitution of 1 to 6 amino acid residues boxed. (D) A 4% to 20% SDS-PAGE with Coomassie blue shows the purified wild-type ADAMTS13 (WT) and various mutants. Arrow indicates molecular weight of approximately 195 kDa.
ADAMTS13 domain organization, sequence alignment of the partial spacer domains, and purified recombinant ADAMTS13 mutants. (A) ADAMTS13 is consisted of a catalytic domain (M) and multiple noncatalytic domains, including the disintegrin domain (D), the first thrombospondin type 1 repeat (1), Cys-rich (C) domain, and spacer domain (S). The more distal C-terminus contains TSP1 2-8 repeats and 2 CUB domains (C1 and C2). (B) Sequence alignment of the partial ADAMTS13 (A13) spacer domains (amino acid residues between Ala606 and Ala686) of human (h), mouse (m), and zebrafish (z), as well as the ADAMTS1 spacer domains of human and mouse (AD1). The boxed area indicates the potential motif that may interact with VWF-A2 domain and autoantibodies against ADAMTS13 in some patients with acquired TTP. (C) Site-directed mutagenesis generates the full-length ADAMTS13 mutants with deletion or substitution of 1 to 6 amino acid residues boxed. (D) A 4% to 20% SDS-PAGE with Coomassie blue shows the purified wild-type ADAMTS13 (WT) and various mutants. Arrow indicates molecular weight of approximately 195 kDa.
Amino acid residues in ADAMTS13 spacer domain required for cleavage of VWF73 peptides
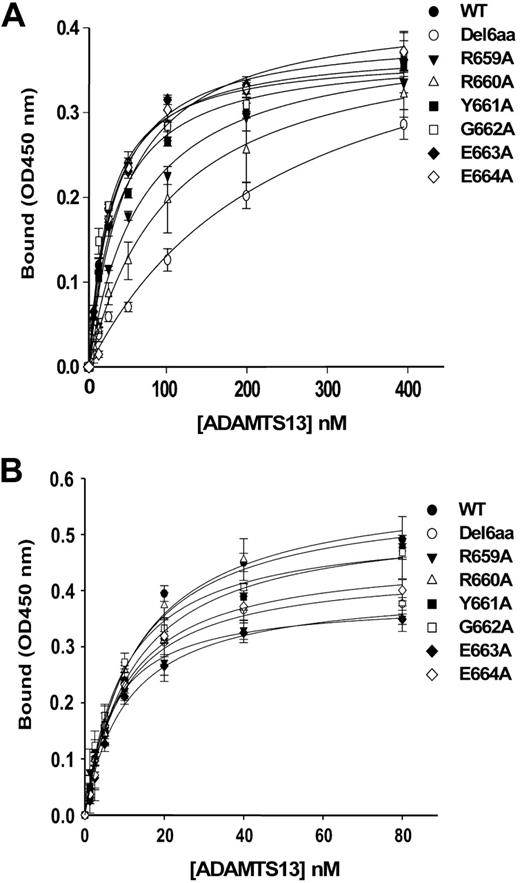
The ADAMTS13 spacer domain is required for the cleavage of small peptidyl substrates such as GST-tagged VWF73 or fluorescein-labeled VWF73.11-13 Therefore, mutations in the ADAMTS13 spacer domain are expected to have a direct impact on recognition and cleavage of these peptidyl substrates. To address this question, GST-VWF73 (50nM) was incubated with wild-type ADAMTS13 or mutants (2.5nM) at 37°C for 1 hour (Figure 2A) and 3 hours (Figure 2B). Construct lacking all 6 amino acid residues (Arg659-Glu664) in the spacer domain of ADATMS13 (del6aa) exhibited no or minimal proteolytic activity toward GST-VWF73 at 1 hour (Figure 2A) and 3 hours of incubation (Figure 2B). Wild-type ADAMTS13 at the same concentration cleaved GST-VWF73 detectably at 1 hour (Figure 2A) and completely at 3 hours, generating a product of 34.5 kDa (Figure 2B). The mutants Arg659Ala, Arg660Ala, and Tyr661Ala exhibited various degrees of reduction in proteolytic activity toward GST-VWF73 under the same conditions (Figure 2A-B). However, other ADAMTS13 mutants Gly662Ala, Glu663Ala, and Glu664Ala had normal activity compared with wild-type ADAMTS13 (Figure 2A-B). These results were consistent with those obtained with recombinant ADAMTS13 and mutants in the concentrated conditioned medium without purification (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). For better kinetic analyses, we used rF-VWF73 as a substrate. By incubating rF-VWF73 (0-12μM) with a fixed concentration (2.5nM) of ADAMTS13 and mutants for 60 minutes, we showed that the construct del6aa cleaved rF-VWF73 with the ratio of kcat/Km of only 75M−1s−1, approximately 10-fold less efficiently than wild-type ADAMTS13 (the kcat [/Km ratio = 730M−1s−1) under the same conditions (Table 2; Figure 2C). The cleavage efficiency of the Arg659Ala and Arg660Ala toward rF-VWF73 substrate reduced by approximately 5.2, and approximately 3.3 fold, respectively, compared with that of wild-type ADAMTS13 (Table 2; Figure 2C). However, a substitution of the residues Tyr661, Gly662, Glu663, and Glu664 with Ala did not appear to alter cleavage efficiency toward the rF-VWF73 substrate under these conditions (Table 2; Figure 2C). Together, these data indicate that the amino acid residues Arg659, Arg660, and perhaps Tyr661 in the ADAMTS13 spacer domain may participate in recognition of the unfolded central VWF-A2 domain.
Cleavage of GST-VWF73 and rF-VWF73 by wild-type ADAMTS13 and various mutants. Purified GST-VWF73 (50nM) was incubated with the wild-type ADAMTS13 or mutants (2.5nM) in the absence (−) or presence (+) of EDTA (20mM) at 37°C for 1 hour (A) and 3 hours (B). The proteolytic cleavage product (open arrow) was determined by Western blotting. *The preexisting nonspecific bands in the purified GST-VWF73 substrate. (C) Purified recombinant rF-VWF73 at various concentrations (0-12μM) was incubated at 37°C with wild-type (WT) ADAMTS13 or mutants (2.5nM). The rate of fluorescent generation was determined at the ex 485 nm and em 535 nm. The maximal rate of fluorescent generation (Vmax) with D[b]-Phe[b]-Pro[b]-Arg[b]-chloromethylketone-treated chymotrypsin (20nM) that cleaves specifically at the methionyl bond was used for a calibration. The relative mean product formation (nanomoles per second), compared with chymotrypsin at the same concentration, is expressed as a function of the substrate concentrations (micromole). The data represent the means ± SD (n = 3).
Cleavage of GST-VWF73 and rF-VWF73 by wild-type ADAMTS13 and various mutants. Purified GST-VWF73 (50nM) was incubated with the wild-type ADAMTS13 or mutants (2.5nM) in the absence (−) or presence (+) of EDTA (20mM) at 37°C for 1 hour (A) and 3 hours (B). The proteolytic cleavage product (open arrow) was determined by Western blotting. *The preexisting nonspecific bands in the purified GST-VWF73 substrate. (C) Purified recombinant rF-VWF73 at various concentrations (0-12μM) was incubated at 37°C with wild-type (WT) ADAMTS13 or mutants (2.5nM). The rate of fluorescent generation was determined at the ex 485 nm and em 535 nm. The maximal rate of fluorescent generation (Vmax) with D[b]-Phe[b]-Pro[b]-Arg[b]-chloromethylketone-treated chymotrypsin (20nM) that cleaves specifically at the methionyl bond was used for a calibration. The relative mean product formation (nanomoles per second), compared with chymotrypsin at the same concentration, is expressed as a function of the substrate concentrations (micromole). The data represent the means ± SD (n = 3).
Kinetic cleavage of rF-VWF73 by ADAMTS13 and spacer domain mutants
| . | K (M × 10−6) . | k (s−1) . | k/K (×109M−1s−1) . | Fold change . |
|---|---|---|---|---|
| WT | 1.81 ± 0.14 | 1.32 ± 0.03 | 733.8 | 1.00 |
| del6aa | 21.94 ± 6.32 | 1.56 ± 0.33 | 74.6 | 0.10 |
| R659A | 10.20 ± 5.03 | 1.43 ± 0.40 | 140.1 | 0.19 |
| R660A | 4.96 ± 0.79 | 1.09 ± 0.08 | 219.2 | 0.30 |
| Y661A | 1.70 ± 0.38 | 0.76 ± 0.05 | 447.9 | 0.61 |
| G662A | 4.90 ± 0.43 | 2.05 ± 0.08 | 418.4 | 0.57 |
| E663A | 3.48 ± 0.51 | 1.55 ± 0.09 | 444.3 | 0.61 |
| E664A | 3.36 ± 0.53 | 1.63 ± 0.10 | 487.7 | 0.66 |
| . | K (M × 10−6) . | k (s−1) . | k/K (×109M−1s−1) . | Fold change . |
|---|---|---|---|---|
| WT | 1.81 ± 0.14 | 1.32 ± 0.03 | 733.8 | 1.00 |
| del6aa | 21.94 ± 6.32 | 1.56 ± 0.33 | 74.6 | 0.10 |
| R659A | 10.20 ± 5.03 | 1.43 ± 0.40 | 140.1 | 0.19 |
| R660A | 4.96 ± 0.79 | 1.09 ± 0.08 | 219.2 | 0.30 |
| Y661A | 1.70 ± 0.38 | 0.76 ± 0.05 | 447.9 | 0.61 |
| G662A | 4.90 ± 0.43 | 2.05 ± 0.08 | 418.4 | 0.57 |
| E663A | 3.48 ± 0.51 | 1.55 ± 0.09 | 444.3 | 0.61 |
| E664A | 3.36 ± 0.53 | 1.63 ± 0.10 | 487.7 | 0.66 |
The data are expressed in means ± SD (n = 3).
Km indicates the Michaelis-Menten constant; kcat equals to Vmax/[E]; M, molar concentration; and s, second.
Amino acid residues in ADAMTS13 spacer domain required for cleavage of multimeric VWF or UL-VWF under various conditions
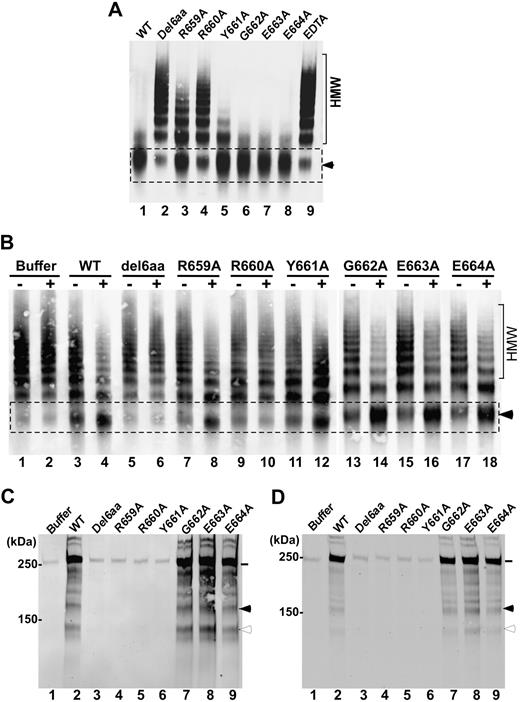
The VWF73 peptides (both GST-VWF73 and rF-VWF73) are devoid of many ancillary sites necessary for ADAMTS13 binding.26,30 To assess whether the mutations in the spacer domain of ADAMTS13 affect proteolytic cleavage of multimeric VWF, we first performed experiments with purified multimeric VWF under denaturing conditions. When incubated with guanidine-HCl–denatured VWF (150nM) for 8 hours, wild-type ADAMTS13 (20nM) almost completely cleaved high-molecular-weight multimeric VWF, generating a cleavage product of approximately 350 kDa (the dimer of 2 C-terminal fragments) as shown by the agarose gel electrophoresis and Western blot under nonreducing conditions (Figure 3A lane 1). The addition of EDTA (20mM) to the same reaction completely blocked the proteolytic cleavage of multimeric VWF by wild-type ADAMTS13 (Figure 3A lane 9). The construct del6aa and Arg660Ala were unable to cleave predenatured VWF under the same conditions (Figure 3A lanes 2 and 4). The other mutants Arg659Ala and Tyr661Ala exhibited moderately and slightly reduced activity for cleavage of predenatured VWF, respectively (Figure 3A lanes 3 and 5). In contrast, ADAMTS13 mutants Gly662Ala, Glu663Ala, and Glu664Ala showed normal proteolytic activity toward the predenatured VWF (Figure 3A lanes 6-8) compared with the wild-type ADAMTS13. These results suggest that the amino acid residues Arg659 and Arg660 and Tyr661 may be important for recognition of multimeric VWF under static and denaturing conditions.
Cleavage of multimeric VWF by wild-type ADAMTS13 or mutants under various conditions. (A) Guanidine-HCl denatured VWF (37.5 μg/mL) was incubated at 37°C for 8 hours with wild-type ADAMTS13 (WT; lane 1) or various mutants (lanes 2-8; final concentration, 20nM) in the absence or presence of EDTA (20mM; lane 9) in 50mM Tris-HCl, 50mM NaCl, 5mM CaCl2, pH 8.0. (B) Native VWF (37.5 μg/mL) was incubated for 15 minutes with WT ADAMTS13 or various mutants (final concentration, 50nM) in the absence (−) or presence (+) of recombinant factor VIII (20nM) and lyophilized platelets (∼ 200 × 103/μL) in 50mM HEPES, pH 7.5, containing 1 mg/mL BSA, 150mM NaCl, and 5mM CaCl2. The reaction mixtures were subjected to vortexing at 2500 rpm for 5 minutes and then quenched by heating for 20 minutes at 60°C in the presence of SDS sample buffer. The proteolytic cleavage product (dimer of C-terminal fragments, ∼ 350 kDa) was determined by 1% agarose gel and Western blotting. HMW indicates the area with high-molecular-weight VWF multimers, whereas the arrowhead indicates the proteolytic cleavage product (∼ 350 kDa). (C-D) Human umbilical vascular endothelial cells cultured on 6-well plates or a gelatin-coated glass cover slides were stimulated with histamine (100μM) for 2 minutes and washed with PBS. The washed cells were treated for 5 minutes with a buffer alone, wild-type ADAMTS13, or mutants (10nM) in the absence of fluid shear stress (C) or in the presence of 2.5 dyne/cm2 of fluid shear stress generated (D). The conditioned media or perfusion buffers collected from the 6-well plate or flow chamber were concentrated by filtration. The UL-VWF antigen and degradation products in the conditioned media were determined by Western blot. The intact VWF polypeptide (∼ 250 kDa) is indicated by a solid line. The proteolytic cleavage products (176 kDa and 140 kDa) are marked by a closed and open arrowhead, respectively. Other bands between 176 kDa and 250 kDa, seen in panels C and D (lane 2 and lanes 7-9), may be the result of proteolysis by ADAMTS13 and/or other proteases in the conditioned medium.
Cleavage of multimeric VWF by wild-type ADAMTS13 or mutants under various conditions. (A) Guanidine-HCl denatured VWF (37.5 μg/mL) was incubated at 37°C for 8 hours with wild-type ADAMTS13 (WT; lane 1) or various mutants (lanes 2-8; final concentration, 20nM) in the absence or presence of EDTA (20mM; lane 9) in 50mM Tris-HCl, 50mM NaCl, 5mM CaCl2, pH 8.0. (B) Native VWF (37.5 μg/mL) was incubated for 15 minutes with WT ADAMTS13 or various mutants (final concentration, 50nM) in the absence (−) or presence (+) of recombinant factor VIII (20nM) and lyophilized platelets (∼ 200 × 103/μL) in 50mM HEPES, pH 7.5, containing 1 mg/mL BSA, 150mM NaCl, and 5mM CaCl2. The reaction mixtures were subjected to vortexing at 2500 rpm for 5 minutes and then quenched by heating for 20 minutes at 60°C in the presence of SDS sample buffer. The proteolytic cleavage product (dimer of C-terminal fragments, ∼ 350 kDa) was determined by 1% agarose gel and Western blotting. HMW indicates the area with high-molecular-weight VWF multimers, whereas the arrowhead indicates the proteolytic cleavage product (∼ 350 kDa). (C-D) Human umbilical vascular endothelial cells cultured on 6-well plates or a gelatin-coated glass cover slides were stimulated with histamine (100μM) for 2 minutes and washed with PBS. The washed cells were treated for 5 minutes with a buffer alone, wild-type ADAMTS13, or mutants (10nM) in the absence of fluid shear stress (C) or in the presence of 2.5 dyne/cm2 of fluid shear stress generated (D). The conditioned media or perfusion buffers collected from the 6-well plate or flow chamber were concentrated by filtration. The UL-VWF antigen and degradation products in the conditioned media were determined by Western blot. The intact VWF polypeptide (∼ 250 kDa) is indicated by a solid line. The proteolytic cleavage products (176 kDa and 140 kDa) are marked by a closed and open arrowhead, respectively. Other bands between 176 kDa and 250 kDa, seen in panels C and D (lane 2 and lanes 7-9), may be the result of proteolysis by ADAMTS13 and/or other proteases in the conditioned medium.
The conformations of VWF predenatured by urea or guanidine-HCl may be quite different from those under physiologic fluid shear stress.31,32 To determine proteolytic activity of the various ADAMTS13 mutants under more physiologic conditions, we incubated multimeric VWF (150nM) with wild-type ADAMTS13 or various mutants in the presence of purified recombinant factor VIII (10nM) and lyophilized platelets (210 × 103/μL) under constant vortexing at 2500 rpm for 5 minutes. This vortexing rate generated approximately ∼54 dyne/cm2 of fluid shear stress on the basis of the mathematical modeling and experimental determination of the fold increase in cleavage product with or without FVIII (20nM) compared with that in a cone-plate viscometer (C.G.S., Wenjing Cao, and X.L.Z., unpublished data). In the absence of FVIII and platelets, very little proteolytic cleavage product could be detected with any of the constructs under the conditions used (Figure 3B lanes 3, 5, 7, 9, 11, 13, 15, and 17). Addition of recombinant factor VIII and platelets dramatically increased the proteolytic cleavage of multimeric VWF by the wild-type ADAMTS13 and the mutants Gly662Ala, Glu663Ala, and Glu664Ala, accompanied by the dramatic reduction of high-molecular-weight multimers (Figure 3B lanes 4, 14, 16, and 18). However, the mutant del6aa and Arg660Ala demonstrated no detectable proteolytic activity even in the presence of physiologic cofactors (Figure 3B lanes 6 and 10). The other mutants Arg659Ala and Tyr661Ala exhibited moderately reduced proteolytic activity in the absence of FVIII and platelets, but their activity appeared to be restored to some degree in the presence of FVIII and platelets (Figure 3B lanes 8 and 12). These findings support an idea that the amino acid residues Arg659, Arg660, and Tyr661 in the spacer domain of ADAMTS13 may play a role in recognition and proteolytic cleavage of multimeric VWF under more physiologically relevant conditions.
The removal of newly released UL-VWF strings or bundles anchored on endothelial cells by ADAMTS13 occurs rapidly and efficiently in the presence33-35 and in the absence of fluid shear stress,17,36 suggesting that the cell-bound UL-VWF polymers may be preferred substrates for ADAMTS13. To determine whether these amino acid residues in the ADAMTS13 spacer domain play a role in recognition of UL-VWF at the cellular level, histamine-stimulated and washed endothelial cells were treated for 5 minutes with the wild-type ADAMTS13 or various mutants (10nM) in the absence and the presence of laminar flow (2.5 dyne/cm2). The amount of UL-VWF released from endothelial cells and proteolytic cleavage products (176 kDa and 140 kDa) in the conditioned medium were determined by Western blotting with anti-VWF IgG under reduced conditions. As shown, the constructs del6aa, Arg659Ala, Arg660Ala, and Tyr661Ala exhibited dramatically reduced activity, cleaving the cell-bound UL-VWF polymers in the absence (Figure 3C lanes 3-6) and the presence (Figure 3D lanes 3-6) of fluid shear stress compared with the wild-type ADAMTS13. The amount of soluble UL-VWF antigen (∼250 kDa, under reduced conditions) in the conditioned medium released from cells treated with the mutants del6aa, Arg659Ala, Arg660Ala, and Tyr661Ala was significantly lower than that in the conditioned medium from cells treated with the wild-type ADAMTS13 either in the absence (Figure 3C lane 2) or in the presence (Figure 3D lane 2) of fluid shear stress. In contrast, the mutants Gly662Ala, Glu663Ala, and Glu664Ala did not exhibit any functional defect in cleavage of the cell-bound UL-VWF polymers under the same conditions (Figure 3C; lanes 7-9 and Figure 3D, lanes 7-9). The proteolytic cleavage products (176 kDa and 140 kDa) were only detected in the conditioned media collected from the cells treated with the wild-type ADAMTS13 (Figure 3C-D; lane 2) and the mutants Gly662Ala, Glu663Ala, and Glu664Ala (Figure 3C-3D; lanes 7-8), but not from those with buffer alone (Figure 3C-D; lane 1) and the mutants del6aa, Arg559Ala, Arg660, and Tyr661Ala (Figure 3C-D; lanes 3-6) even after normalization of VWF antigen in each lane (data not shown). These results suggest the amino acid residues Arg659, Arg660, and Tyr661 in the ADAMTS13 spacer domain also may play a role in recognition of the cell-bound UL-VWF polymers in the absence and the presence of fluid shear stress.
The spacer domain and synthetic peptides derived from the spacer domain inhibit cleavage of VWF by ADAMTS13 under fluid shear stress
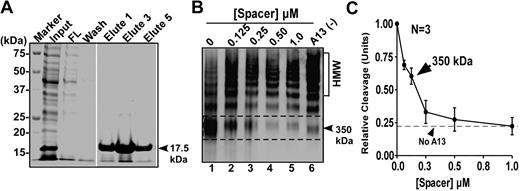
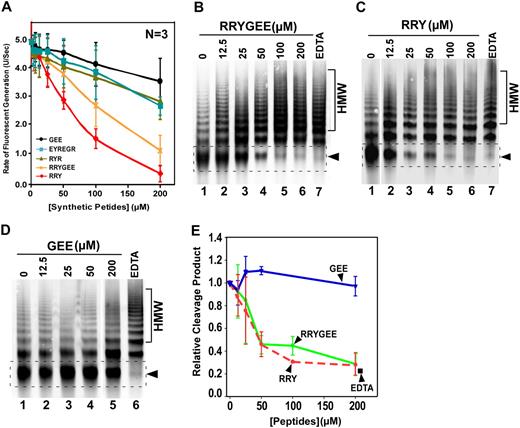
To rule out the possibility that the deletion or mutations in the ADAMTS13 spacer domain may alter the protein structure or folding, we performed competitive inhibition assays. Recombinant spacer domain of ADAMTS13 was expressed and purified from Escherichia coli and refolded, which exhibited a molecular weight of 17.5 kDa on a 15% SDS-PAGE with Coomassie blue staining (Figure 4A lanes 5-7). When added into the reaction consisting of purified VWF (150nM) and purified wild-type ADAMTS13 (20nM), the isolated ADAMTS13 spacer domain dramatically inhibited the proteolytic cleavage of multimeric VWF by ADAMTS13 in a concentration-dependent manner (Figure 4B). The concentration of the spacer domain that inhibits ∼50% of the wild-type ADAMTS13 activity (IC50) under these conditions was estimated to be less than 0.10μM (Figure 4C). Moreover, the peptides derived from the ADATMS13 spacer domain, including RRYGEE and RRY, also inhibited proteolytic cleavage of rF-VWF73 (Figure 5A) and VWF (Figure 5B-5C) by ADAMTS13. The inhibitory activity of these peptides was concentration dependent (Figure 5A-C,E). At the concentration of 200μM, the peptides RRYGEE and RRY blocked proteolytic cleavage of rF-VWF73 by ADAMTS13 activity by approximately 85% and approximately 90%, respectively (Figure 5A). The estimated IC50 (S) for RRYGEE and RRY was approximately 165μM and approximately 86μM, respectively. The matched peptide GEE or scrambled peptides RYR and EGREYR, however, exhibited little inhibitory activity on the cleavage of rF-VWF73 by ADAMTS13 even at the greatest concentration (200μM) tested (Figure 5A). Similar results were obtained with these peptides for inhibition of the proteolytic cleavage of multimeric VWF by ADAMTS13 under fluid shear stress (Figure 5D). At the concentration of 100μM, both peptides (RRYGEE and RRY) almost completely inhibited proteolytic cleavage of multimeric VWF by ADAMTS13 (Figure5B,C,E), whereas the peptide GEE at the same concentration or greater showed minimal inhibitory activity in this reaction (Figure 5D-E). These results suggest that the amino acid residues Arg659, Arg660, and Tyr661 in ADAMTS13 spacer domain may directly bind the VWF-A2 domain that can be exposed under fluid shear stress. An approximately 10-fold difference in the IC50 was observed between the isolated spacer domain (0.1μM) and the synthetic peptides (RRYGEE, ∼ 160μM and RRY, ∼ 86μM) for the inhibition of proteolytic cleavage of multimeric VWF by ADAMTS13, suggesting a role of some other amino acid residues in the spacer domain in substrate recognition.
Recombinant spacer domain inhibits proteolytic cleavage of multimeric VWF by ADAMTS13 under fluid shear stress. (A) SDS-PAGE with Coomassie blue shows the various fractions of recombinant ADAMTS13 spacer domain before and after Ni-affinity chromatography. (B) Purified plasma-derived VWF (37.5 μg/mL) was incubated at 37°C with 0-1.0μM of purified spacer domain in the absence or presence of 20mM EDTA for 60 minutes. Then, ADAMTS13 (20nM) along with recombinant FVIII (10nM) and lyophilized platelets (∼ 200 × 103/μL) was added into the reaction (total volume, 20 μL). The reaction mixture was subjected to vortexing (2500 rpm) for 5 minutes. The proteolysis of VWF was determined by multimer analysis on 1% agarose gel electrophoresis and Western blot. HMW indicates high-molecular-weight multimer. The proteolytic cleavage product (350 kDa) is indicated with dashed frame and arrowhead. (C) Quantification of the 350-kDa cleavage product by densitometry is plotted in the y-axis against the concentrations of purified recombinant spacer domain (micromole) in x-axis.
Recombinant spacer domain inhibits proteolytic cleavage of multimeric VWF by ADAMTS13 under fluid shear stress. (A) SDS-PAGE with Coomassie blue shows the various fractions of recombinant ADAMTS13 spacer domain before and after Ni-affinity chromatography. (B) Purified plasma-derived VWF (37.5 μg/mL) was incubated at 37°C with 0-1.0μM of purified spacer domain in the absence or presence of 20mM EDTA for 60 minutes. Then, ADAMTS13 (20nM) along with recombinant FVIII (10nM) and lyophilized platelets (∼ 200 × 103/μL) was added into the reaction (total volume, 20 μL). The reaction mixture was subjected to vortexing (2500 rpm) for 5 minutes. The proteolysis of VWF was determined by multimer analysis on 1% agarose gel electrophoresis and Western blot. HMW indicates high-molecular-weight multimer. The proteolytic cleavage product (350 kDa) is indicated with dashed frame and arrowhead. (C) Quantification of the 350-kDa cleavage product by densitometry is plotted in the y-axis against the concentrations of purified recombinant spacer domain (micromole) in x-axis.
Synthetic peptides derived from ADAMTS13 spacer domain block proteolytic cleavage of rF-VWF73 and multimeric VWF by ADAMTS13. (A) Purified rF-VWF73 (1.0μM) was incubated with various concentrations (0-200μM) of synthetic peptides as indicated for 10 minutes. Then, recombinant ADATMS13 (2.5nM) was added and proteolytic cleavage of rF-VWF73 was monitored at 37°C on a fluorescent microtiter plate reader (ex 485 nm and em 540 nm). The maximal rates of fluorescent generation per second (units/second) were determined and plotted against the log concentrations of synthetic peptides used. The data presented are means ± SD of 3 independent measurements. (B-D) Purified plasma-derived VWF (37.5 μg/mL) was incubated for 60 minutes at 37°C with various concentrations (0-200μM) of synthetic peptides RRYGEE (B), RRY (C), and GEE (D). ADAMTS13 (50nM) along with recombinant FVIII (10nM) and lyophilized platelets (210 × 103/μL) was added into the reaction (total volume, 20μL). The reaction mixture was subjected to vortexing (2500 rpm) for 5 minutes. The proteolytic cleavage product was determined by 1% agarose gel and Western blot. (E) Quantification of the 350-kDa cleavage product (indicated by arrowheads in panels B, C, and D) by densitometry is plotted against the concentrations of synthetic peptides (micromole).
Synthetic peptides derived from ADAMTS13 spacer domain block proteolytic cleavage of rF-VWF73 and multimeric VWF by ADAMTS13. (A) Purified rF-VWF73 (1.0μM) was incubated with various concentrations (0-200μM) of synthetic peptides as indicated for 10 minutes. Then, recombinant ADATMS13 (2.5nM) was added and proteolytic cleavage of rF-VWF73 was monitored at 37°C on a fluorescent microtiter plate reader (ex 485 nm and em 540 nm). The maximal rates of fluorescent generation per second (units/second) were determined and plotted against the log concentrations of synthetic peptides used. The data presented are means ± SD of 3 independent measurements. (B-D) Purified plasma-derived VWF (37.5 μg/mL) was incubated for 60 minutes at 37°C with various concentrations (0-200μM) of synthetic peptides RRYGEE (B), RRY (C), and GEE (D). ADAMTS13 (50nM) along with recombinant FVIII (10nM) and lyophilized platelets (210 × 103/μL) was added into the reaction (total volume, 20μL). The reaction mixture was subjected to vortexing (2500 rpm) for 5 minutes. The proteolytic cleavage product was determined by 1% agarose gel and Western blot. (E) Quantification of the 350-kDa cleavage product (indicated by arrowheads in panels B, C, and D) by densitometry is plotted against the concentrations of synthetic peptides (micromole).
Binding of ADAMTS13 and mutants to the immobilized GST-VWF73 and multimeric VWF
To determine whether the reduced proteolytic activity of various ADAMTS13 mutants resulted from a decrease in substrate binding, we assessed the binding affinities between ADATMS13 mutants and substrates. When incubated with the immobilized GST-VWF73 (0.2 μg/well) or plasma-derived VWF (1.25 μg/well), wild-type ADAMTS13 bound to the GST-VWF73 and the multimeric VWF with the apparent dissociation constants (KD; app) of approximately 28nM and approximately 13nM, respectively (Table 3). These values are quite similar to those reported in the literature.11,37 However, the binding affinity of the ADATMS13 mutants, that is, del6aa, Arg659Ala, and Arg660Ala to GST-VWF73 were decreased by approximately 7-, 2.3-, and 3.4-fold, respectively (Figure 6A; Table 3). No binding defect to GST-VWF73 was observed with the mutants Tyr661Ala, Gly662Ala, Glu663Ala, and Glu664Ala when compared with wild-type ADAMTS13 (Figure 6A; Table 3), which is consistent with the kinetic analyses by rF-VWF73 assay. As expected, the binding affinities between ADAMTS13 spacer mutants and multimeric VWF immobilized were not significantly reduced compared with that of wild-type ADAMTS13 (Figure 6B; Table 3), which is consistent with the previous findings showing that multiple binding sites exist between the C-terminus of ADATMS13 and the multimeric VWF immobilized. These results also reassured that the overall structure of the C-terminal domains of ADAMTS13 was normal despite the deletion or mutations generated in the spacer domain. The data also suggest that not all binding events between ADAMTS13 and VWF are necessary or will result in VWF proteolysis.
Binding of ADATMS13 and mutants to GST-VWF73 and multimeric vWF
| Constructs . | GST-VWF73 . | Multimeric VWF . | ||||
|---|---|---|---|---|---|---|
| BMax . | KD (×10−6M) . | BMax/KD (×103M−1) . | BMax . | KD (×10−6M) . | BMax/KD (×103M−1) . | |
| WT | 0.38 ± 0.01 | 28.4 ± 2.4 | 13.3 | 0.59 ± 0.02 | 12.8 ± 1.2 | 45.9 |
| del6aa | 0.46 ± 0.04 | 245.7 ± 45.7 | 1.9 | 0.44 ± 0.02 | 9.8 ± 1.6 | 45.2 |
| R659A | 0.39 ± 0.01 | 66.0 ± 26.3 | 5.9 | 0.38 ± 0.02 | 7.1 ± 1.3 | 53.8 |
| R660A | 0.40 ± 0.04 | 103.6 ± 26.3 | 3.9 | 0.57 ± 0.01 | 12.1 ± 1.6 | 47.2 |
| Y661A | 0.37 ± 0.01 | 35.5 ± 3.8 | 10.5 | 0.52 ± 0.03 | 12.3 ± 2.3 | 43.0 |
| G662A | 0.37 ± 0.01 | 25.2 ± 3.2 | 14.6 | 0.51 ± 0.02 | 9.7 ± 1.0 | 52.8 |
| E663A | 0.39 ± 0.01 | 32.3 ± 2.7 | 12.2 | 0.40 ± 0.02 | 10.6 ± 1.3 | 38.2 |
| E664A | 0.42 ± 0.02 | 47.6 ± 8.4 | 8.9 | 0.46 ± 0.03 | 10.2 ± 2.2 | 45.4 |
| Constructs . | GST-VWF73 . | Multimeric VWF . | ||||
|---|---|---|---|---|---|---|
| BMax . | KD (×10−6M) . | BMax/KD (×103M−1) . | BMax . | KD (×10−6M) . | BMax/KD (×103M−1) . | |
| WT | 0.38 ± 0.01 | 28.4 ± 2.4 | 13.3 | 0.59 ± 0.02 | 12.8 ± 1.2 | 45.9 |
| del6aa | 0.46 ± 0.04 | 245.7 ± 45.7 | 1.9 | 0.44 ± 0.02 | 9.8 ± 1.6 | 45.2 |
| R659A | 0.39 ± 0.01 | 66.0 ± 26.3 | 5.9 | 0.38 ± 0.02 | 7.1 ± 1.3 | 53.8 |
| R660A | 0.40 ± 0.04 | 103.6 ± 26.3 | 3.9 | 0.57 ± 0.01 | 12.1 ± 1.6 | 47.2 |
| Y661A | 0.37 ± 0.01 | 35.5 ± 3.8 | 10.5 | 0.52 ± 0.03 | 12.3 ± 2.3 | 43.0 |
| G662A | 0.37 ± 0.01 | 25.2 ± 3.2 | 14.6 | 0.51 ± 0.02 | 9.7 ± 1.0 | 52.8 |
| E663A | 0.39 ± 0.01 | 32.3 ± 2.7 | 12.2 | 0.40 ± 0.02 | 10.6 ± 1.3 | 38.2 |
| E664A | 0.42 ± 0.02 | 47.6 ± 8.4 | 8.9 | 0.46 ± 0.03 | 10.2 ± 2.2 | 45.4 |
BMax indicates the maximal binding; KD is the dissociation constant; and M is molar concentration. R, Y, G and E are single letter abbreviations of amino acids argine, tyrosine, glycine and glutamic acid.
Binding of ADAMTS13 and mutants to immobilized VWF73 and multimeric VWF. The microtiter plate was coated without (nonspecific binding controls) or with 100 μL of purified GST-VWF73 (2.0 μg/mL; panel A) or plasma-derived VWF (12.5 μg/mL; panel B). After being blocked with 2.5% BSA in 20mM Tris-HCl, 150mM NaCl for 30 minutes, 100 μL of wild-type ADAMTS13 or mutants at the concentration indicated diluted with PBS containing 10mM EDTA, 0.5% BSA, and 0.05% Tween 20 were added for 2 hours. The plate was washed 3 times, and the bound wild-type (WT) ADAMTS13 and mutants were detected by anti-V5 IgG, peroxidase conjugated (1:1000). The data in both panels A and B represent the means of 3 independent experiments. The apparent dissociation constants (KD; app) were determined by fitting the data into the Michaelis-Menten equation with SigmaPlot software.
Binding of ADAMTS13 and mutants to immobilized VWF73 and multimeric VWF. The microtiter plate was coated without (nonspecific binding controls) or with 100 μL of purified GST-VWF73 (2.0 μg/mL; panel A) or plasma-derived VWF (12.5 μg/mL; panel B). After being blocked with 2.5% BSA in 20mM Tris-HCl, 150mM NaCl for 30 minutes, 100 μL of wild-type ADAMTS13 or mutants at the concentration indicated diluted with PBS containing 10mM EDTA, 0.5% BSA, and 0.05% Tween 20 were added for 2 hours. The plate was washed 3 times, and the bound wild-type (WT) ADAMTS13 and mutants were detected by anti-V5 IgG, peroxidase conjugated (1:1000). The data in both panels A and B represent the means of 3 independent experiments. The apparent dissociation constants (KD; app) were determined by fitting the data into the Michaelis-Menten equation with SigmaPlot software.
Discussion
ADAMTS13 cleaves VWF precisely at the scissile bond (Tyr1605-Met1606) on the central A2 domain.6,7 This remarkable substrate specificity is largely achieved by the extensive interactions between VWF and C-terminal noncatalytic domains of ADAMTS13.12-15 We11 and others12-15 have previously demonstrated that the metalloprotease domain alone is not sufficient to cleave VWF substrate at a significant rate. Additions of the disintegrin domain, the first TSP1 repeat, the Cys-rich domain, and the spacer domain gradually restore proteolytic activity toward both peptidyl substrates and multimeric VWF under static/denaturing conditions.11-15 Therefore, there appeared to be a linear correlation between the proteolytic activity and the number of the proximal C-terminal domains of ADAMTS13 being sequentially added.11,13 An internal deletion of either disintegrin domain or the first TSP1 repeat or disintegrin/TSP1 from the construct MDTCS retains some proteolytic activity toward GST-VWF73 but has minimal or no activity toward multimeric VWF under denaturing conditions.11,16
A recent study by de Groot et al38 has identified several amino acid residues within the disintegrin domain of ADAMTS13 (ie, Arg349, Leu350, and Val352) that may be required for proteolytic cleavage of VWF substrates under static conditions. Moreover, there appeared to be a role of the middle and distal C-terminal domains of ADAMTS13 in recognition of globular VWF under more physiologic conditions.27,39 Direct binding interactions between a C-terminal fragment containing TSP1 fifth and eighth repeats and/or CUB domains and VWF can be demonstrated by the use of surface plasmon resonance27,39 or by competitive inhibition39 or immunoprecipitation assay.40 This fragment appears to specifically bind the C-terminus of VWF, that is, D4CK domains.39 Disruption of the interactions between VWF and ADAMTS13 by purified C-terminal fragment of VWF39 or by purified C-terminal fragment of ADAMT1327 dramatically reduced the cleavage of multimeric VWF by ADAMTS13 under fluid shear stress. These fragments, however, have no inhibitory activity toward the cleavage of predenatured VWF by ADAMTS13 under static conditions.27,39 Collectively, these results suggest that multiple domain-domain interactions are necessary for proteolytic cleavage of VWF by ADAMTS13 under more physiologic conditions.
Despite the possible involvement of multiple C-domains of ADAMTS13 for function, the spacer domain appears to play a critical role in recognition of the central A2 domain of soluble VWF or cell-bound UL-VWF under various static and fluid shear conditions. For example, the C-terminal truncated ADAMTS13 variants lacking the spacer domain (ie, constructs MDT and MDTC) exhibited a dramatic reduction in proteolytic activity toward the peptidyl substrates, that is, GST-VWF73 or FRETS-VWF73, and multimeric VWF under denaturing conditions14,15 as well as UL-VWF anchored on endothelial cell surface.17 The construct MDTC demonstrated very low proteolytic activity toward guanidine-HCl denatured VWF even at the 10-fold physiologic concentration.16 In the present study, we show that the deletion of a hexapeptide (Arg659-Glu664) from the spacer domain dramatically reduced the efficiency cleaving peptidyl substrates (Figure 2), predenatured multimeric VWF (Figure 3A), and native VWF after being exposed to fluid shear stress (Figure 3B), as well as UL-VWF anchored on endothelial cells (in the absence and the presence of fluid shear stress; Figure 3C-D). Such a dramatic loss of ADAMTS13 activity in construct del6aa appeared to be directly related to the reduced binding affinity toward the central VWF-A2 domain (Figure 6A). The binding interactions between the other C-terminal domains of ADAMTS13 and the other parts of multimeric VWF remain intact (Figure 6B). These results suggest that despite the multiple interactions that occur between VWF and ADATMS13, the high-affinity binding of the spacer domain to the central A2 domain of VWF appears to be necessary for proteolytic cleavage of VWF by ADAMTS13 under various conditions.
The importance of the spacer domain and the amino acid residues Arg659, Arg660, and Tyr661 in the spacer domain in substrate recognition was supported by the site-directed mutagenesis results (Figures 1,Figure 2–3) and the concentration-dependent inhibition of proteolytic cleavage of soluble VWF under fluid shear stress by ADAMTS13 with purified ADAMTS13 spacer domain (Figure 4) or synthetic peptides derived from the ADAMTS13 spacer domain (Figure 5). Kinetic analyses show that the spacer mutants (del6aa, Arg659Ala, and Arg660Ala) exhibit dramatic reduction in catalytic efficiency toward the peptidyl substrates. These mutants also had reduced proteolytic cleavage of predenatured VWF, native multimeric VWF exposed to shear stress, and cell-bound UL-VWF under static and fluid shear conditions (Figures 2–3). The difference observed for the mutant Tyr661Ala in cleaving various substrates (Figures 2–3) suggests the differential function of this amino acid residue in substrate recognition under various conditions. However, the assay sensitivity and kinetics may also contribute to the variation observed.
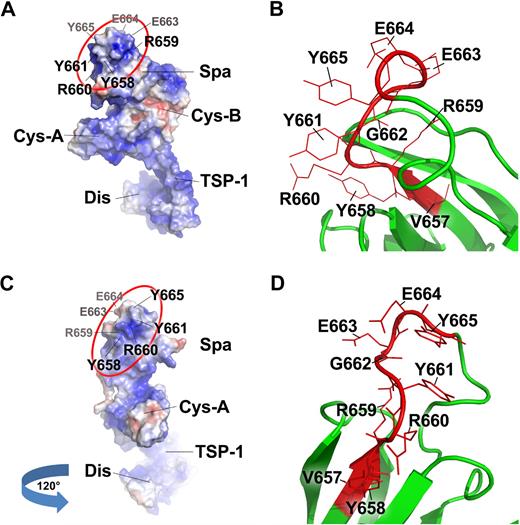
Recent data from Akiyama et al41 have shown that a substitution of both tyrosine residues at the positions of 661 and 665 with glutamine (Tyr661Gln/Tyr665Gln) results in a dramatic reduction in proteolytic cleavage of FRETS-VWF73, suggesting a synergistic role of the residues Tyr661 and Tyr665 in substrate recognition. Crystal structure of ADAMTS13-DTCS fragment demonstrates that the amino acid residues Arg659, Arg660, Tyr661, and Tyr665 form a surface loop facing the solvent (Figure 7; supplemental Figure 2).41 This loop may directly interact with the central A2 domain, presumably the C-terminal end of VWF73 peptide (Gly1660-Arg1668) as the deletion of this C-terminal end from the VWF73 peptide resulted in an approximately 20-fold reduction of the cleavage rates by wild-type ADAMTS13.13 Such a rate reduction appears to be similar to that after the deletion of ADAMTS13 spacer domain11,13 or the deletion of the amino acid residues (Arg659-Glu664) from the ADAMTS13 spacer domain (Table 2).
Surface representation of ADAMTS13-DTCS and close view of the Tyr658-Tyr665 motif in the spacer domain. Panels A and C are the surface representations of ADAMTS13-DTCS fragment on the basis of the crystal structure reported by Akiyama et al.41 The amino acids facing away are labeled in gray, whereas those facing toward are in black. Panels B and D are the close views of the motif (Tyr658-Tyr665) within which the residues Arg659, Arg660, Tyr661 are located. These residues may directly interact with the central A2 domain of VWF. The Gly662 is buried, but Glu663 and Glu664 point to the opposite direction of the residues Arg659, Arg660, Tyr661 (also see the video in supplemental Figure 2). R, Y, G, and E are single-letter abbreviations of the amino acids arginine, tyrosine, glycine, and glutamic acid.
Surface representation of ADAMTS13-DTCS and close view of the Tyr658-Tyr665 motif in the spacer domain. Panels A and C are the surface representations of ADAMTS13-DTCS fragment on the basis of the crystal structure reported by Akiyama et al.41 The amino acids facing away are labeled in gray, whereas those facing toward are in black. Panels B and D are the close views of the motif (Tyr658-Tyr665) within which the residues Arg659, Arg660, Tyr661 are located. These residues may directly interact with the central A2 domain of VWF. The Gly662 is buried, but Glu663 and Glu664 point to the opposite direction of the residues Arg659, Arg660, Tyr661 (also see the video in supplemental Figure 2). R, Y, G, and E are single-letter abbreviations of the amino acids arginine, tyrosine, glycine, and glutamic acid.
It remains to be elucidated, however, how ADAMTS13 spacer domain gains the access to the central A2 domain of the UL-VWF polymers anchored on endothelial cells. The newly released UL-VWF polymers from human endothelial cells after histamine stimulation appeared to be highly susceptible to ADATMS13 whether in the presence33,34 or in the absence17,36 of fluid shear stress. ADAMTS13 variant truncated after the eighth TSP1 repeat (construct delCUB) or the spacer domain (construct MDTCS) was able to cleave the cell-bound UL-VWF as efficiently as the wild-type ADAMTS13,17,18 but the further removal of the Cys-rich and spacer domain17,18 or deletion of the motif Arg659-Glu664 from the spacer domain or a substitution of residues Arg659, Arg660, and Tyr661 with alanine (Figure 3C-D) markedly reduced the proteolytic activity toward the cell-bound UL-VWF polymers, suggesting that the ADAMTS13 spacer domain, particularly the residues Arg659, Arg660, and Tyr661, play a role in recognition of cell-bound UL-VWF polymers.
The importance of the ADAMTS13 spacer domain, particularly the amino acid residues Arg659, Arg660, and Tyr661 in substrate recognition in vivo, is highlighted by the results of antibody mapping. Luken et al22,24 have shown that anti-ADAMTS13 IgG isolated from several patients with acquired idiopathic TTP directly targets against the amino acid residues between Val657 and Gly666, which contains the triple peptide (Arg-Arg-Tyr). However, the role of this triple peptide in substrate recognition does not undermine the importance of the other C-terminal noncatalytic domains of ADAMTS13. For instance, the precise engagement of amino acid residues Arg349, Leu350, and Val352 in the disintegrin domain with VWF-A2 domain38 and interactions between the VWF-D4CK domain with the more distal C-terminal domains of ADAMTS1339 also may play a role for productive cleavage of multimeric VWF in solution under physiologically relevant conditions. The results from genetic studies in mice and humans may further support the role of more distal C-terminal domain for ADATMS13 function.
For example, Banno et al42 have reported that a naturally occurring mutation in certain strains of mice that truncates murine ADATMS13 after the sixth TSP1 repeat results in significantly reduced efficacy for systemic antithrombosis.42 A deletion mutation in the TSP1 sixth repeat of ADAMTS13 was identified in a family with hereditary TTP.43 Furthermore, a homozygous mutation (W688X) found in a patient with hereditary TTP, which truncates ADAMTS13 4 amino acids after the spacer domain, was reported to have comparable activity to catalyze VWF73 peptides and predenatured VWF with wild-type ADAMTS13 but exhibited much less activity in cleaving native VWF under fluid shear stress.44 Finally, autoantibodies against the TSP1 second to eighth repeats and the CUB domains of ADAMTS13 were identified in many patients with acquired TTP, although the functional consequence of these anti-ADAMTS13 IgGs remains to be determined.20
We conclude that the amino acid residues Arg659, Arg660, and Tyr661 in the ADAMTS13 spacer domain play a critical role in recognition and proteolytic cleavage of peptidyl substrates, soluble VWF, and cell-bound VWF polymers under both static and fluid shear conditions. The reduced proteolytic activity of the spacer mutants in many cases is associated with the impaired binding interactions with the central A2 domain of VWF. Our findings may provide novel insight into the structure-function relationship of ADAMTS13 and shed some light on the pathogenesis of TTP and perhaps other arterial thrombotic disorders related to compromised VWF proteolysis by ADAMTS13.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Sriram Krishnaswamy at The Children's Hospital of Philadelphia for providing recombinant FVIII, Dr John Owen at Wake Forest University for rF-VWF73 construct, and Dr Douglas Cines (University of Pennsylvania Medical Center, Philadelphia) for providing human umbilical cord to isolate endothelial cells. We also thank Erica Falls (The Children's Hospital of Philadelphia) for reading the manuscript before submission.
This work was supported in part by National Institutes of Health grants (R01HL079027 and P50HL30954), American Heart Association-Established Investigator (EIA), and Bayer Hemophilia Awards.
National Institutes of Health
Authorship
Contribution: S.-Y.J. and X.L.Z. designed and performed research, analyzed results, and wrote the paper; and C.G.S. performed some experiments and revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: X. Long Zheng, MD, PhD, Department of Pathology and Laboratory Medicine, The Children's Hospital of Philadelphia and The University of Pennsylvania Medical Center, 816G Abramson Research Center, 34th St and Civic Center Blvd, Philadelphia, PA 19104; e-mail: zheng@email.chop.edu.

![Figure 2. Cleavage of GST-VWF73 and rF-VWF73 by wild-type ADAMTS13 and various mutants. Purified GST-VWF73 (50nM) was incubated with the wild-type ADAMTS13 or mutants (2.5nM) in the absence (−) or presence (+) of EDTA (20mM) at 37°C for 1 hour (A) and 3 hours (B). The proteolytic cleavage product (open arrow) was determined by Western blotting. *The preexisting nonspecific bands in the purified GST-VWF73 substrate. (C) Purified recombinant rF-VWF73 at various concentrations (0-12μM) was incubated at 37°C with wild-type (WT) ADAMTS13 or mutants (2.5nM). The rate of fluorescent generation was determined at the ex 485 nm and em 535 nm. The maximal rate of fluorescent generation (Vmax) with D[b]-Phe[b]-Pro[b]-Arg[b]-chloromethylketone-treated chymotrypsin (20nM) that cleaves specifically at the methionyl bond was used for a calibration. The relative mean product formation (nanomoles per second), compared with chymotrypsin at the same concentration, is expressed as a function of the substrate concentrations (micromole). The data represent the means ± SD (n = 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/11/10.1182_blood-2009-07-235101/4/m_zh89991049910002.jpeg?Expires=1769388734&Signature=rXNCbpw1s01C6W-MOiuEq3PemK~hEq7EMqf8D9nOba-5PCkgnN94Ik9lYkOEpusTRrdUIXRuapJVOWVfhZhxfJlkkx2fsvYpbsOjx8oLUDzPQqe-i~0BAJttRp-e2-b~-GBLhKUiK3Qz0rU-PQ7KaMrMxycUU5XPlZHc~SwUA5UW1jPo9-JNlJCZ~qaoH615FW0kYMtU~FjtxMgbe2Eu~mmrS5YC68QcjlQsB8R-vnsloCUiCZtKHrVGGymcTsr-D4C~IiVwc-B3qxu4~hmW9mgyictywT2yOMC5Z6am-62hF9ZU8hREWQmYDjkWp5qJIlIh45vV4wIEV8pkpyIehw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal