Abstract
Landmark analyses are used to investigate the importance for survival of achieving complete response (CR), an important initial goal of myeloma therapy. With median times to CR in Total Therapy (TT) trials of approximately 1 year, this approach excludes a sizeable fraction of patients dying before such a landmark. To permit inclusion of all trial participants, we investigated the prognostic implications of both onset and duration of CR as time-dependent variables. Superseding the adverse effects of cytogenetic abnormalities and other standard prognostic parameters, both failure to achieve CR (non-CR) and, especially, loss of CR (los-CR) were independently associated with inferior survival in TT1, TT2, and TT3 protocols. In the context of gene array–defined risk, available in TT2 and TT3 subsets, both los-CR and non-CR terms were retained in the survival model as dominant adverse variables, stressing the prognostic importance of sustaining CR status, especially in high-risk disease.
Introduction
As result of a substantial increase in the rate of complete response (CR)1,2 from less than 5% to more than 60%, the median myeloma survival has been significantly extended from less than 3 years with standard melphalan-prednisone to more than 10 years on the experimental arm of Total Therapy 2 (TT2) with thalidomide.3 Recent reports of novel agent combination trials using immunomodulatory agents (thalidomide or lenalidomide), together with bortezomib or melphalan, have demonstrated CR rates as high as 40% without resorting to transplantation.4-6 Follow-up time, however, is too short to judge the long-term efficacy of such novel therapies.
In addressing the fallacies of CR,7 we noted that a fraction of 10-year survivors had never achieved CR,8 which was attributed to myeloma evolving from a smoldering phase9 or having gene expression profiling (GEP) characteristics of monoclonal gammopathy of undetermined significance (MGUS).10 Conversely, high CR rates, especially in GEP-defined high-risk myeloma, were critical to extended survival.11 In the context of TT2, the median survival of the 15% with high-risk myeloma was only 2 years as opposed to longer than 10 years in the remainder.12 Sustaining CR status for at least 3 years (sus-CR) was associated with superior survival versus not achieving CR (non-CR) and especially attaining and losing CR (los-CR).13 Patients not surviving the 3-year landmark had to be dropped from the analysis. Here we examine a different statistical approach that accounts for all patients and permits investigation of prognosis in the context of precise timing of onset and loss of CR. By applying time-dependent variable status to posttreatment events, such as achieving CR status or losing such designation, the full complexity of CR and its duration can be examined in patients with high-risk myeloma who, despite high CR rates, often have a short survival as a result of rapid disease recurrence.
Methods
The details of TT1, TT2, and TT3 regimens have been published previously.3,14-17 All protocols had been approved by the Institutional Review Board of the University of Arkansas, and all patients had signed a written informed consent acknowledging the investigational nature of the trials and appreciating the existence of other treatment options, in accordance with Food and Drug Administration guidelines and the Declaration of Helsinki. Nearly 70% of all charts had been reviewed by independent auditor teams, verifying protocol compliance as well as efficacy and toxicity data.
GEP analyses were performed to define molecular subgroups,18 prognostic risk,12 and MGUS likeness.19
Our continuously updated multiple myeloma database was interrogated to determine, among 231 patients enrolled in TT1, 668 patients accrued to TT2, and 303 to TT3, the timing of onset of stringently defined CR.1,2 CR duration was measured from its onset until the recurrence of the original monoclonal protein, documented by immunofixation analysis on at least 2 successive occasions at least 2 months apart. Relapse from CR was defined as persistent reemergence, on at least 3 successive occasions over the course of 3 months, of the original monoclonal band on immunofixation analysis; other criteria of relapse from CR included the documentation of serum-M or urine-M on standard electrophoresis, recurrence or increase in monoclonal bone marrow plasmacytosis to greater than 10%, development of cytogenetic abnormalities (CAs), or of imaging abnormalities, such as focal lesions on magnetic resonance imaging or positron emission tomography scans.
We then applied multivariate Cox regression modeling20 to determine which baseline parameters, along with time-dependent onset of and duration of CR, significantly affected overall survival. Treating onset and duration of CR as time-dependent variables21 in the Cox regression models enabled us to analyze the impact of these outcomes on the entire population, unlike landmarking techniques, which only consider a subset of the population. Stepwise selection and Cox proportional hazard regression modeling were applied to the multivariate analyses. Kaplan-Meier methods were used to generate survival distribution graphs,22 and comparisons were made using the log-rank test.23 Categorical comparisons were made using the χ2 test.24
Results
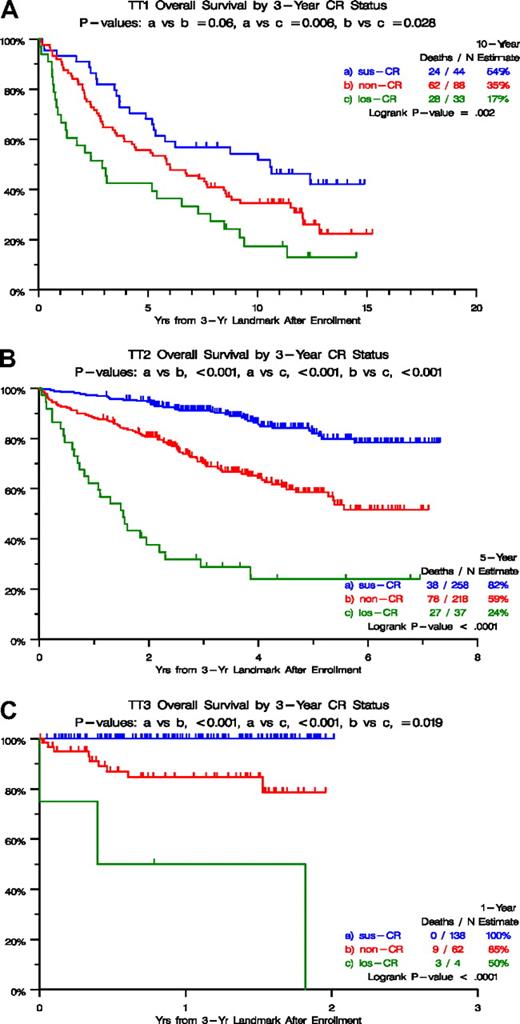
Figure 1 depicts survival outcomes according to CR categories from a 3-year landmark after initiation of TT1 (Figure 1A), TT2 (Figure 1B), and TT3 protocols (Figure 1C). In all 3 trials, survival was particularly poor in the los-CR category, and non-CR was significantly inferior to sus-CR in TT2 and TT3. These marked survival differences could be traced to differences in baseline characteristics (Table 1), which were least favorable in the los-CR category, in which high lactate dehydrogenase (LDH) was overrepresented in TT1 (36% vs 16% in sus-CR vs 14% in non-CR; P = .015) and TT2 (46% vs 30% in sus-CR vs 21% in non-CR; P = .003). In TT2, albumin less than 3.5 g/dL and beta-2-microglobulin (B2M) more than 3.5 mg/L were present in 30% and 51%, respectively, of los-CR patients, compared with 14% and 31% in sus-CR and 16% and 34% in non-CR (P = .043, P = .043). In TT3, B2M more than 5.5 mg/L and creatinine more than 2 mg/dL were observed in 50% and 25%, respectively, of patients with los-CR compared with 12% and 3% in sus-CR and 24% and 15% in non-CR (P = .022, P = .004). With access to GEP risk information, available for 273 patients on TT2 and 185 patients on TT3 protocols, high-risk designation was present in 22% and 67% of los-CR versus 8% and 10% of sus-CR versus 3% and 9% in non-CR (P = .005, P = .005). Similarly, the MF (MAF/MAFB) subgroup was overrepresented in the los-CR (17% and 67%) compared with sus-CR (4% and 7%) and non-CR (3% and 7%; P = .047, P < .001).
Overall survival by 3-year CR status. (A) For TT1, survival was superior in case complete response (CR) status had been sustained for 3 years (sus-CR), compared with not having attained CR (non-CR) and especially to having attained and lost CR (los-CR) within the 3-year time frame. (B) For TT2, survival was superior in case complete response (CR) status had been sustained for 3 years (sus-CR), compared with not having attained CR (non-CR) and especially to having attained and lost CR (los-CR) within the 3-year time frame. (C) For TT3, survival was superior in case complete response (CR) status had been sustained for 3 years (sus-CR), compared with not having attained CR (non-CR) and especially to having attained and lost CR (los-CR) within the 3-year time frame.
Overall survival by 3-year CR status. (A) For TT1, survival was superior in case complete response (CR) status had been sustained for 3 years (sus-CR), compared with not having attained CR (non-CR) and especially to having attained and lost CR (los-CR) within the 3-year time frame. (B) For TT2, survival was superior in case complete response (CR) status had been sustained for 3 years (sus-CR), compared with not having attained CR (non-CR) and especially to having attained and lost CR (los-CR) within the 3-year time frame. (C) For TT3, survival was superior in case complete response (CR) status had been sustained for 3 years (sus-CR), compared with not having attained CR (non-CR) and especially to having attained and lost CR (los-CR) within the 3-year time frame.
Baseline characteristics of patients enrolled in TT1, TT2, and TT3 according to response category*
| Factor . | TT1 . | TT2 . | TT3 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sus-CR . | los-CR . | non-CR . | P . | sus-CR . | los-CR . | non-CR . | P . | sus-CR . | los-CR . | non-CR . | P . | |
| Age ≥ 65 y | 3/44 (7) | 0/33 (0) | 8/88 (9) | .203 | 42/258 (16) | 10/37 (27) | 44/218 (20) | .224 | 33/138 (24) | 0/4 (0) | 20/62 (32) | .225 |
| Albumin < 3.5 g/dL | 9/44 (20) | 7/33 (21) | 26/88 (30) | .434 | 35/257 (14) | 11/37 (30) | 35/216 (16) | .043 | 21/138 (15) | 0/4 (0) | 12/62 (19) | .515 |
| B2M ≥ 3.5 mg/L | 12/44 (27) | 14/33 (42) | 32/86 (37) | .350 | 79/258 (31) | 19/37 (51) | 75/218 (34) | .043 | 55/138 (40) | 2/4 (50) | 27/62 (44) | .830 |
| B2M > 5.5 mg/L | 6/44 (14) | 2/33 (6) | 13/86 (15) | .412 | 37/258 (14) | 9/37 (24) | 32/218 (15) | .275 | 17/138 (12) | 2/4 (50) | 15/62 (24) | .022 |
| CRP ≥ 4 mg/L | 17/43 (40) | 18/33 (55) | 29/84 (35) | .138 | 144/255 (56) | 24/37 (65) | 105/213 (49) | .118 | 78/138 (57) | 4/4 (100) | 33/61 (54) | .200 |
| CRP ≥ 8 mg/L | 11/43 (26) | 10/33 (30) | 19/84 (23) | .685 | 111/255 (44) | 15/37 (41) | 68/213 (32) | .035 | 39/138 (28) | 2/4 (50) | 19/61 (31) | .610 |
| Creatinine ≥ 2 mg/dL | 3/44 (7) | 1/33 (3) | 8/88 (9) | .515 | 22/251 (9) | 3/36 (8) | 16/214 (7) | .880 | 4/138 (3) | 1/4 (25) | 9/62 (15) | .004 |
| LDH ≥ 190 U/L | 7/43 (16) | 12/33 (36) | 12/88 (14) | .015 | 77/257 (30) | 17/37 (46) | 46/217 (21) | .003 | 29/138 (21) | 2/4 (50) | 15/62 (24) | .366 |
| CAs† | 10/43 (23) | 11/33 (33) | 24/79 (30) | .588 | 54/257 (21) | 9/37 (24) | 59/214 (28) | .252 | 41/138 (30) | 1/4 (25) | 14/61 (23) | .612 |
| GEP high-risk | NA | NA | NA | NA | 11/139 (8) | 4/18 (22) | 3/116 (3) | .005 | 12/124 (10) | 2/3 (67) | 5/58 (9) | .005 |
| GEP CD-1 subgroup | NA | NA | NA | NA | 13/139 (9) | 2/18 (11) | 4/116 (3) | .141 | 8/124 (6) | 1/3 (33) | 1/58 (2) | .041 |
| GEP CD-2 subgroup | NA | NA | NA | NA | 17/139 (12) | 0/18 (0) | 21/116 (18) | .085 | 7/124 (6) | 0/3 (0) | 11/58 (19) | .016 |
| GEP HY subgroup | NA | NA | NA | NA | 37/139 (27) | 3/18 (17) | 41/116 (35) | .145 | 36/124 (29) | 0/3 (0) | 20/58 (34) | .391 |
| GEP LB subgroup | NA | NA | NA | NA | 16/139 (12) | 0/18 (0) | 11/116 (9) | .300 | 15/124 (12) | 0/3 (0) | 8/58 (14) | .764 |
| GEP MF subgroup | NA | NA | NA | NA | 6/139 (4) | 3/18 (17) | 4/116 (3) | .047 | 9/124 (7) | 2/3 (67) | 4/58 (7) | <.001 |
| GEP MS subgroup | NA | NA | NA | NA | 9/139 (6) | 3/18 (17) | 16/116 (14) | .103 | 15/124 (12) | 0/3 (0) | 7/58 (12) | .814 |
| GEP MY subgroup | NA | NA | NA | NA | 27/139 (19) | 5/18 (28) | 15/116 (13) | .185 | 23/124 (19) | 0/3 (0) | 6/58 (10) | .275 |
| GEP PR subgroup | NA | NA | NA | NA | 14/139 (10) | 2/18 (11) | 4/116 (3) | .106 | 11/124 (9) | 0/3 (0) | 1/58 (2) | .170 |
| GEP MGUS-like | NA | NA | NA | NA | 45/139 (32) | 2/18 (11) | 44/116 (38) | .076 | 32/124 (26) | 1/3 (33) | 15/58 (26) | .958 |
| Factor . | TT1 . | TT2 . | TT3 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sus-CR . | los-CR . | non-CR . | P . | sus-CR . | los-CR . | non-CR . | P . | sus-CR . | los-CR . | non-CR . | P . | |
| Age ≥ 65 y | 3/44 (7) | 0/33 (0) | 8/88 (9) | .203 | 42/258 (16) | 10/37 (27) | 44/218 (20) | .224 | 33/138 (24) | 0/4 (0) | 20/62 (32) | .225 |
| Albumin < 3.5 g/dL | 9/44 (20) | 7/33 (21) | 26/88 (30) | .434 | 35/257 (14) | 11/37 (30) | 35/216 (16) | .043 | 21/138 (15) | 0/4 (0) | 12/62 (19) | .515 |
| B2M ≥ 3.5 mg/L | 12/44 (27) | 14/33 (42) | 32/86 (37) | .350 | 79/258 (31) | 19/37 (51) | 75/218 (34) | .043 | 55/138 (40) | 2/4 (50) | 27/62 (44) | .830 |
| B2M > 5.5 mg/L | 6/44 (14) | 2/33 (6) | 13/86 (15) | .412 | 37/258 (14) | 9/37 (24) | 32/218 (15) | .275 | 17/138 (12) | 2/4 (50) | 15/62 (24) | .022 |
| CRP ≥ 4 mg/L | 17/43 (40) | 18/33 (55) | 29/84 (35) | .138 | 144/255 (56) | 24/37 (65) | 105/213 (49) | .118 | 78/138 (57) | 4/4 (100) | 33/61 (54) | .200 |
| CRP ≥ 8 mg/L | 11/43 (26) | 10/33 (30) | 19/84 (23) | .685 | 111/255 (44) | 15/37 (41) | 68/213 (32) | .035 | 39/138 (28) | 2/4 (50) | 19/61 (31) | .610 |
| Creatinine ≥ 2 mg/dL | 3/44 (7) | 1/33 (3) | 8/88 (9) | .515 | 22/251 (9) | 3/36 (8) | 16/214 (7) | .880 | 4/138 (3) | 1/4 (25) | 9/62 (15) | .004 |
| LDH ≥ 190 U/L | 7/43 (16) | 12/33 (36) | 12/88 (14) | .015 | 77/257 (30) | 17/37 (46) | 46/217 (21) | .003 | 29/138 (21) | 2/4 (50) | 15/62 (24) | .366 |
| CAs† | 10/43 (23) | 11/33 (33) | 24/79 (30) | .588 | 54/257 (21) | 9/37 (24) | 59/214 (28) | .252 | 41/138 (30) | 1/4 (25) | 14/61 (23) | .612 |
| GEP high-risk | NA | NA | NA | NA | 11/139 (8) | 4/18 (22) | 3/116 (3) | .005 | 12/124 (10) | 2/3 (67) | 5/58 (9) | .005 |
| GEP CD-1 subgroup | NA | NA | NA | NA | 13/139 (9) | 2/18 (11) | 4/116 (3) | .141 | 8/124 (6) | 1/3 (33) | 1/58 (2) | .041 |
| GEP CD-2 subgroup | NA | NA | NA | NA | 17/139 (12) | 0/18 (0) | 21/116 (18) | .085 | 7/124 (6) | 0/3 (0) | 11/58 (19) | .016 |
| GEP HY subgroup | NA | NA | NA | NA | 37/139 (27) | 3/18 (17) | 41/116 (35) | .145 | 36/124 (29) | 0/3 (0) | 20/58 (34) | .391 |
| GEP LB subgroup | NA | NA | NA | NA | 16/139 (12) | 0/18 (0) | 11/116 (9) | .300 | 15/124 (12) | 0/3 (0) | 8/58 (14) | .764 |
| GEP MF subgroup | NA | NA | NA | NA | 6/139 (4) | 3/18 (17) | 4/116 (3) | .047 | 9/124 (7) | 2/3 (67) | 4/58 (7) | <.001 |
| GEP MS subgroup | NA | NA | NA | NA | 9/139 (6) | 3/18 (17) | 16/116 (14) | .103 | 15/124 (12) | 0/3 (0) | 7/58 (12) | .814 |
| GEP MY subgroup | NA | NA | NA | NA | 27/139 (19) | 5/18 (28) | 15/116 (13) | .185 | 23/124 (19) | 0/3 (0) | 6/58 (10) | .275 |
| GEP PR subgroup | NA | NA | NA | NA | 14/139 (10) | 2/18 (11) | 4/116 (3) | .106 | 11/124 (9) | 0/3 (0) | 1/58 (2) | .170 |
| GEP MGUS-like | NA | NA | NA | NA | 45/139 (32) | 2/18 (11) | 44/116 (38) | .076 | 32/124 (26) | 1/3 (33) | 15/58 (26) | .958 |
Values are n/N (%), where n is number with factor and N is number with valid data for factor. NA indicates not applicable.
CR indicates complete response; sus-CR, attained and sustained through at least 3 years; los-CR, attained and lost in 3 years; non-CR, never obtained response in 3 years.
CAs at any time before enrollment.
We then examined the time dependence of achieving and losing CR status for the entire population of each TT protocol in the context of baseline variables (Table 2). According to multivariate analyses, in the absence of GEP data, los-CR was the dominant variable associated with inferior survival, regardless of protocol, with hazard ratio (HR) values of 7.71 in TT1 (P < .001), 8.89 in TT2 (P < .001) and 23.01 in TT3 (P < .001; Table 2). Non-CR was also associated with inferior prognosis in all 3 trials, as was the presence of CA. High LDH imparted short survival in TT2 and TT3. In the context of GEP data, available together with all other variables in 334 patients of TT2 and 274 of TT3, los-CR and non-CR both retained independent adverse consequences in both TT2 and TT3. MGUS-like myeloma and both HY (hyperdiploidy) and LB (low bone disease) subgroups implied good prognosis in TT2, whereas no molecular subgroup was found to be significant in TT3. In both trials, high-risk status, the presence of CA, and elevated LDH levels retained independent adverse consequences for survival. A combined look at all 3 protocols in the context of standard variables only revealed that both los-CR and non-CR survived the multivariate model with higher HR values than observed for the remaining independently significant variables such as CA, B2M, creatinine, LDH, and C-reactive protein (CRP; Table 3). In the context of GEP data, available for TT2 and TT3 protocols, both los-CR and non-CR were associated with higher HR values than GEP-defined high risk. MGUS-like myeloma and both HY and LB subgroups implied good prognosis, whereas high LDH and presence of CA conferred short survival.
Univariate and multivariate analyses of parameters associated with overall survival
| Variable . | TT1 . | TT2 . | TT3 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | |
| Univariate | |||||||||
| Age ≥ 65 y | 21/231 (9) | 2.04 (1.29, 3.22) | .002 | 136/668 (20) | 1.44 (1.10, 1.88) | .008 | 84/303 (28) | 1.29 (0.77, 2.16) | .338 |
| Female | 88/231 (38) | 0.92 (0.68, 1.25) | .592 | 272/668 (41) | 0.83 (0.65, 1.05) | .122 | 110/303 (36) | 1.41 (0.87, 2.28) | .166 |
| White | 206/231 (89) | 0.98 (0.62, 1.57) | .944 | 580/668 (87) | 1.27 (0.89, 1.83) | .193 | 268/303 (88) | 0.63 (0.33, 1.20) | .156 |
| Albumin < 3.5 g/dL | 62/231 (27) | 1.09 (0.78, 1.51) | .608 | 119/664 (18) | 1.65 (1.25, 2.17) | <.001 | 72/303 (24) | 1.74 (1.03, 2.92) | .037 |
| B2M ≥ 3.5 mg/L | 95/229 (41) | 1.55 (1.15, 2.08) | .004 | 243/668 (36) | 1.77 (1.40, 2.23) | <.001 | 136/303 (45) | 2.04 (1.25, 3.32) | .004 |
| B2M > 5.5 mg/L | 43/229 (19) | 1.69 (1.17, 2.43) | .005 | 122/668 (18) | 2.03 (1.56, 2.65) | <.001 | 65/303 (21) | 3.32 (2.04, 5.39) | <.001 |
| CRP ≥ 8 mg/L | 70/223 (31) | 1.62 (1.19, 2.22) | .002 | 263/658 (40) | 1.32 (1.04, 1.66) | .021 | 100/302 (33) | 1.66 (1.03, 2.70) | .039 |
| Creatinine ≥ 2 mg/dL | 22/231 (10) | 1.80 (1.13, 2.87) | .013 | 62/654 (9) | 1.82 (1.29, 2.56) | <.001 | 23/303 (8) | 2.38 (1.21, 4.65) | .012 |
| Hb < 10 g/dL | 78/231 (34) | 1.53 (1.13, 2.08) | .006 | 161/667 (24) | 1.31 (1.01, 1.69) | .039 | 94/303 (31) | 2.07 (1.28, 3.35) | .003 |
| LDH ≥ 190 U/L | 49/230 (21) | 1.60 (1.13, 2.27) | .008 | 204/666 (31) | 1.65 (1.30, 2.09) | <.001 | 81/303 (27) | 2.88 (1.78, 4.65) | <.001 |
| CAs | 74/221 (33) | 1.78 (1.30, 2.43) | <.001 | 197/661 (30) | 2.17 (1.71, 2.74) | <.001 | 100/302 (33) | 3.02 (1.85, 4.91) | <.001 |
| GEP high-risk | NA | NA | NA | 46/351 (13) | 4.00 (2.70, 5.93) | <.001 | 40/275 (15) | 4.54 (2.69, 7.65) | <.001 |
| GEP CD-1 subgroup | NA | NA | NA | 23/351 (7) | 0.71 (0.33, 1.52) | .382 | 15/275 (5) | 0.88 (0.27, 2.80) | .822 |
| GEP CD-2 subgroup | NA | NA | NA | 45/351 (13) | 0.79 (0.45, 1.37) | .400 | 27/275 (10) | 0.65 (0.24, 1.80) | .409 |
| GEP HY subgroup | NA | NA | NA | 98/351 (28) | 0.59 (0.39, 0.90) | .013 | 80/275 (29) | 0.73 (0.40, 1.33) | .306 |
| GEP LB subgroup | NA | NA | NA | 30/351 (9) | 0.43 (0.19, 0.98) | .045 | 33/275 (12) | 0.24 (0.06, 0.98) | .046 |
| GEP MF subgroup | NA | NA | NA | 19/351 (5) | 1.89 (1.02, 3.49) | .044 | 22/275 (8) | 2.58 (1.34, 4.98) | .005 |
| GEP MS subgroup | NA | NA | NA | 44/351 (13) | 2.08 (1.37, 3.18) | <.001 | 33/275 (12) | 0.77 (0.33, 1.78) | .538 |
| GEP MY subgroup | NA | NA | NA | 58/351 (17) | 0.98 (0.63, 1.54) | .943 | 38/275 (14) | 0.98 (0.46, 2.06) | .954 |
| GEP PR subgroup | NA | NA | NA | 34/351 (10) | 2.03 (1.26, 3.26) | .004 | 27/275 (10) | 2.83 (1.50, 5.34) | .001 |
| GEP MF or PR subgroups | NA | NA | NA | 53/351 (15) | 2.11 (1.41, 3.15) | <.001 | 49/275 (18) | 3.22 (1.91, 5.42) | <.001 |
| GEP MF, MS or PR subgroups | NA | NA | NA | 97/351 (28) | 2.51 (1.78, 3.53) | <.001 | 82/275 (30) | 2.31 (1.39, 3.84) | .001 |
| GEP HY or LB subgroups | NA | NA | NA | 128/351 (36) | 0.50 (0.34, 0.73) | <.001 | 113/275 (41) | 0.50 (0.28, 0.89) | .018 |
| GEP MGUS-like | NA | NA | NA | 101/351 (29) | 0.41 (0.26, 0.65) | <.001 | 65/275 (24) | 0.63 (0.32, 1.24) | .179 |
| Randomized to thalidomide | NA | NA | NA | 323/668 (48) | 0.79 (0.62, 1.00) | .047 | NA | NA | NA |
| Los-CR* | 2.24 (1.61, 3.11) | <.001 | 4.51 (3.36, 6.06) | <.001 | 16.28 (7.22, 36.70) | <.001 | |||
| Did not achieve CR* | 1.22 (0.90, 1.65) | . 209 | 1.86 (1.45, 2.38) | <.001 | 3.24 (1.84, 5.72) | <.001 | |||
| Multivariate without GEP | |||||||||
| Age ≥ 65 y | 20/214 (9) | 2.01 (1.24, 3.27) | .005 | NS | NS | NS | NS | NS | NS |
| CAs | 74/214 (35) | 1.74 (1.25, 2.42) | <.001 | 188/634 (30) | 1.77 (1.39, 2.26) | <.001 | 100/301 (33) | 2.77 (1.69, 4.56) | <.001 |
| B2M > 5.5 mg/L | NS | NS | NS | 115/634 (18) | 1.54 (1.16, 2.05) | .003 | 65/301 (22) | 2.01 (1.21, 3.33) | .007 |
| CRP ≥ 8 mg/L | 70/214 (33) | 1.47 (1.06, 2.03) | .021 | NS | NS | NS | NS | NS | NS |
| Creatinine ≥ 2 mg/dL | 19/214 (9) | 2.23 (1.34, 3.70) | .002 | NS | NS | NS | NS | NS | NS |
| LDH ≥ 190 U/L | NS | NS | NS | 197/634 (31) | 1.35 (1.04, 1.74) | .025 | 80/301 (27) | 1.88 (1.13, 3.14) | .015 |
| Los-CR* | 7.71 (4.09, 14.53) | <.001 | 8.89 (5.93, 13.33) | <.001 | 23.01 (8.64, 61.23) | <.001 | |||
| Did not achieve CR* | 3.77 (2.06, 6.90) | <.001 | 4.03 (2.83, 5.73) | <.001 | 5.35 (2.69, 10.64) | <.001 | |||
| Multivariate with GEP | |||||||||
| LDH ≥ 190 U/L | NA | NA | NA | 114/334 (34) | 1.51 (1.05, 2.16) | .026 | 74/274 (27) | 1.82 (1.06, 3.15) | .031 |
| CAs | NA | NA | NA | 108/334 (32) | 1.70 (1.19, 2.44) | .004 | 95/274 (35) | 2.83 (1.65, 4.87) | <.001 |
| GEP high-risk | NA | NA | NA | 44/334 (13) | 2.16 (1.38, 3.38) | <.001 | 40/274 (15) | 2.27 (1.26, 4.09) | .006 |
| GEP HY or LB subgroups | NA | NA | NA | 123/334 (37) | 0.49 (0.32, 0.76) | .001 | NS | NS | NS |
| GEP MGUS-like | NA | NA | NA | 93/334 (28) | 0.47 (0.28, 0.74) | .004 | NS | NS | NS |
| Los-CR* | NA | NA | NA | 8.66 (4.66, 16.09) | <.001 | 19.06 (6.88, 52.78) | <.001 | ||
| Did not achieve CR* | NA | NA | NA | 5.60 (3.21, 9.76) | <.001 | 5.05 (2.50, 10.20) | <.001 | ||
| Variable . | TT1 . | TT2 . | TT3 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | |
| Univariate | |||||||||
| Age ≥ 65 y | 21/231 (9) | 2.04 (1.29, 3.22) | .002 | 136/668 (20) | 1.44 (1.10, 1.88) | .008 | 84/303 (28) | 1.29 (0.77, 2.16) | .338 |
| Female | 88/231 (38) | 0.92 (0.68, 1.25) | .592 | 272/668 (41) | 0.83 (0.65, 1.05) | .122 | 110/303 (36) | 1.41 (0.87, 2.28) | .166 |
| White | 206/231 (89) | 0.98 (0.62, 1.57) | .944 | 580/668 (87) | 1.27 (0.89, 1.83) | .193 | 268/303 (88) | 0.63 (0.33, 1.20) | .156 |
| Albumin < 3.5 g/dL | 62/231 (27) | 1.09 (0.78, 1.51) | .608 | 119/664 (18) | 1.65 (1.25, 2.17) | <.001 | 72/303 (24) | 1.74 (1.03, 2.92) | .037 |
| B2M ≥ 3.5 mg/L | 95/229 (41) | 1.55 (1.15, 2.08) | .004 | 243/668 (36) | 1.77 (1.40, 2.23) | <.001 | 136/303 (45) | 2.04 (1.25, 3.32) | .004 |
| B2M > 5.5 mg/L | 43/229 (19) | 1.69 (1.17, 2.43) | .005 | 122/668 (18) | 2.03 (1.56, 2.65) | <.001 | 65/303 (21) | 3.32 (2.04, 5.39) | <.001 |
| CRP ≥ 8 mg/L | 70/223 (31) | 1.62 (1.19, 2.22) | .002 | 263/658 (40) | 1.32 (1.04, 1.66) | .021 | 100/302 (33) | 1.66 (1.03, 2.70) | .039 |
| Creatinine ≥ 2 mg/dL | 22/231 (10) | 1.80 (1.13, 2.87) | .013 | 62/654 (9) | 1.82 (1.29, 2.56) | <.001 | 23/303 (8) | 2.38 (1.21, 4.65) | .012 |
| Hb < 10 g/dL | 78/231 (34) | 1.53 (1.13, 2.08) | .006 | 161/667 (24) | 1.31 (1.01, 1.69) | .039 | 94/303 (31) | 2.07 (1.28, 3.35) | .003 |
| LDH ≥ 190 U/L | 49/230 (21) | 1.60 (1.13, 2.27) | .008 | 204/666 (31) | 1.65 (1.30, 2.09) | <.001 | 81/303 (27) | 2.88 (1.78, 4.65) | <.001 |
| CAs | 74/221 (33) | 1.78 (1.30, 2.43) | <.001 | 197/661 (30) | 2.17 (1.71, 2.74) | <.001 | 100/302 (33) | 3.02 (1.85, 4.91) | <.001 |
| GEP high-risk | NA | NA | NA | 46/351 (13) | 4.00 (2.70, 5.93) | <.001 | 40/275 (15) | 4.54 (2.69, 7.65) | <.001 |
| GEP CD-1 subgroup | NA | NA | NA | 23/351 (7) | 0.71 (0.33, 1.52) | .382 | 15/275 (5) | 0.88 (0.27, 2.80) | .822 |
| GEP CD-2 subgroup | NA | NA | NA | 45/351 (13) | 0.79 (0.45, 1.37) | .400 | 27/275 (10) | 0.65 (0.24, 1.80) | .409 |
| GEP HY subgroup | NA | NA | NA | 98/351 (28) | 0.59 (0.39, 0.90) | .013 | 80/275 (29) | 0.73 (0.40, 1.33) | .306 |
| GEP LB subgroup | NA | NA | NA | 30/351 (9) | 0.43 (0.19, 0.98) | .045 | 33/275 (12) | 0.24 (0.06, 0.98) | .046 |
| GEP MF subgroup | NA | NA | NA | 19/351 (5) | 1.89 (1.02, 3.49) | .044 | 22/275 (8) | 2.58 (1.34, 4.98) | .005 |
| GEP MS subgroup | NA | NA | NA | 44/351 (13) | 2.08 (1.37, 3.18) | <.001 | 33/275 (12) | 0.77 (0.33, 1.78) | .538 |
| GEP MY subgroup | NA | NA | NA | 58/351 (17) | 0.98 (0.63, 1.54) | .943 | 38/275 (14) | 0.98 (0.46, 2.06) | .954 |
| GEP PR subgroup | NA | NA | NA | 34/351 (10) | 2.03 (1.26, 3.26) | .004 | 27/275 (10) | 2.83 (1.50, 5.34) | .001 |
| GEP MF or PR subgroups | NA | NA | NA | 53/351 (15) | 2.11 (1.41, 3.15) | <.001 | 49/275 (18) | 3.22 (1.91, 5.42) | <.001 |
| GEP MF, MS or PR subgroups | NA | NA | NA | 97/351 (28) | 2.51 (1.78, 3.53) | <.001 | 82/275 (30) | 2.31 (1.39, 3.84) | .001 |
| GEP HY or LB subgroups | NA | NA | NA | 128/351 (36) | 0.50 (0.34, 0.73) | <.001 | 113/275 (41) | 0.50 (0.28, 0.89) | .018 |
| GEP MGUS-like | NA | NA | NA | 101/351 (29) | 0.41 (0.26, 0.65) | <.001 | 65/275 (24) | 0.63 (0.32, 1.24) | .179 |
| Randomized to thalidomide | NA | NA | NA | 323/668 (48) | 0.79 (0.62, 1.00) | .047 | NA | NA | NA |
| Los-CR* | 2.24 (1.61, 3.11) | <.001 | 4.51 (3.36, 6.06) | <.001 | 16.28 (7.22, 36.70) | <.001 | |||
| Did not achieve CR* | 1.22 (0.90, 1.65) | . 209 | 1.86 (1.45, 2.38) | <.001 | 3.24 (1.84, 5.72) | <.001 | |||
| Multivariate without GEP | |||||||||
| Age ≥ 65 y | 20/214 (9) | 2.01 (1.24, 3.27) | .005 | NS | NS | NS | NS | NS | NS |
| CAs | 74/214 (35) | 1.74 (1.25, 2.42) | <.001 | 188/634 (30) | 1.77 (1.39, 2.26) | <.001 | 100/301 (33) | 2.77 (1.69, 4.56) | <.001 |
| B2M > 5.5 mg/L | NS | NS | NS | 115/634 (18) | 1.54 (1.16, 2.05) | .003 | 65/301 (22) | 2.01 (1.21, 3.33) | .007 |
| CRP ≥ 8 mg/L | 70/214 (33) | 1.47 (1.06, 2.03) | .021 | NS | NS | NS | NS | NS | NS |
| Creatinine ≥ 2 mg/dL | 19/214 (9) | 2.23 (1.34, 3.70) | .002 | NS | NS | NS | NS | NS | NS |
| LDH ≥ 190 U/L | NS | NS | NS | 197/634 (31) | 1.35 (1.04, 1.74) | .025 | 80/301 (27) | 1.88 (1.13, 3.14) | .015 |
| Los-CR* | 7.71 (4.09, 14.53) | <.001 | 8.89 (5.93, 13.33) | <.001 | 23.01 (8.64, 61.23) | <.001 | |||
| Did not achieve CR* | 3.77 (2.06, 6.90) | <.001 | 4.03 (2.83, 5.73) | <.001 | 5.35 (2.69, 10.64) | <.001 | |||
| Multivariate with GEP | |||||||||
| LDH ≥ 190 U/L | NA | NA | NA | 114/334 (34) | 1.51 (1.05, 2.16) | .026 | 74/274 (27) | 1.82 (1.06, 3.15) | .031 |
| CAs | NA | NA | NA | 108/334 (32) | 1.70 (1.19, 2.44) | .004 | 95/274 (35) | 2.83 (1.65, 4.87) | <.001 |
| GEP high-risk | NA | NA | NA | 44/334 (13) | 2.16 (1.38, 3.38) | <.001 | 40/274 (15) | 2.27 (1.26, 4.09) | .006 |
| GEP HY or LB subgroups | NA | NA | NA | 123/334 (37) | 0.49 (0.32, 0.76) | .001 | NS | NS | NS |
| GEP MGUS-like | NA | NA | NA | 93/334 (28) | 0.47 (0.28, 0.74) | .004 | NS | NS | NS |
| Los-CR* | NA | NA | NA | 8.66 (4.66, 16.09) | <.001 | 19.06 (6.88, 52.78) | <.001 | ||
| Did not achieve CR* | NA | NA | NA | 5.60 (3.21, 9.76) | <.001 | 5.05 (2.50, 10.20) | <.001 | ||
The multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the .05 level. A multivariate P value greater than .05 indicates variable forced into model with significant variables chosen using stepwise selection.
HR indicates hazard ratio; CI, confidence interval; P, P value from Wald χ2 test in Cox regression; NA, not applicable; and NS, multivariate results not statistically significant at .05 level. All univariate P values are reported regardless of significance.
Treated as a time-dependent variable.
Univariate and multivariate analyses of parameters associated with overall survival with combination therapy
| Variable . | TT1, TT2, and TT3 combined . | TT2 and TT3 combined . | ||||
|---|---|---|---|---|---|---|
| n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | |
| Univariate | ||||||
| Age ≥ 65 y | 241/1202 (20) | 1.37 (1.11, 1.68) | .004 | 220/971 (23) | 1.38 (1.08, 1.75) | .009 |
| Female | 470/1202 (39) | 0.92 (0.77, 1.10) | .357 | 382/971 (39) | 0.93 (0.75, 1.15) | .484 |
| White | 1054/1202 (88) | 1.08 (0.83, 1.40) | .575 | 848/971 (87) | 1.10 (0.80, 1.50) | .571 |
| Albumin < 3.5 g/dL | 253/1198 (21) | 1.48 (1.22, 1.80) | <.001 | 191/967 (20) | 1.65 (1.30, 2.10) | <.001 |
| B2M ≥ 3.5 mg/L | 474/1200 (40) | 1.72 (1.45, 2.03) | <.001 | 379/971 (39) | 1.79 (1.45, 2.21) | <.001 |
| B2M > 5.5 mg/L | 230/1200 (19) | 2.03 (1.68, 2.47) | <.001 | 187/971 (19) | 2.25 (1.79, 2.83) | <.001 |
| CRP ≥ 8 mg/L | 433/1183 (37) | 1.40 (1.18, 1.66) | <.001 | 363/960 (38) | 1.39 (1.13, 1.72) | .002 |
| Creatinine ≥ 2 mg/dL | 107/1188 (9) | 1.86 (1.44, 2.40) | <.001 | 85/957 (9) | 1.92 (1.42, 2.60) | <.001 |
| Hb < 10 g/dL | 333/1201 (28) | 1.50 (1.25, 1.79) | <.001 | 255/970 (26) | 1.44 (1.15, 1.80) | .001 |
| LDH ≥ 190 U/L | 334/1199 (28) | 1.71 (1.43, 2.05) | <.001 | 285/969 (29) | 1.85 (1.50, 2.30) | <.001 |
| CAs | 371/1184 (31) | 2.14 (1.80, 2.55) | <.001 | 297/963 (31) | 2.30 (1.87, 2.84) | <.001 |
| GEP high-risk | NA | NA | NA | 86/626 (14) | 4.12 (3.02, 5.63) | <.001 |
| GEP CD-1 subgroup | NA | NA | NA | 38/626 (6) | 0.76 (0.40, 1.44) | .407 |
| GEP CD-2 subgroup | NA | NA | NA | 72/626 (12) | 0.76 (0.47, 1.24) | .271 |
| GEP HY subgroup | NA | NA | NA | 178/626 (28) | 0.63 (0.45, 0.89) | .008 |
| GEP LB subgroup | NA | NA | NA | 63/626 (10) | 0.35 (0.17, 0.72) | .004 |
| GEP MF subgroup | NA | NA | NA | 41/626 (7) | 2.11 (1.35, 3.29) | .001 |
| GEP MS subgroup | NA | NA | NA | 77/626 (12) | 1.59 (1.09, 2.31) | .016 |
| GEP MY subgroup | NA | NA | NA | 96/626 (15) | 0.99 (0.67, 1.45) | .956 |
| GEP PR subgroup | NA | NA | NA | 61/626 (10) | 2.29 (1.56, 3.34) | <.001 |
| GEP MF, MS, or PR subgroups | NA | NA | NA | 179/626 (29) | 2.42 (1.83, 3.22) | <.001 |
| GEP HY or LB subgroups | NA | NA | NA | 241/626 (38) | 0.50 (0.36, 0.68) | <.001 |
| GEP MGUS-like | NA | NA | NA | 166/626 (27) | 0.48 (0.33, 0.69) | <.001 |
| TT1 protocol | 231/1202 (19) | 1.54 (1.27, 1.86) | <.001 | NA | NA | NA |
| TT2 protocol | 668/1202 (56) | 0.83 (0.70, 1.00) | .045 | 668/971 (69) | 1.26 (0.95, 1.66) | .106 |
| TT3 protocol | 303/1202 (25) | 0.73 (0.56, 0.95) | .020 | 303/971 (31) | 0.80 (0.60, 1.05) | .106 |
| Los-CR* | 3.71 (3.00, 4.60) | <.001 | 5.13 (3.87, 6.79) | <.001 | ||
| Did not achieve CR* | 1.80 (1.50, 2.16) | <.001 | 2.06 (1.64, 2.59) | <.001 | ||
| Multivariate without GEP | ||||||
| CAs | 362/1149 (32) | 1.85 (1.54, 2.21) | <.001 | 288/935 (31) | 1.93 (1.55, 2.40) | <.001 |
| B2M > 5.5 mg/L | 221/1149 (19) | 1.63 (1.32, 2.00) | <.001 | 180/935 (19) | 1.63 (1.28, 2.08) | <.001 |
| CRP ≥ 8 mg/L | 426/1149 (37) | 1.22 (1.02, 1.47) | .032 | NS | NS | NS |
| LDH ≥ 190 U/L | 323/1149 (28) | 1.35 (1.11, 1.64) | .003 | 277/935 (30) | 1.45 (1.15, 1.82) | .002 |
| Los-CR* | 9.26 (6.79, 12.63) | <.001 | 10.09 (6.95, 14.65) | <.001 | ||
| Did not achieve CR* | 4.45 (3.38, 5.86) | <.001 | 4.31 (3.15, 5.90) | <.001 | ||
| Multivariate with GEP | ||||||
| LDH ≥ 190 U/L | NA | NA | NA | 188/608 (31) | 1.61 (1.19, 2.18) | <.001 |
| CAs | NA | NA | NA | 203/608 (33) | 2.13 (1.58, 2.86) | <.001 |
| GEP high-risk | NA | NA | NA | 84/608 (14) | 1.31 (0.74, 2.30) | <.001 |
| GEP HY or LB subgroups | NA | NA | NA | 236/608 (39) | 0.53 (0.37, 0.76) | <.001 |
| GEP MGUS-like | NA | NA | NA | 157/608 (26) | 0.63 (0.41, 0.97) | .034 |
| Los-CR* | NA | NA | NA | 10.12 (6.03, 16.99) | <.001 | |
| Did not achieve CR* | NA | NA | NA | 5.41 (3.52, 8.33) | <.001 | |
| Variable . | TT1, TT2, and TT3 combined . | TT2 and TT3 combined . | ||||
|---|---|---|---|---|---|---|
| n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | |
| Univariate | ||||||
| Age ≥ 65 y | 241/1202 (20) | 1.37 (1.11, 1.68) | .004 | 220/971 (23) | 1.38 (1.08, 1.75) | .009 |
| Female | 470/1202 (39) | 0.92 (0.77, 1.10) | .357 | 382/971 (39) | 0.93 (0.75, 1.15) | .484 |
| White | 1054/1202 (88) | 1.08 (0.83, 1.40) | .575 | 848/971 (87) | 1.10 (0.80, 1.50) | .571 |
| Albumin < 3.5 g/dL | 253/1198 (21) | 1.48 (1.22, 1.80) | <.001 | 191/967 (20) | 1.65 (1.30, 2.10) | <.001 |
| B2M ≥ 3.5 mg/L | 474/1200 (40) | 1.72 (1.45, 2.03) | <.001 | 379/971 (39) | 1.79 (1.45, 2.21) | <.001 |
| B2M > 5.5 mg/L | 230/1200 (19) | 2.03 (1.68, 2.47) | <.001 | 187/971 (19) | 2.25 (1.79, 2.83) | <.001 |
| CRP ≥ 8 mg/L | 433/1183 (37) | 1.40 (1.18, 1.66) | <.001 | 363/960 (38) | 1.39 (1.13, 1.72) | .002 |
| Creatinine ≥ 2 mg/dL | 107/1188 (9) | 1.86 (1.44, 2.40) | <.001 | 85/957 (9) | 1.92 (1.42, 2.60) | <.001 |
| Hb < 10 g/dL | 333/1201 (28) | 1.50 (1.25, 1.79) | <.001 | 255/970 (26) | 1.44 (1.15, 1.80) | .001 |
| LDH ≥ 190 U/L | 334/1199 (28) | 1.71 (1.43, 2.05) | <.001 | 285/969 (29) | 1.85 (1.50, 2.30) | <.001 |
| CAs | 371/1184 (31) | 2.14 (1.80, 2.55) | <.001 | 297/963 (31) | 2.30 (1.87, 2.84) | <.001 |
| GEP high-risk | NA | NA | NA | 86/626 (14) | 4.12 (3.02, 5.63) | <.001 |
| GEP CD-1 subgroup | NA | NA | NA | 38/626 (6) | 0.76 (0.40, 1.44) | .407 |
| GEP CD-2 subgroup | NA | NA | NA | 72/626 (12) | 0.76 (0.47, 1.24) | .271 |
| GEP HY subgroup | NA | NA | NA | 178/626 (28) | 0.63 (0.45, 0.89) | .008 |
| GEP LB subgroup | NA | NA | NA | 63/626 (10) | 0.35 (0.17, 0.72) | .004 |
| GEP MF subgroup | NA | NA | NA | 41/626 (7) | 2.11 (1.35, 3.29) | .001 |
| GEP MS subgroup | NA | NA | NA | 77/626 (12) | 1.59 (1.09, 2.31) | .016 |
| GEP MY subgroup | NA | NA | NA | 96/626 (15) | 0.99 (0.67, 1.45) | .956 |
| GEP PR subgroup | NA | NA | NA | 61/626 (10) | 2.29 (1.56, 3.34) | <.001 |
| GEP MF, MS, or PR subgroups | NA | NA | NA | 179/626 (29) | 2.42 (1.83, 3.22) | <.001 |
| GEP HY or LB subgroups | NA | NA | NA | 241/626 (38) | 0.50 (0.36, 0.68) | <.001 |
| GEP MGUS-like | NA | NA | NA | 166/626 (27) | 0.48 (0.33, 0.69) | <.001 |
| TT1 protocol | 231/1202 (19) | 1.54 (1.27, 1.86) | <.001 | NA | NA | NA |
| TT2 protocol | 668/1202 (56) | 0.83 (0.70, 1.00) | .045 | 668/971 (69) | 1.26 (0.95, 1.66) | .106 |
| TT3 protocol | 303/1202 (25) | 0.73 (0.56, 0.95) | .020 | 303/971 (31) | 0.80 (0.60, 1.05) | .106 |
| Los-CR* | 3.71 (3.00, 4.60) | <.001 | 5.13 (3.87, 6.79) | <.001 | ||
| Did not achieve CR* | 1.80 (1.50, 2.16) | <.001 | 2.06 (1.64, 2.59) | <.001 | ||
| Multivariate without GEP | ||||||
| CAs | 362/1149 (32) | 1.85 (1.54, 2.21) | <.001 | 288/935 (31) | 1.93 (1.55, 2.40) | <.001 |
| B2M > 5.5 mg/L | 221/1149 (19) | 1.63 (1.32, 2.00) | <.001 | 180/935 (19) | 1.63 (1.28, 2.08) | <.001 |
| CRP ≥ 8 mg/L | 426/1149 (37) | 1.22 (1.02, 1.47) | .032 | NS | NS | NS |
| LDH ≥ 190 U/L | 323/1149 (28) | 1.35 (1.11, 1.64) | .003 | 277/935 (30) | 1.45 (1.15, 1.82) | .002 |
| Los-CR* | 9.26 (6.79, 12.63) | <.001 | 10.09 (6.95, 14.65) | <.001 | ||
| Did not achieve CR* | 4.45 (3.38, 5.86) | <.001 | 4.31 (3.15, 5.90) | <.001 | ||
| Multivariate with GEP | ||||||
| LDH ≥ 190 U/L | NA | NA | NA | 188/608 (31) | 1.61 (1.19, 2.18) | <.001 |
| CAs | NA | NA | NA | 203/608 (33) | 2.13 (1.58, 2.86) | <.001 |
| GEP high-risk | NA | NA | NA | 84/608 (14) | 1.31 (0.74, 2.30) | <.001 |
| GEP HY or LB subgroups | NA | NA | NA | 236/608 (39) | 0.53 (0.37, 0.76) | <.001 |
| GEP MGUS-like | NA | NA | NA | 157/608 (26) | 0.63 (0.41, 0.97) | .034 |
| Los-CR* | NA | NA | NA | 10.12 (6.03, 16.99) | <.001 | |
| Did not achieve CR* | NA | NA | NA | 5.41 (3.52, 8.33) | <.001 | |
The multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the .05 level. A multivariate P value greater than .05 indicates variable forced into model with significant variables chosen using stepwise selection.
HR indicates hazard ratio; CI, confidence interval; P, P value from Wald χ2 test in Cox regression; NA, not applicable; and NS, multivariate results not statistically significant at .05 level. All univariate P values are reported regardless of significance.
Treated as a time-dependent variable.
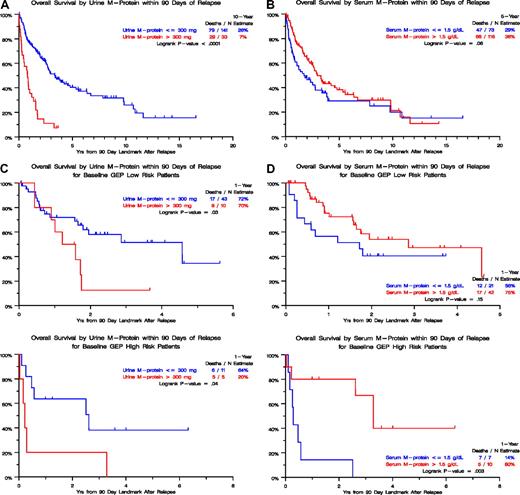
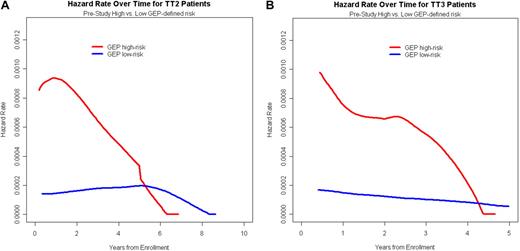
The relapse kinetics in the first 3 months of documentation of relapse from CR had opposite implications for urine-M and serum-M categories. Those with higher levels of urine-M (> 300 mg/day) had a median postrelapse survival (PRS), measured from a 90-day landmark, of only 1 year as opposed to 3 years for the patients with slower relapse kinetics (P < .001; Figure 2A). In the case of serum-M, higher levels (> 1.5 g/dL) were favorable (P = .06; Figure 2B). When put into the context of GEP-defined risk, higher urine-M levels observed in the first 3 months after relapse documentation conferred shorter PRS in both low- and high-risk GEP groups (Figure 2C), whereas, in the case of serum-M, higher levels favored longer PRS in the high-risk group (P = .003; Figure 2D). We interpret these data as being consistent with urine-M secretion representing less and serum-M more differentiated disease relapse. Analysis of hazard rates over time for low-risk and high-risk disease in TT2 and TT3 trials revealed a steep decline in high-risk myeloma to levels of low-risk disease, approximately 5 years for TT2 and 4 years for TT3 (Figure 3).
Postrelapse survival by level of urine-M protein or serum M-protein within 90 days of relapse. (A) In the absence of GEP-defined risk designation (TT1, TT2, and TT3 patients combined), postrelapse survival was significantly shorter when urinary M excretion exceeded 300 mg/day than in case of lower values. (B) In the absence of GEP-defined risk designation (TT1, TT2, and TT3 patients combined), postrelapse survival tended to be longer when serum-M levels exceeded 1.5 g/dL than in case of lower levels. (C) In the presence of GEP-defined risk designation, available in subsets of patients treated with TT2 and TT3, postrelapse survival was significantly shorter when urinary M excretion exceeded 300 mg/day than in case of lower values, regardless of GEP-defined risk (low risk, top panel; high risk, bottom panel). (D) In the presence of GEP-defined risk designation, available in subsets of patients treated with TT2 and TT3, postrelapse survival tended to be longer when serum-M levels exceeded 1.5 g/dL than in case of lower levels in the setting of low-risk myeloma (top panel) whereas, in the high-risk setting, postrelapse survival was significantly prolonged (bottom panel).
Postrelapse survival by level of urine-M protein or serum M-protein within 90 days of relapse. (A) In the absence of GEP-defined risk designation (TT1, TT2, and TT3 patients combined), postrelapse survival was significantly shorter when urinary M excretion exceeded 300 mg/day than in case of lower values. (B) In the absence of GEP-defined risk designation (TT1, TT2, and TT3 patients combined), postrelapse survival tended to be longer when serum-M levels exceeded 1.5 g/dL than in case of lower levels. (C) In the presence of GEP-defined risk designation, available in subsets of patients treated with TT2 and TT3, postrelapse survival was significantly shorter when urinary M excretion exceeded 300 mg/day than in case of lower values, regardless of GEP-defined risk (low risk, top panel; high risk, bottom panel). (D) In the presence of GEP-defined risk designation, available in subsets of patients treated with TT2 and TT3, postrelapse survival tended to be longer when serum-M levels exceeded 1.5 g/dL than in case of lower levels in the setting of low-risk myeloma (top panel) whereas, in the high-risk setting, postrelapse survival was significantly prolonged (bottom panel).
Time-dependent hazard rate of death in TT2 and TT3 protocols according to GEP-defined risk. (A) For TT2, the hazard rate of death declined steeply in high-risk myeloma to reach levels of low-risk disease at 5 to 6 years. (B) For TT3, the hazard rate of death declined steeply in high-risk myeloma to reach levels of low-risk disease at 4 to 5 years.
Time-dependent hazard rate of death in TT2 and TT3 protocols according to GEP-defined risk. (A) For TT2, the hazard rate of death declined steeply in high-risk myeloma to reach levels of low-risk disease at 5 to 6 years. (B) For TT3, the hazard rate of death declined steeply in high-risk myeloma to reach levels of low-risk disease at 4 to 5 years.
Discussion
We have previously addressed the issue of prognostic implications of CR in myeloma. CR was not critical to the outcomes of patients with a documented preceding course of smoldering disease25 or with an MGUS-like GEP signature,18 but was of great prognostic importance in the setting of GEP-defined high-risk myeloma.11 The topic was also the subject of 2 Inside Blood commentaries7,26 and a letter to the editor of Leukemia.27 Here we have addressed, for the first time, the importance not only of attaining CR per se but also the issues of timing of onset and duration of CR. Applying time-dependent variable methodology permitted the inclusion of all trial subjects. Independent of traditional prognostic baseline parameters (CA, LDH, B2M, albumin) and even GEP-defined high-risk designation, survival was dominantly favorably affected both by achieving CR early in the treatment course and by sustaining such status for prolonged time durations.
Of interest was the observation that, in the case of relapse from CR, a higher magnitude of M-protein increase in the first 3 months had opposite PRS implications for serum-M (favorable) and urine-M (adverse) levels. Whereas the latter was GEP-risk-independent, higher serum-M levels were associated with superior PRS only in the high-risk setting. We interpret these findings as indicative of urinary light chains being reflective of a less differentiated type of myeloma compared with complete immunoglobulin secretion.
The consistency of data in TT2 and TT3, in the absence and presence of GEP data, provides validation of our CR model. Given its dire prognosis, the high-risk setting requires major emphasis on (1) maximizing the odds of achieving CR status promptly and (2) sustaining such CR for a minimum of 4 to 5 years, beyond which hazard rates decline to levels of low-risk disease.28 Our results provide a basis for assessing the issue of CR consolidation, especially in high-risk disease, to determine, prospectively, the critical time required for such CR status to be actively sustained.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the National Cancer Institute (Program Project Grant CA 55819), Bethesda, MD.
National Institutes of Health
Authorship
Contribution: A.H., J.C., J.D.S., and B.B. designed the study; A.H. and B.B. wrote the manuscript; K.H., Y.A., S.W., B.N., F.v.R., E.A., and B.B. accrued patients and provided clinical care; and A.H., J.C., and J.S. performed statistical analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bart Barlogie, Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, Slot 816, 4301 West Markham, Little Rock, AR 72205; e-mail: barlogiebart@uams.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal