Abstract
To more comprehensively assess the pathogenic contribution of the PTEN-PI3K-AKT pathway to T-cell acute lymphoblastic leukemia (T-ALL), we examined diagnostic DNA samples from children with T-ALL using array comparative genomic hybridization and sequence analysis. Alterations of PTEN, PI3K, or AKT were identified in 47.7% of 44 cases. There was a striking clustering of PTEN mutations in exon 7 in 12 cases, all of which were predicted to truncate the C2 domain without disrupting the phosphatase domain of PTEN. Induction chemotherapy failed to induce remission in 3 of the 4 patients whose lymphoblasts harbored PTEN deletions at the time of diagnosis, compared with none of the 12 patients with mutations of PTEN exon 7 (P = .007), suggesting that PTEN deletion has more adverse therapeutic consequences than mutational disruptions that preserve the phosphatase domain. These findings add significant support to the rationale for the development of therapies targeting the PTEN-PI3K-AKT pathway in T-ALL.
Introduction
Despite recent improvements in therapy, approximately 25% of children and 50% to 70% of adults with T-cell acute lymphoblastic leukemia (T-ALL) develop treatment-resistant disease,1,2 which carries a dire prognosis.3 Molecularly targeted agents hold considerable promise for the treatment of T-ALL, although limits in our current understanding of the key pathways that drive T-ALL pathogenesis restrict our ability to use these agents effectively
PTEN is a negative regulator of oncogenic PI3K-AKT signaling,4 and recent studies have demonstrated the inactivation of PTEN in human T-ALL cell lines and primary samples.5-8 Furthermore, the inactivation of PTEN has been shown to play a prominent role in resistance to NOTCH inhibition in T-ALL cell lines, an effect that appears to be mediated by AKT.7 The activation of PI3K-AKT signaling can also occur by mutation of PI3K or AKT genes, which have not previously been assessed in T-ALL. Finally, the spectrum of PTEN mutations has not been extensively analyzed in clinical samples of primary T-ALL. Here we investigated the frequency and prognostic implications of PTEN, PI3K, and AKT abnormalities in childhood T-ALL, using array comparative genomic hybridization (CGH), fluorescence in situ hybridization (FISH), and sequence analysis.
Methods
T-ALL diagnostic specimens were collected with informed consent obtained in accordance with the Declaration of Helsinki and Institutional Review Board approval from children treated on Children's Oncology Group 9404 or Dana-Farber Cancer Institute 00-001 clinical trials.9,10 Complete materials and methods are available in supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results and discussion
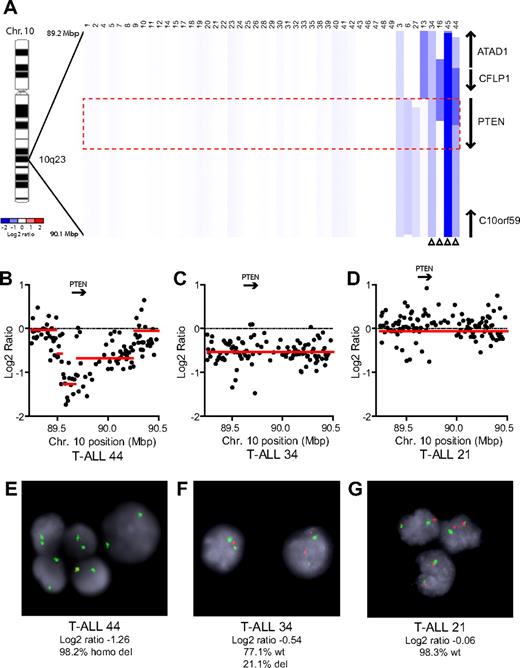
We performed array CGH with genomic DNAs from 47 pediatric T-ALL diagnostic specimens, 7 of which were reported previously.5 Homozygous deletions of PTEN were identified in 2 cases (cases 44 and 45), and heterozygous deletions in 2 others (cases 34 and 16; Figure 1). An additional case, T-ALL 13, harbored a heterozygous deletion that spanned a locus immediately upstream of PTEN, with no CGH evidence of deletion involving PTEN coding sequence. Because this deletion may or may not have disrupted upstream gene regulatory elements, we considered the PTEN status of this case to be indeterminate. FISH analysis with a commercial PTEN probe was used to validate our CGH results in cases with sufficient cells (Figure 1B-G). Overall, PTEN deletions were identified in 8.7% (n = 4 of 46) of primary T-ALL samples.
PTEN deletions in T-ALL. (A) Array CGH was performed with genomic DNA from diagnostic specimens collected from 47 children with T-ALL. The data are shown as a dChip plot of CGH segmented log2 copy number ratios at the PTEN locus. The red box denotes the location of the PTEN coding sequence. White arrowheads point to cases with segmented log2 copy number ratios of less than −0.5 involving the PTEN coding sequence. Two samples on which CGH was unsuccessful (T-ALL 36 and 37) were excluded from analysis. (B-D) Raw CGH data from representative patient samples. Red lines represent the segmented log2 copy number ratio shown in panel A. (E-G) FISH analysis of representative cases confirmed the deletions identified by CGH. Orange, PTEN probe; green, centromere 10 probe. Images were obtained with an Axio Imager A1 fluorescence microscope (Carl Zeiss) using a 100× Alpha Apochromatic Plan oil-immersion objective (Carl Zeiss), a JAI CV-M4+CL progressive scan camera (JAI Inc), and Genus acquisition software version 3.92 build 7 (Genetix USA). Note that the Genus cytogenetic image acquisition software applies an automated “thresholding” algorithm that sets a signal intensity threshold below which any signal is considered background and thus excluded from the final composite image. During image acquisition, all images generated by the software were compared to the view from the microscope to confirm that they were fully representative. (B,E) Homozygous deletions had log2 ratios of −1.26 (case 44) and −4.11 (case 45). Cells were available for FISH on case 44 and clearly showed homozygous loss of PTEN. (C,F) The CGH detection of a heterozygous deletion in case 34 (log2 ratio, −0.54) correlated with the detection of PTEN deletion by FISH on 1 allele in 21% of the cells examined. (D,G) Case 21 retained both PTEN alleles intact by FISH and CGH.
PTEN deletions in T-ALL. (A) Array CGH was performed with genomic DNA from diagnostic specimens collected from 47 children with T-ALL. The data are shown as a dChip plot of CGH segmented log2 copy number ratios at the PTEN locus. The red box denotes the location of the PTEN coding sequence. White arrowheads point to cases with segmented log2 copy number ratios of less than −0.5 involving the PTEN coding sequence. Two samples on which CGH was unsuccessful (T-ALL 36 and 37) were excluded from analysis. (B-D) Raw CGH data from representative patient samples. Red lines represent the segmented log2 copy number ratio shown in panel A. (E-G) FISH analysis of representative cases confirmed the deletions identified by CGH. Orange, PTEN probe; green, centromere 10 probe. Images were obtained with an Axio Imager A1 fluorescence microscope (Carl Zeiss) using a 100× Alpha Apochromatic Plan oil-immersion objective (Carl Zeiss), a JAI CV-M4+CL progressive scan camera (JAI Inc), and Genus acquisition software version 3.92 build 7 (Genetix USA). Note that the Genus cytogenetic image acquisition software applies an automated “thresholding” algorithm that sets a signal intensity threshold below which any signal is considered background and thus excluded from the final composite image. During image acquisition, all images generated by the software were compared to the view from the microscope to confirm that they were fully representative. (B,E) Homozygous deletions had log2 ratios of −1.26 (case 44) and −4.11 (case 45). Cells were available for FISH on case 44 and clearly showed homozygous loss of PTEN. (C,F) The CGH detection of a heterozygous deletion in case 34 (log2 ratio, −0.54) correlated with the detection of PTEN deletion by FISH on 1 allele in 21% of the cells examined. (D,G) Case 21 retained both PTEN alleles intact by FISH and CGH.
To identify other genetic lesions that could activate PI3K-AKT signaling, we carefully examined the CGH data but did not find focal copy number alterations involving the PI3K or AKT genes, or the PDK1 and p70s6k genes, which encode other components of the pathway known to be amplified in other types of human cancers. We then sequenced the entire PTEN coding region in 44 of the 47 samples on which CGH arrays were performed, as well as selected AKT and PI3K exons known to harbor oncogenic mutations in human cancers. These included exons 9 and 20 of PIK3CA (encoding the catalytic subunit of class IA PI3K),11,12 exons 12 and 13 of PIK3RA (encoding the regulatory subunit of class IA PI3K),13 and exon 2 of the AKT1-3 genes.14 We additionally sequenced exons 1 and 2 of NRAS and KRAS and exons 3 and 13 of PTPN11, which act upstream of PI3K-AKT signaling and are each known to harbor mutations in some cases of ALL,15-17 as well as exons 26, 27, and 34 of NOTCH1, and exons 9 and 10 of FBXW7.18,19
Nonsynonymous sequence alterations in PTEN were identified in 12 (27.3%) of the 44 primary T-ALL patient samples that were sequenced. Each mutation consisted of a unique nonsense or frame-shift mutation in exon 7, most of which resulted from small insertions or insertion/deletions that were predicted to cause truncation of the protein resulting from premature termination of translation. Of the 12 T-ALL cases with PTEN sequence alterations, 3 harbored biallelic alterations resulting from compound heterozygous mutations, whereas the other 9 harbored simple heterozygous mutations. No mutations were identified in cases with heterozygous PTEN deletions. Strikingly, all of the mutations identified were predicted to disrupt the PTEN protein within an 18-amino acid region of the C2 domain, leading to a carboxy-terminal truncation (Figure 2A; supplemental Table 1). Importantly, the PTEN phosphatase core domain, encoded by exon 5 and targeted by half of the PTEN mutations described in other types of primary human tumor samples,20 was not disrupted by mutations identified in our T-ALL samples. Although PTEN mutations can occur in exon 7 in other cancers, including glioblastoma, endometrial, breast, and prostate carcinomas,20 detection of mutations exclusively in this region has not been described in other types of human cancer.20-22 Interestingly, the clustering of mutations within exon 7 is specific to primary T-ALL samples, as PTEN mutations in T-ALL cell lines frequently disrupt the phosphatase domain.5,7,23
Mutations of PTEN and the PI3K-AKT pathway in T-ALL. (A) Sequencing of PTEN in 44 of the primary samples shown in Figure 1 identified nonsynonymous sequence alterations in 12 of these samples, all of which were predicted to disrupt the PTEN protein within an 18-amino acid region of the C2 domain. Note that the specific mutations in cases 14 and 27 were impossible to determine because of the presence of 2 simultaneous frameshift sequences. (B) Targeted sequencing of PIK3R1, PIK3CA, and AKT1-3 exons known to be mutated in human cancer identified nonsynonymous sequence alterations in PTEN and the PI3K-AKT pathway in 47.7% of primary T-ALL cases. Lesions within the PTEN-PI3K-AKT pathway were mutually exclusive. Abnormalities in the NF1 and RAS genes were also identified but were not solely associated with PTEN-PI3K-AKT pathway abnormalities. *Novel in-frame insertion/deletions. (C-D) Kaplan-Meier event-free survival curves for the 44 cases analyzed by CGH and sequencing demonstrate that, overall, genetic alterations of the PTEN-PI3K-AKT pathway did not predict event-free survival, whereas deletions of PTEN were significantly associated with early treatment failure.
Mutations of PTEN and the PI3K-AKT pathway in T-ALL. (A) Sequencing of PTEN in 44 of the primary samples shown in Figure 1 identified nonsynonymous sequence alterations in 12 of these samples, all of which were predicted to disrupt the PTEN protein within an 18-amino acid region of the C2 domain. Note that the specific mutations in cases 14 and 27 were impossible to determine because of the presence of 2 simultaneous frameshift sequences. (B) Targeted sequencing of PIK3R1, PIK3CA, and AKT1-3 exons known to be mutated in human cancer identified nonsynonymous sequence alterations in PTEN and the PI3K-AKT pathway in 47.7% of primary T-ALL cases. Lesions within the PTEN-PI3K-AKT pathway were mutually exclusive. Abnormalities in the NF1 and RAS genes were also identified but were not solely associated with PTEN-PI3K-AKT pathway abnormalities. *Novel in-frame insertion/deletions. (C-D) Kaplan-Meier event-free survival curves for the 44 cases analyzed by CGH and sequencing demonstrate that, overall, genetic alterations of the PTEN-PI3K-AKT pathway did not predict event-free survival, whereas deletions of PTEN were significantly associated with early treatment failure.
Nonsynonymous sequence alterations were also identified in the PI3K and AKT genes (supplemental Table 1). An E17K AKT1 activating mutation was identified in one case, whereas 2 others harbored activating mutations of PIK3CA, encoding the catalytic subunit of class IA PI3K. Two additional cases harbored novel in-frame insertions/deletions in the PI3K regulatory subunit PIK3R1, in a region of the gene frequently mutated in human cancers.13,24 Alterations of PTEN, AKT, and PI3K were mutually exclusive and occurred in 21 (47.7%) of the 44 primary T-ALL patient samples analyzed by both array CGH and sequencing (Figure 2B). When analyzed together, genetic alterations in the PTEN-PI3K-AKT pathway did not predict event-free survival (Figure 2C), in contrast to PTEN deletions, which were significantly associated with early treatment failure (Figure 2D). This suggests that deletions and truncating mutations may have different implications for clinical outcome in T-ALL. Indeed, induction chemotherapy failed in 3 of the 4 patients with PTEN deletions, including both cases with homozygous deletions, compared with none of the 12 cases with PTEN exon 7 mutations (P = .007). Nevertheless, the number of patients with PTEN deletions we have identified is small, and it will be important to confirm the prognostic utility of PTEN deletions in a sufficient number of additional cases before incorporating this finding into clinical decision-making.
We also identified activating mutations of NRAS in 4 cases, including 3 without genetic alterations in the PTEN-PI3K-AKT pathway and one with a PTEN mutation (Figure 2B). One of these cases harbored a heterozygous NF1 deletion (supplemental Table 1). An activating KRAS mutation was identified in a case that also had an activating NRAS mutation. There was no apparent correlation between alterations of the PTEN-PI3K-AKT or RAS-NF1 pathways and known T-ALL oncogenic abnormalities, including NOTCH1 or FBXW7 mutation, MYB duplication, or CDKN2A gene deletion (supplemental Table 1). Finally, 2 cases had a homozygous RB1 deletion (supplemental Table 1), a genomic aberration not previously described in primary T-ALL samples.
The detection of abnormalities in the PTEN, PI3K, and AKT genes in a large fraction of primary T-ALL samples demonstrates a prominent role for oncogenic PI3K-AKT signaling in the pathogenesis of T-ALL. Moreover, PTEN deletions appeared to impart a high risk of induction failure with contemporary chemotherapy. Our findings add significant support to the rationale for clinical trials of small molecule inhibitors of PI3K, AKT, and mTOR, now in development,25 as therapeutic agents for T-ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the children with T-ALL and their families, as well as members of the Children's Oncology Group and Dana-Farber Cancer Institute Acute Lymphoblastic Leukemia Consortium member institutions, for the samples analyzed in these studies; and John Gilbert for editorial assistance.
This work was supported by the National Institutes of Health (grant 5P01CA68484; S.E.S., A.T.L.) and by an award from the Wine Advocate Fund for Philanthropy of the Community Foundation of the National Capital Region. Children's Oncology Group cell banking and sample distribution were supported by the Children's Oncology Group 9900 cell biology study and grants CA98543 and CA114766. A.G. is supported by the William Lawrence Foundation, the American Society of Hematology-Amos Medical Faculty Development program, and by the National Institutes of Health (grant 1K08CA133103). S.P.H. is the Ergen Family Chair in Pediatric Cancer at the University of Colorado. The array CGH analyses were performed and supported in part by the Center for Applied Cancer Science of the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science.
A list of Children's Oncology Group and Dana-Farber Cancer Institute Acute Lymphoblastic Leukemia Consortium participants appears in the supplemental Appendix.
National Institutes of Health
Authorship
Contribution: A.G. designed, performed, and analyzed research and wrote the paper; T.S., R.G., Y.A., and L.A.M. performed research and analyzed data; S.D. and D.N. analyzed data; S.S.W., R.L., L.B.S., S.P.H., and S.E.S. provided vital reagents and analyzed data; J.Z., A.P., and L.C. developed vital CGH analytical tools and analyzed data; A.C., L.S., and P.P.P. analyzed data; and A.T.L. supervised research and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. Thomas Look, Department of Pediatric Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: Thomas_look@dfci.harvard.edu.