Abstract
Alternative splicing of pre-mRNAs is a central process to the generation of proteome complexity. However, many alternative mRNA isoforms carry premature termination codons (PTCs) rendering them possible targets for the nonsense-mediated mRNA decay (NMD) pathway. The F11 gene consists of 15 exons spanning approximately 23 kb on chromosome 4q35 and codes for coagulation factor XI (FXI), a 160-kDa dimeric zymogen composed of 4 apple domains and a serine protease domain. Here, we characterized the F11 splicing pattern in human liver and platelets identifying multiple in-frame and out-of-frame splicing events. Inhibition of NMD resulted in the up-regulation of all unproductively spliced F11 transcripts, thus providing evidence that these PTC-containing mRNAs are under the control of NMD. Among in-frame alternatively spliced transcripts, the one skipping exons 6 and 7 would lead to the synthesis of a FXI protein lacking 1 apple domain (FXI-Δ6/7). Ex vivo expression in mammalian cells demonstrated that FXI-Δ6/7 is mostly retained intracellularly, and secreted only in low amounts. Traces of this FXI isoform were detectable in human plasma. Our results suggest that the coupling of alternative splicing and NMD may play a role in regulating F11 expression, and point to the existence of a novel FXI isoform.
Introduction
Factor XI (FXI) is a trypsin-like serine protease required for normal hemostasis. It is present in plasma in its precursor form noncovalently bound to high-molecular-weight kininogen. This zymogen is unique among plasma coagulation enzymes because it exists as a homodimer of 160 kDa composed of 2 identical polypeptide chains held together by both noncovalent interactions and an intersubunit disulfide bond. Each monomer is composed of a light chain, harboring a serine protease catalytic domain, and a heavy chain, consisting of 4 tandem repeats of approximately 90 residues (termed apple domains, A1-A4, from the N-terminus), which plays critical roles in protein-protein interactions.1
FXI can be converted to activated FXI by thrombin, activated factor XII, or by autoactivation. Cleavage at the Arg369-Ile370 bond in each monomer produces an N-terminal noncatalytic heavy chain and a disulfide-linked C-terminal trypsin-like light chain with a typical catalytic triad His413-Asp462-Ser557. Once activated, FXI contributes to the contact phase of blood coagulation through limited proteolysis of its physiologic substrate, factor IX.2-4
Human FXI is produced primarily by hepatocytes, although low levels of the transcript are also detectable in platelets, blood mononuclear cells, and granulocytes.5 Well-washed platelets contain small amounts of FXI, accounting for approximately 0.5% of the total FXI activity in blood.6,7 Three independent groups have reported the partial purification from platelet extracts of a 220-kDa protein (55 kDa in reducing conditions), recognized by anti-FXI polyclonal antibodies and showing FXI activity.7-9 The site of synthesis and nature of this platelet variant still remain a matter of debate.6,10 The existence of a platelet transcript, originating from the skipping of exon 5, was thought for a long time to account for the “platelet-specific” FXI isoform,10 even though the not-in-frame nature of this exon skipping, and the subsequent introduction of a premature termination codon (PTC), was not easily reconcilable with translation after exon 5. Indeed, subsequent studies did not confirm the presence of an alternatively spliced transcript lacking exon 5 in human platelets, in which it was only possible to detect full-length correctly spliced FXI mRNA.5,11
Hereditary FXI deficiency is rare in most populations (estimated prevalence of 1 in 1 million) but is particularly common among Ashkenazi and Iraqi Jews, in whom 2 prevalent mutations (Glu117stop and Phe283Leu) account for 98% of alleles. To date, more than 180 mutations, all located in the FXI gene (F11), have been identified, and approximately 30% of them are predicted to cause the introduction of a PTC.12-14
It has been estimated that one-third of hereditary genetic diseases, as well as many forms of cancer, are caused by mutations that lead to the generation of transcripts bearing a PTC (PTC+ transcripts).15 The evolutionarily conserved posttranscriptional mechanism by which PTC+ transcripts are selectively detected and degraded has been called nonsense-mediated mRNA decay (NMD), and is responsible for the elimination of aberrant PTC+ mRNAs generated as a consequence of routine errors in gene expression (eg, inefficient or faulty splicing and errors introduced by RNA polymerase II).16-20 Furthermore, NMD, through a mechanism called regulated unproductive splicing and translation, has been experimentally shown to physiologically regulate the expression of a wide variety of genes in many organisms from yeast to humans.21-23
Concerning the F11 gene, which is located on chromosome 4q35 and is composed of 15 exons (http://genome.ucsc.edu; March 2006 release of the human genome, hg18),24 our group demonstrated the selective degradation of PTC+ mRNAs in FXI-deficient patients bearing the Glu117stop mutation.25 Moreover, we recently characterized 2 splicing mutations (325G>A and IVS6+3A>G), both resulting in a frameshift introducing a PTC, which were expected to trigger NMD. Real-time reverse-transcription polymerase chain reaction (RT-PCR) assays, performed to study the effect of these mutations on mRNA stability, revealed the occurrence of multiple aberrant splicing events involving the wild-type F11 pre-mRNA, suggesting the existence of a complex pattern of alternative splicing (AS).26
In this work, we demonstrate that multiple aberrant splicing events involving the wild-type FXI pre-mRNA normally occur in human liver and platelets and are physiologically down-regulated by NMD. Moreover, we show that at least one of the identified in-frame ASs gives rise to a novel, although minor, coagulation FXI isoform.
Methods
Human sample preparation
This study was approved by the Institutional Review Board of the University of Milan and was conducted in accordance with the Declaration of Helsinki. Samples were obtained from the analyzed persons after acquiring appropriate informed consent.
Liver samples were obtained from an Italian female patient with normal FXI levels in plasma, affected by Kasabach-Merrit syndrome, who received a liver transplantation. Three biopsies were taken from the healthy parenchyma of the removed liver.
Analysis of the F11 splicing pattern
Total RNA was isolated from HepG2 (human hepatocellular carcinoma) cells, platelets, and human liver using the RNAWIZ reagent (Applied Biosystems).
Random nonamers and ImProm-II Reverse Transcriptase System (Promega) were used to perform first-strand complementary DNA (cDNA) synthesis starting from 1 μg of total RNA, according to the manufacturer's instructions. Of a total of 20 μL, 1 μL was used as template for PCR reactions with gene-specific primers. PCRs were carried out under standard conditions using either the FastStart Taq DNA Polymerase (Roche Diagnostics) or the AmpliTaq Gold DNA polymerase (Applied Biosystems) on a Mastercycler EPgradient (Eppendorf AG).
The PCR products were cloned into pGEM-T easy vector (Promega) by the TA cloning procedure,29 and selected recombinant plasmids were sequenced using the BigDye Terminator Cycle Sequencing Kit, Version 1.1, and an automated ABI-3130XL DNA sequencer (Applied Biosystems).
All primers used in these and in the following experiments are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
NMD analysis
HepG2 cells were cultured in RPMI 1640 (EuroClone) supplemented with sodium pyruvate (1mM; Sigma-Aldrich), antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin; EuroClone), glutamine (2mM; EuroClone), and 10% fetal bovine serum (HyClone), and grown at 37°C in a humidified atmosphere of 5% CO2 and 95% air, according to standard procedures.
HepG2 were plated at a density of 5.5 × 106 per 10-cm dish and, after 72 hours, treated for 8 hours with puromycin (300 μg/mL; Sigma-Aldrich), a drug that inhibits NMD by prematurely terminating translation.
After the treatment, cells were washed twice with phosphate-buffered saline (PBS) and total RNA was extracted and reverse-transcribed as described in “Analysis of the F11 splicing pattern.” Real-time PCR assays were carried out under standard conditions using the iQ5 real-time PCR detection system (Bio-Rad) and the iQ SYBR Green Supermix kit (Bio-Rad). In each reaction, 1.5 μL of cDNA from treated or untreated HepG2 cells was used. Variations in the expression levels of the alternatively spliced transcripts on puromycin treatment were quantified by the ΔΔCT method,30 using as reference a transcript not subjected to NMD modulation (ie, the mRNA for human connexin 32, whose coding sequence is all contained in a single exon).
Quantitation of the Δ6/7 transcript in human liver and platelets
Absolute quantitative real-time RT-PCR was performed to determine the physiologic expression levels of the splice variant lacking exons 6 and 7 in human liver and platelets. The ratio between exon 6+7–containing and exon 6+7–lacking transcripts was calculated by measuring the absolute quantities of the 2 mRNAs. To this purpose, to generate appropriate standard curves, a pcDNA3/FXI-wt+Δ6/7 vector (containing 1 copy of the full-length FXI cDNA and 1 copy of the Δ6/7 splicing isoform) was constructed. In particular, a plasmid containing the FXI cDNA lacking exons 6 and 7 (pcDNA3/FXI-Δ6/7) was used as template in a PCR reaction with the primer couple FXIex3/4F and FXIex9R. To generate the pcDNA3/FXI-wt+Δ6/7 construct, the obtained PCR product was cloned into the EcoRV site of the pcDNA3/FXI vector (kindly provided by Dr A. Zivelin, Institute of Thrombosis and Hemostasis, Chaim Sheba Medical Center, Tel-Hashomer, Israel) by the TA cloning procedure.29 All PCR products were checked by direct sequencing.
Absolute standard curves (spanning at least 3 orders of magnitude) were constructed using serial dilutions of the pcDNA3/FXI-wt+Δ6/7 plasmid.
Expression and quantitation of recombinant Δ6/7 FXI
The deletion of exons 6 and 7 (270 bp) was introduced in pcDNA3/FXI by a modification of the QuickChange Site-Directed Mutagenesis Kit protocol (Stratagene), as described.31 The mutant plasmid pcDNA3/FXI-Δ6/7 was verified by sequencing the whole FXI cDNA insert as well as 200 bp of the flanking DNA on both sides of the cloning site. Large-scale plasmid preparations were obtained using the EndoFree Plasmid Maxi Kit (Sigma-Aldrich).
African green monkey kidney COS-1 cells were cultured in Dulbecco modified Eagle medium (DMEM; EuroClone) supplemented with 10% fetal bovine serum, antibiotics, and glutamine, according to standard procedures.
In each transfection experiment, an equal number of cells (400 000) were transiently transfected in 6-well plates with the Lipofectamine 2000 reagent (Invitrogen) and 4 μg of plasmid DNA (pcDNA3/FXI, or pcDNA3/FXI-Δ6/7, or equimolar amounts of both plasmids, ie, 2 μg each) following the manufacturer's instructions.
Twenty-four hours after transfection, cells were washed twice with DMEM lacking methionine and cysteine (MP Biomedicals) and incubated in 1.5 mL/well of methionine- and cysteine-free DMEM supplemented with 200 μCi (7.4 MBq) [35S]-labeled methionine and cysteine (Translable; MP Biomedicals), 10% dialyzed fetal calf serum (HyClone), 2mM l-glutamine, 2.5mM CaCl2, and 5 mg/mL bovine serum albumin. Labeled proteins were immunoprecipitated from conditioned media and lysates after 16 hours. Briefly, conditioned media were collected in prechilled tubes containing a protease inhibitor mixture (Complete; Roche), and centrifuged to remove cell debris. To obtain cell lysates, cells were washed 3 times with prechilled PBS and incubated for 1 hour on ice with lysis buffer (1× PBS, 1.5% Triton X-100, 1× Complete).
Cell lysates and media were added to 50 μg of goat anti–human FXI antibody (catalog no. GAFXI-IG) from Affinity Biologicals, previously adsorbed for 40 minutes at room temperature to 14 μL of magnetic bead-protein G (Dynal Biotech), and incubated for 1 hour at 4°C. Subsequently, beads were washed with 70 volumes of lysis buffer and resuspended in either reducing or nonreducing Laemmli loading buffer. The immunoprecipitated proteins were released from protein-coated beads by boiling for 5 minutes in loading buffer, and samples were analyzed by 6% and 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Gels were dried under vacuum at 80°C for 1 hour, exposed to a storage phosphor screen (GE Healthcare), and analyzed using a Typhoon 9200 phosphor imager and the ImageQuant software (GE Healthcare).
For Δ6/7 FXI measurements in conditioned media and in cell lysates, 24 hours after transfection, COS-1 cells were washed twice with PBS and cultured for additional 48 hours in 1 mL of serum-free medium supplemented with glutamine, antibiotics, and 5 mg/mL bovine serum albumin. For each experiment (performed 3 times in duplicate), a mock sample, with the empty pcDNA3 plasmid, was set up.
FXI antigen levels were evaluated by an enzyme-linked immunosorbent assay (ELISA) based on a goat anti–human FXI affinity-purified IgG as capture antibody and a goat anti–human FXI peroxidase-conjugated IgG as detecting antibody (Affinity Biologicals), both in conditioned media and in cell lysates. Standard curves were constructed with reference plasma diluted 1:100 to 1:6400 in Tris-buffered saline (50mM Tris, 150mM NaCl, pH 7.5).
FXI coagulant activity was measured in media (collected without protease inhibitor) by a one-stage method based on a modified partial thromboplastin time, using FXI-deficient plasma as substrate (Hemoliance). FXI levels were expressed in both tests as percentages of pooled plasma from 30 normal persons. The detection limits of the FXI functional and immunologic assays were 1% and 0.1%, respectively.
Western blotting analysis
Western blotting analysis on plasma samples, conditioned media, and cell lysates of COS-1–transfected cells was performed on immunoprecipitated proteins (obtained using either the polyclonal goat anti–human FXI and the mouse monoclonal anti–human FXI antibodies, catalog no. 001ER147; Enzyme Research Laboratories). Samples were resolved by nonreducing 6% SDS-PAGE and electroblotted onto 0.45-μm pore-size nitrocellulose membranes (Schleicher & Schuell) overnight at 100 mA by a Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). Membranes were soaked in a blocking solution (5% skim milk, 10mM Tris-HCl pH 7.4, 150mM NaCl, 0.1% Tween-20) for 1 hour at room temperature and incubated with goat anti–human FXI peroxidase-conjugated IgG (catalog no. GAFXI-HRP; Affinity Biologicals) diluted 1:500 in blocking solution at room temperature for 2 hours. Proteins were detected using a highly sensitive enhanced chemiluminescent substrate (SuperSignal West Dura Extended Duration Substrate; Pierce Chemical).
Isolation of the FXI Δ6/7 isoform
Commercial human FXI (Enzyme Research Laboratories) is purified from plasma of healthy donors by affinity chromatography using a monoclonal antibody anti–human FXI, not available to customers. Approximately 80 μg of FXI was separated under reducing condition on an 8% SDS-PAGE. Bands, visualized using Bio-Safe Coomassie (Bio-Rad), were excised from the gel for the matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry analysis (customized service from Nurex). Briefly, excised bands were destained with 50% acetonitrile in 5mM ammonium bicarbonate. Gel pieces were then dried for 30 minutes in acetonitrile and subjected to protein digestion by trypsin. Mass spectrometry analysis of peptides was performed by a MALDI micro MX (Micromass) equipped with a delayed extraction unit, according to the tuning procedures suggested by the manufacturer. Samples were loaded onto MALDI target using 2 μL of the tryptic digest mixed 1:1 with a solution of α-cyano 4-hydroxycinnamic acid (10 mg/mL in 40% acetonitrile, 0.1% vol/vol trifluoroacetic acid). Peak lists were generated with ProteinLynx Data Preparation using the following parameters: external calibration with lock mass, using mass 2465.1989 Da of adrenocorticotropic hormone; background subtract type adaptive combining all scans; deisotoping using a threshold of 5%. Peak lists were then sent to PLGS 2.2.5 and compared with the Swiss-Prot database (release 54.0 of January 2008), allowing 1 missed cleavage of the trypsin enzyme, peptide tolerance of 0.2 Da, and limiting the search to human proteins.
Results
Characterization of the F11 splicing pattern
A preliminary characterization of the splicing pattern of the F11 gene was performed starting from total RNA isolated from HepG2 cells. To this aim, a set of RT-PCR assays was designed to cover the majority of possible AS events (Figure 1A).
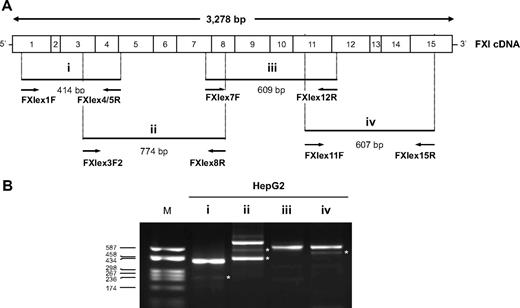
Global screening of the F11 splicing pattern. The existence of physiologic F11 alternatively spliced transcripts was first assayed in HepG2 cells. (A) Schematic representation of the FXI cDNA. The position of the primer couples used for each RT-PCR assay (indicated with i-iv), and the expected length of the corresponding amplification products are also indicated. (B) RT-PCR products obtained with each primer couple (i-iv) separated on a 2% agarose gel. Lane M indicates molecular weight marker (pUC8-HaeIII). *AS products whose existence was confirmed by at least one sequenced recombinant clone. No clone with an insert compatible with the length of the faint upper band in lane ii was found.
Global screening of the F11 splicing pattern. The existence of physiologic F11 alternatively spliced transcripts was first assayed in HepG2 cells. (A) Schematic representation of the FXI cDNA. The position of the primer couples used for each RT-PCR assay (indicated with i-iv), and the expected length of the corresponding amplification products are also indicated. (B) RT-PCR products obtained with each primer couple (i-iv) separated on a 2% agarose gel. Lane M indicates molecular weight marker (pUC8-HaeIII). *AS products whose existence was confirmed by at least one sequenced recombinant clone. No clone with an insert compatible with the length of the faint upper band in lane ii was found.
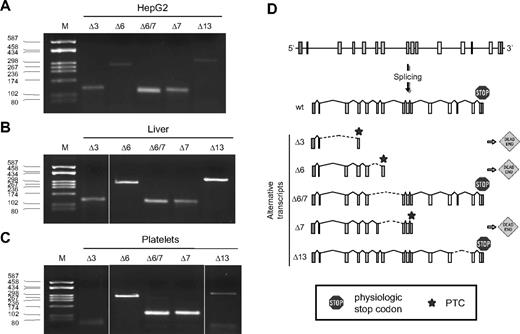
Multiple AS events were detected (Figure 1B, bands marked with an asterisk). In particular, RT-PCR products, on the basis of their electrophoretic mobility, were compatible with an alternatively spliced product lacking exon 3 (assay i), the skipping of exons 4 (or 6) and 6+7 (assay ii), and a transcript lacking exon 13 (assay iv).
To confirm these predicted skipping events, amplified products were TA-cloned into a plasmid vector and 20 selected recombinant clones for each fragment (assays i-iv) were sequenced: 5 skipping events, involving exons 3, 6, 7, 6+7, and 13 were identified. RT-PCR assays were hence designed to specifically detect each AS product (supplemental Table 1) and performed on RNA from human liver, platelets, and HepG2 cells (Figure 2A-C). All analyzed AS events were confirmed. The skipping of exons 3, 6, and 7 would determine the introduction of a PTC, located in exons 4, 7, and 10, respectively, whereas, the skipping of exons 6+7 and 13 does not alter the reading frame (Figure 2D).
Characterization of F11 alternatively spliced transcripts. AS of the F11 pre-mRNA was analyzed in HepG2 cells (A), human liver (B), and platelets (C) by RT-PCR using primer couples specific for each identified splicing isoforms (Δ3-Δ13). Lane M indicates molecular weight marker (pUC8-HaeIII). (D) Schematic representation of the F11 gene and of its splicing pattern. Boxes represent exons; lines, introns; broken lines, splicing events; hatched lines, ASs; dead-end sing, out-of-frame splicings leading to the introduction of a PTC (star); and stop sign, physiologic stop codons.
Characterization of F11 alternatively spliced transcripts. AS of the F11 pre-mRNA was analyzed in HepG2 cells (A), human liver (B), and platelets (C) by RT-PCR using primer couples specific for each identified splicing isoforms (Δ3-Δ13). Lane M indicates molecular weight marker (pUC8-HaeIII). (D) Schematic representation of the F11 gene and of its splicing pattern. Boxes represent exons; lines, introns; broken lines, splicing events; hatched lines, ASs; dead-end sing, out-of-frame splicings leading to the introduction of a PTC (star); and stop sign, physiologic stop codons.
Up-regulation of alternatively spliced transcripts by NMD inhibition
Given that 3 of 5 F11 skipping events gave rise to a frameshift, eventually leading to the introduction of a PTC, the hypothesis that these alternatively spliced transcripts were targets of NMD was tested by pharmacologic inhibition of this pathway. In particular, protein synthesis inhibitors, such as puromycin, have been proven to block the NMD pathway and are widely used for this purpose.32
Real-time RT-PCR assays with exon-skipping specific primers were performed on RNA extracted from puromycin-treated HepG2 cells (Figure 3A). These experiments demonstrated that the expression level of all unproductively spliced F11 transcripts was up-regulated after treatment with puromycin.
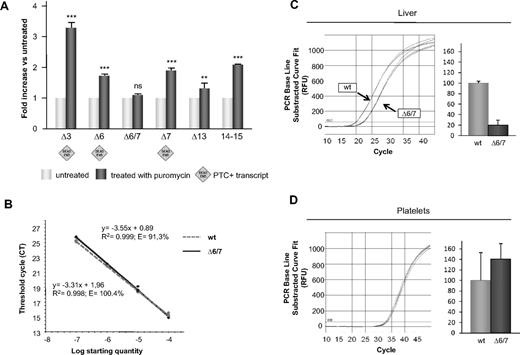
Impact of pharmacologic inhibition of the NMD pathway on F11 splicing and quantification of the alternative transcript lacking exons 6+7. (A) The NMD pathway was blocked in HepG2 cells using puromycin (“NMD analysis”). Variations in the expression levels of splicing isoforms (compared with nontreated samples, set as 1) were quantified using the ΔΔCT method; in all experiments, connexin 32 was used as reference. Bars represent mean ± SEM of 3 independent experiments, each performed in triplicate on different days in different cell batches. The results were analyzed by unpaired t test. **P < .01; ***P < .001; ns indicates not significant. (B) Standard curves for both PCR assays (ie, for the amplification of the wild-type transcript and of the one lacking exons 6+7) are generated from dilution series constructed from the fusion plasmid pcDNA3/FXI-wt+Δ6/7. The quantification of wild-type and Δ6/7 FXI transcripts in liver (C) and platelets (D) was performed by real-time RT-PCR. Amplification curves and schematic visualization of the abundance of the 2 transcripts in liver and platelets are represented. The absolute quantities of the mRNA containing exon 6+7 (wild-type) was set as 100%. Bars represent mean ± SD of 3 independent experiments, each performed in triplicate on different cDNA preparations.
Impact of pharmacologic inhibition of the NMD pathway on F11 splicing and quantification of the alternative transcript lacking exons 6+7. (A) The NMD pathway was blocked in HepG2 cells using puromycin (“NMD analysis”). Variations in the expression levels of splicing isoforms (compared with nontreated samples, set as 1) were quantified using the ΔΔCT method; in all experiments, connexin 32 was used as reference. Bars represent mean ± SEM of 3 independent experiments, each performed in triplicate on different days in different cell batches. The results were analyzed by unpaired t test. **P < .01; ***P < .001; ns indicates not significant. (B) Standard curves for both PCR assays (ie, for the amplification of the wild-type transcript and of the one lacking exons 6+7) are generated from dilution series constructed from the fusion plasmid pcDNA3/FXI-wt+Δ6/7. The quantification of wild-type and Δ6/7 FXI transcripts in liver (C) and platelets (D) was performed by real-time RT-PCR. Amplification curves and schematic visualization of the abundance of the 2 transcripts in liver and platelets are represented. The absolute quantities of the mRNA containing exon 6+7 (wild-type) was set as 100%. Bars represent mean ± SD of 3 independent experiments, each performed in triplicate on different cDNA preparations.
Conversely, the in-frame Δ6/7 transcript was insensitive to NMD inhibition. The slight up-regulation of Δ13 transcript could be instead explained by the association of exon 13 skipping with other upstream out-of-frame skipping events. This hypothesis was confirmed by the results of RT-PCR assays performed by combining Δ6 (FXIex5/7F) or Δ7 (FXIex6/8F) specific forward primers with a Δ13 (FXIex12/14R) specific reverse primer (supplemental Figure 1; supplemental Table 1), which demonstrated that indeed the skipping of exon 13 is associated, at least in some transcripts, with the skipping of exons 6 or 7. These data were further corroborated by the results of real-time RT-PCR on the mRNA region corresponding to the last 2 exons (14 and 15, Figure 3A last bars), which should be present in each splicing variant and therefore should reflect the effect of “all” AS events. The observed up-regulation also suggests that NMD might have a role in the regulation of F11 expression.
Quantitation of the Δ6/7 transcript
Among the 2 in-frame alternatively spliced transcripts, the one skipping exons 6+7 (FXI-Δ6/7) is not affected by NMD and hence may lead to the synthesis of a FXI isoform with a predicted molecular weight of 140 kDa and differing from the wild-type by the lack of one apple domain (ie, the second half of the A2 and the first half of A3 domain).
Absolute expression levels of the alternative transcript lacking exons 6+7 in physiologic human samples (liver and platelets) were obtained by real-time RT-PCR. Two specific assays were designed to selectively amplify either the Δ6/7 or the wild-type mRNA (supplemental Table 1). To generate standard curves for both PCR assays, a new plasmid containing both amplicons (pcDNA3/FXI-wt+Δ6/7) was created and used to prepare dilution series (Figure 3B). These experiments showed that, whereas in liver FXI-Δ6/7 mRNA levels were 19% of the wild-type transcript, in platelets the expression levels of the 2 isoforms were comparable, even though much lower than those detected in the liver (∼ 1:1000; Figure 3C-D).
In vitro expression of wild-type and Δ6/7 FXIs
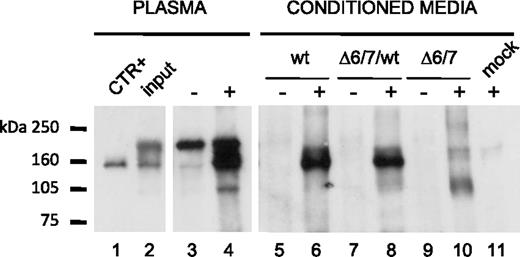
To evaluate whether Δ6/7 FXI is synthesized, assembled, and secreted, wild-type and Δ6/7 FXIs were transiently expressed in COS-1 cells (not expressing FXI). As shown in Figure 4, immunoprecipitated Δ6/7 FXI was detectable both intracellularly and extracellularly under nonreducing (top panels) and reducing (bottom panels) conditions as a band with an apparent molecular weight of approximately 140 and 70 kDa, respectively. In particular, in conditioned media from cells coexpressing wild-type and Δ6/7 FXIs, run under nonreducing conditions, the band corresponding to the wt/wt homodimer was largely prevalent compared with the faint smear corresponding to the heterodimer and to the Δ6/7 homodimer. This could be the result of the combined effect of low efficiency in heterodimer formation and significantly reduced secretion of both homodimers and heterodimers containing Δ6/7 FXI.
Metabolic labeling of wild-type and Δ6/7 mutant recombinant FXI in transiently transfected COS-1 cells. Twenty-four hours after transfection, cells were labeled with [35S]-cysteine and methionine mixture as described in “Expression and quantitation of recombinant Δ6/7 FXI.” Radiolabeled FXIs were immunoprecipitated from cell lysates and media using goat anti–human FXI. Immunoprecipitated FXI was analyzed on 8% reducing (bottom panel) and 6% nonreducing (top panel) SDS-PAGE and visualized by scanning phosphor image screens. The pcDNA3 empty vector was used as negative control (mock). + indicates presence of goat anti–human FXI antibody; −, absence of goat anti–human FXI antibody; and arrowheads, homodimeric molecules in nonreducing panels. In the same panels, the smears corresponding to the heterodimers and the Δ6/7 homodimers are indicated by square brackets.
Metabolic labeling of wild-type and Δ6/7 mutant recombinant FXI in transiently transfected COS-1 cells. Twenty-four hours after transfection, cells were labeled with [35S]-cysteine and methionine mixture as described in “Expression and quantitation of recombinant Δ6/7 FXI.” Radiolabeled FXIs were immunoprecipitated from cell lysates and media using goat anti–human FXI. Immunoprecipitated FXI was analyzed on 8% reducing (bottom panel) and 6% nonreducing (top panel) SDS-PAGE and visualized by scanning phosphor image screens. The pcDNA3 empty vector was used as negative control (mock). + indicates presence of goat anti–human FXI antibody; −, absence of goat anti–human FXI antibody; and arrowheads, homodimeric molecules in nonreducing panels. In the same panels, the smears corresponding to the heterodimers and the Δ6/7 homodimers are indicated by square brackets.
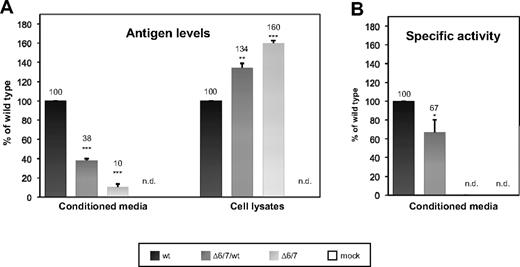
As a confirmation of the immunoprecipitation results, a quantitative analysis, based on ELISA measurements, was performed: Δ6/7 FXI was 10% and 38% of the wild-type protein (set as 100%; 70 ng/mL and 260 ng/mL vs 680 ng/mL, Figure 5A left) in media of cells expressing mutant FXI in the homozygous and heterozygous condition, respectively. The corresponding levels of immunoreactive FXI in cell lysates (Figure 5A right) were approximately 76 and 90 ng/mL versus 57 ng/mL. These results demonstrated that Δ6/7 FXI can be secreted, even though at reduced levels (Figure 5A left), and that unsecreted Δ6/7 FXI accumulates intracellularly (Figure 5A right).
Quantitation by ELISA and measurement of the specific activity of wild-type and Δ6/7 recombinant FXIs. pcDNA3/FXI, or pcDNA3/FXI-Δ6/7, or equimolar amounts of both plasmids (heterozygous condition) were transiently transfected in COS-1 cells. Equal numbers of cells and equal amounts of plasmids were used in transfection experiments, as described in “Expression and quantitation of recombinant Δ6/7 FXI.” (A) Antigen levels of recombinant FXI were measured in both conditioned media and the corresponding cell lysates using an ELISA assay. Bars represent relative concentrations of protein in media and cell lysates compared with the mean antigen level measured in the wild-type, set as 100%. Results are given as mean ± SEM. (B) The specific activities of recombinant proteins were determined by calculating the ratio between FXI activity (measured using a one-stage method based on a modified partial thromboplastin time) and FXI antigen levels. Bars represent mean ± SEM of 3 independent experiments, each performed in duplicate on different days in different cell batches. The mean value of wild-type FXI was set as 100%. The results were analyzed by unpaired t test. *P < .05; **P < .01; ***P < .001; n.d. indicates not detected. The detection limits of the FXI functional and immunologic assays were 1% and 0.1%, respectively.
Quantitation by ELISA and measurement of the specific activity of wild-type and Δ6/7 recombinant FXIs. pcDNA3/FXI, or pcDNA3/FXI-Δ6/7, or equimolar amounts of both plasmids (heterozygous condition) were transiently transfected in COS-1 cells. Equal numbers of cells and equal amounts of plasmids were used in transfection experiments, as described in “Expression and quantitation of recombinant Δ6/7 FXI.” (A) Antigen levels of recombinant FXI were measured in both conditioned media and the corresponding cell lysates using an ELISA assay. Bars represent relative concentrations of protein in media and cell lysates compared with the mean antigen level measured in the wild-type, set as 100%. Results are given as mean ± SEM. (B) The specific activities of recombinant proteins were determined by calculating the ratio between FXI activity (measured using a one-stage method based on a modified partial thromboplastin time) and FXI antigen levels. Bars represent mean ± SEM of 3 independent experiments, each performed in duplicate on different days in different cell batches. The mean value of wild-type FXI was set as 100%. The results were analyzed by unpaired t test. *P < .05; **P < .01; ***P < .001; n.d. indicates not detected. The detection limits of the FXI functional and immunologic assays were 1% and 0.1%, respectively.
Finally, to analyze the effects of the Δ6/7 deletion (corresponding to residues 145-234) on the functional properties of FXI, the specific activity of secreted FXI was calculated in conditioned media as the ratio between FXI:C and FXI:Ag levels. In supernatants of cells coexpressing wild-type and Δ6/7 FXI, the calculated specific activity was reduced (to ∼ 70%) compared with the wild-type one (Figure 5B), whereas in the case of cells transfected with the mutant construct alone, no specific activity was detectable.
Isolation of the Δ6/7 FXI isoform from human plasma
Western blotting experiments on samples immunoprecipitated from human plasma, using both a monoclonal (data not shown) and a polyclonal antihuman FXI antibody, revealed that trace amounts of Δ6/7 FXI isoform were also detectable in plasma, as suggested by the presence of a band with a molecular weight compatible with the lacking of one apple domain (Figure 6lane 4).
Immunoprecipitation of FXI Δ6/7 isoform from normal plasma. FXI present in plasma and in media of COS-1 cells transfected with pcDNA3/FXI, or pcDNA3/FXI-Δ6/7 was immunoprecipitated using both goat anti–human FXI antibody and mouse anti–human FXI (data not shown). Samples were analyzed on 6% nonreducing SDS-PAGE, and Western blotting was performed using polyclonal goat anti–human FXI peroxidase-conjugated IgG. The pcDNA3 empty vector was used as negative control (mock). + represents presence of goat anti–human FXI antibody; −, absence of goat anti–human FXI antibody; CTR+, 2 ng of commercially available human FXI; and input, 10 μL of normal plasma before the immunoprecipitation step.
Immunoprecipitation of FXI Δ6/7 isoform from normal plasma. FXI present in plasma and in media of COS-1 cells transfected with pcDNA3/FXI, or pcDNA3/FXI-Δ6/7 was immunoprecipitated using both goat anti–human FXI antibody and mouse anti–human FXI (data not shown). Samples were analyzed on 6% nonreducing SDS-PAGE, and Western blotting was performed using polyclonal goat anti–human FXI peroxidase-conjugated IgG. The pcDNA3 empty vector was used as negative control (mock). + represents presence of goat anti–human FXI antibody; −, absence of goat anti–human FXI antibody; CTR+, 2 ng of commercially available human FXI; and input, 10 μL of normal plasma before the immunoprecipitation step.
To confirm this observation, approximately 80 μg of affinity-purified commercial FXI were separated on a 8% SDS-PAGE. After Coomassie staining, full-length FXI was clearly visualized as a sharp band of approximately 80 kDa; in addition, a very faint band, possibly corresponding to Δ6/7 FXI isoform, was visible. The bands were excised and submitted to mass spectrometry analysis. A list of candidate proteins was obtained on the basis of their calculated posterior probability. Protein identity was reverified by obtaining isoelectric point, molecular weight, and peptide mass fingerprinting data, and by comparing them with the Swiss-Prot/National Center for Biotechnology Information databases. This analysis revealed the presence of tryptic peptides corresponding to human FXI; however, because no fragments distinctive of the deleted isoform are predicted to be generated by tryptic digestion, this analysis could not unequivocally confirm the existence of a FXI Δ6/7 protein isoform.
Discussion
AS represents a main source of transcriptome and proteome diversity and is therefore relevant to disease and therapy. The use of exon-exon junction microarrays revealed that transcripts from at least 74% of all multiexon genes are alternatively spliced in humans.33 More recently, the availability of high-throughput DNA sequencing technologies allowed an unbiased genomic view of AS. However, estimates on the proportion of genes undergoing AS still widely range from 30% to 95%.34,35
Even though it is now perceived that most AS events are nonfunctional and might reflect a stochastic noise in the splicing process, alternative isoforms enriched in some characteristics, such as conservation across evolution, tissue specificity, coding frame preservation, abundance, and changes in functional regions of the protein, probably have a function.36 Judgment on the physiologic role of AS is further complicated by the fact that even prematurely terminated transcripts have been shown to participate in gene expression regulation via coupling to the NMD degradation pathway.21-23
Concerning the F11 gene, the existence of AS isoforms has been known for many years.6,10,26 Unfortunately, information on the splicing pattern of this gene is still unclear, if not misleading. Indeed, most nucleotide and protein databases still report the existence of a “FXI isoform 2,” lacking amino acids from 91 to 144,10,37 notwithstanding that this isoform was not confirmed by more recent literature5,25 and the relevant reference sequence (NM_019559.2) was removed from GenBank (National Center for Biotechnology Information) because of insufficient supporting data. This situation, besides illustrating the laboriousness of the process of amending erroneous or unclear data in the highly redundant biomedical web databases, underlines the need to investigate in-depth the actual splicing pattern of this gene.
A comprehensive analysis of the F11 splicing pattern, performed by an RT-PCR–based strategy covering the whole gene, revealed the existence of at least 5 AS events, all involving the skipping of 1 or 2 exons. All transcripts were confirmed to be present either in human liver or in platelets.
Among these 5 AS transcripts, those skipping exons 6+7 and exon 13 are in frame, and their presence in liver is further supported by at least one expressed sequence tag (DA640741 and CB163164, respectively) in the AS database of the Swiss Institute of Bioinformatics (retrieved from the SIB Alt-Splicing track of the UCSC genome browser, http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=143839514&c=chr4&g=sibTxGraph). The same transcripts were searched in murine and bovine liver RNA by RT-PCR with species-specific primers: no skipping of exons 6+7 or 13 was identified (data not shown), suggesting that these AS events are not evolutionarily conserved.
The skipping of exons 6+7, which was previously reported in Dami cells (A. Zivelin, personal communication in Gailani et al6 ), stood out from the other ones for its quantitative abundance (Figure 1B). Therefore, we decided to measure its relative amount over the “wild-type” splicing by comparing the absolute quantities of the 2 transcripts in liver and platelets. A striking difference was observed between the 2 samples, with the alternative isoform being almost as abundant as the “wild-type” one in platelets (Figure 3D). In the light of the very low level of global F11 transcription in platelets, instead of representing a true tissue-specific AS, the high level of Δ6/7 mRNA might reflect a lower efficiency in the control of accurate splicing. Indeed, very recent data on AS on a global scale suggest that the amount of noisy splicing in a given gene is directly proportional to the number of introns and inversely proportional to its transcription level, in the latter case probably reflecting a tolerance of cells to underexpressed erroneously spliced mRNAs.36
Concerning the 3 out-of-frame AS transcripts, we explored their possible modulation by the NMD pathway. HepG2 cells treated with the NMD inhibitor drug puromycin were used as a cellular model system to study the susceptibility of PTC+F11 mRNAs to NMD degradation. The obtained results confirmed that all transcripts containing a PTC were up-regulated after NMD inhibition. Moreover, treatment with puromycin also caused a significant increase in the total amount of F11 mRNA, as assessed by amplifying an mRNA region spanning the last 2 exons, which are expected to be present in almost all transcripts. These data suggest that the coupling of AS and NMD might participate in the posttranscriptional regulation of the F11 gene expression. Indeed, down-regulation of PTC+ mRNAs can be particularly important for a dimeric protein, for which even heterodimers carrying a single mutant subunit might lead to potentially deleterious dominant-negative effects.
Among F11 AS transcripts, the most interesting at the protein level was by far the one resulting from the skipping of exons 6+7. Indeed, this isoform is not only relatively abundant, but it is also predicted to produce a FXI protein lacking an entire apple domain and having a chimeric A2-A3 domain.
Truncated proteins and chimeras in which the truncation or ligation sites coincide with interfaces between domains are known to be more likely to keep secretion competence.38 In this view, we explored whether recombinant Δ6/7 FXI retains the potential to be secreted by COS-1 cells. In contrast with what is reported in the literature,6 small amounts of Δ6/7 FXI were immunoprecipitated from culture media, demonstrating that this protein isoform can dimerize and be secreted by cells, yet with reduced efficiency. Conversely, it is probable that the wt/Δ6/7 heterodimer is largely retained in the ER, as confirmed by the level of FXI antigen measured in conditioned media of cells coexpressing the 2 FXI isoforms (38% vs the expected 50%; Figure 5A left). The increased intracellular levels of FXI molecules containing the Δ6/7 isoform (Figure 5A right) suggest that the lower secretion efficiency is not completely balanced by intracellular degradation.
The absence of the C-terminal half of the A2 and of the N-terminal half of the A3 domains is predicted to completely abolish FXI binding to its substrate FIX39,40 ; therefore, it was not surprising to find that the recombinant Δ6/7 protein had no measurable activity (Figure 5B). The specific activity measured in media conditioned by cells coexpressing wild-type and Δ6/7 FXIs was significantly higher than 50%. This is compatible with a total absence of activity of the deleted isoform, when considering the higher efficiency of secretion of the wild-type homodimer over the mutant homodimer or heterodimer.
Finally, we tried to address the question whether detectable amounts of Δ6/7 FXI are present in vivo. To this purpose, we immunoprecipitated FXI from human plasma and analyzed it by Western blotting. A band with an apparent molecular weight compatible with the recombinant deleted protein was detected. A similar experiment on washed lysed platelets only identified a faint band of approximately 160 kDa, probably corresponding to the full-length protein (data not shown), thus suggesting that the Δ6/7 protein isoform is present in such a tiny amount that it could not be detected. We also searched for clearer evidence of the presence of the novel FXI isoform in plasma by analyzing commercially available FXI concentrate. A band with a molecular weight of approximately 70 kDa was detected and excised from the gel. Mass spectrometry analysis confirmed that it was composed of FXI; however, no fragment distinctive of the Δ6/7 FXI was found, and we cannot exclude the possibility that this band could represent a degradation product of the full-length protein. Moreover, we cannot rule out that the Δ6/7 FXI was not recognized by the monoclonal antibody used for isolation of FXI in the processing of the commercial preparation.
In conclusion, from the structural and functional characterization of the splicing pattern of a single gene, we could recapitulate most theories presently proposed to explain the widespread diffusion of AS among higher eukaryotes: (1) production of PTC+ transcripts regulating gene expression by NMD (Δ3, Δ6, and Δ7 transcripts), (2) synthesis of protein isoforms with or without functional implications (Δ6/7 protein isoform), and (3) outcome of stochastic noise in the splicing process (potentially all of the above).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Stefano Lancellotti (Hemostasis Research Center, Catholic University School of Medicine, Rome, Italy) for helping in plasma Western blotting experiments, Dr Rossella Bader (Angelo Bianchi Bonomi Hemophilia and Thrombosis Center, Maggiore Hospital, Milan, Italy) for FXI antigen and coagulant activity measurements, and Patricia Corcoran, MSEd (Tucson, AZ) for her meticulous proofreading.
Authorship
Contribution: R.A. and V.R. designed the study, drafted the manuscript, expressed recombinant FXI, and analyzed the FXI isoforms in human plasma and platelets; I.G. and G.S. performed quantitative and qualitative analysis of the F11 splicing pattern; R.D.C. characterized the recombinant Δ6/7 FXI, interpreted the results, and reviewed the manuscript; F.P. measured FXI antigen and activity levels and discussed the results; S.D. wrote the manuscript and supervised the entire study; and all authors participated in the conception and design of the present study, the analysis and interpretation of data, and revision of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefano Duga, Department of Biology and Genetics for Medical Sciences, University of Milan, Via Viotti, 3/5-20133 Milano, Italy; e-mail: stefano.duga@unimi.it.
References
Author notes
R.A. and V.R. contributed equally to this study.

![Figure 4. Metabolic labeling of wild-type and Δ6/7 mutant recombinant FXI in transiently transfected COS-1 cells. Twenty-four hours after transfection, cells were labeled with [35S]-cysteine and methionine mixture as described in “Expression and quantitation of recombinant Δ6/7 FXI.” Radiolabeled FXIs were immunoprecipitated from cell lysates and media using goat anti–human FXI. Immunoprecipitated FXI was analyzed on 8% reducing (bottom panel) and 6% nonreducing (top panel) SDS-PAGE and visualized by scanning phosphor image screens. The pcDNA3 empty vector was used as negative control (mock). + indicates presence of goat anti–human FXI antibody; −, absence of goat anti–human FXI antibody; and arrowheads, homodimeric molecules in nonreducing panels. In the same panels, the smears corresponding to the heterodimers and the Δ6/7 homodimers are indicated by square brackets.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/10/10.1182_blood-2009-09-245092/4/m_zh89990949360004.jpeg?Expires=1769142359&Signature=tF5rVyhwDgHv5e4YMd2S-yF34GRd65qt0w2K47IgDT6QtCbxC8UGmASHnIovTDTSt-n-Aetz184PI5pUvu2C1pr6Yldr2Z9OBa2zr9VANnVC6jNnWhGnIDBhTGIl59EiqfiJ0Ch3E0QIODQikGoRjtZaMWvLTONqRoHzrFFQSOWgmSWLg4c~CbZ3Jh1lmKofM7EmXv0OHv8kUwSYgc~gpjYuIg8fAE3MmjiCuGuwtyOOEOyM1LiIIaofp6aaG3s0L0bkmdYr1nrqREJX0DNY-1Psj3HNaeFipAeZ30hJkSR2gYwuYLeKednPDCF4Hd7sajP6ipVKYnF80x2uA~HlUw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal