Abstract
Deregulated cell survival programs are a classic hallmark of cancer. We have previously identified a serine residue (Ser585) in the βc subunit of the granulocyte-macrophage colony-stimulating factor receptor that selectively and independently promotes cell survival. We now show that Ser585 phosphorylation is constitutive in 20 (87%) of 23 acute myeloid leukemia (AML) patient samples, indicating that this survival-only pathway is frequently deregulated in leukemia. We performed a global expression screen to identify gene targets of this survival pathway and report a 138-gene βc Ser585-regulated transcriptome. Pathway analysis defines a gene network enriched for PI3-kinase target genes and a cluster of genes involved in cancer and cell survival. We show that one such gene, osteopontin (OPN), is a functionally relevant target of the Ser585-survival pathway as shown by siRNA-mediated knockdown of OPN expression that induces cell death in both AML blasts and CD34+CD38−CD123+ leukemic progenitors. Increased expression of OPN at diagnosis is associated with poor prognosis with multivariate analysis indicating that it is an independent predictor of overall patient survival in normal karyotype AML (n = 60; HR = 2.2; P = .01). These results delineate a novel cytokine-regulated Ser585/PI3-kinase signaling network that is deregulated in AML and identify OPN as a potential prognostic and therapeutic target.
Introduction
Many hemopoietic cytokines are potent regulators of both cell survival and proliferation. However, although cell survival and proliferation are often viewed as inextricably linked and overlapping cellular fates, they are subject to independent regulation and can be controlled by distinct signaling pathways.1 For example, cytokines such as granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-3 (IL-3) have been long recognized to promote cell survival in the absence of other biologic responses such as proliferation, differentiation, or activation.2 This ability to regulate cell survival alone is particularly important in the hemopoietic compartment where many cell types require the continuous presence of cytokines to survive and rapidly execute apoptosis programs on cytokine withdrawal.3 The corollary of this is that cells receiving a mitogenic signal in the absence of a concomitant growth factor–mediated prosurvival signal attempt to undergo cell-cycle progression but die by apoptosis.4 Segregating the cytokine-mediated signals that promote hemopoietic cell survival from those that regulate proliferation has important biologic advantages in that it enforces at least 2 obligate steps for cellular transformation and leukemogenesis: one in which deregulated cell proliferation programs are activated and the other in which deregulated survival signals override cell death programs.
Mutations that result in the activation of components of cytokine signaling pathways have been identified in a wide range of human leukemias. These mutations have been found in tyrosine kinase receptors (eg, FLT3-ITD, FLT3-D835, c-KIT–D816, c-KIT–ITD), tyrosine kinases (BCR-ABL, TEL-JAK2), and other components of cytokine signaling pathways (N-RAS, K-RAS).5 Others have shown that the expression of activating mutants of the GM-CSF and IL-3 receptor β subunit (βc) or deregulation of GM-CSF signaling pathways can promote myeloproliferative disease or leukemia in mice,6 and loss of the neurofibromatosis type 1 gene results in hypersensitivity to GM-CSF and juvenile chronic myelogenous leukemia.7 Furthermore, deregulation of phosphoinositide 3-kinase (PI3-kinase) signaling has been observed in a range of cancers, including myeloproliferative diseases and leukemia.8,9 Collectively, what these findings show is that subverting the normal cytokine receptor signaling pathways promotes deregulated cell survival and proliferation leading to cellular transformation.
Although some cytokines are able to regulate cell survival independently of other cellular responses,2,3 cytokine receptor signaling motifs specifically dedicated to cell survival only have not been identified. Our previous studies identified 2 cytokine receptor pathways leading to cell survival. Those studies identified a specific motif in the cytoplasmic domain of the βc subunit of the GM-CSF receptor composed of Ser585 and Tyr577 that functions as a phospho-binary switch whereby Ser585 phosphorylation specifically regulates cell survival only, whereas Tyr577 phosphorylation integrates both cell survival and proliferation.10-12 To better define the survival response initiated by Ser585 of βc, we have performed microarray analysis and identified a 138-gene transcriptome of which a major component consists of PI3-kinase target genes. We have previously shown in preliminary studies that the phospho-Tyr577/phospho-Ser585 binary switch may be subject to deregulation in at least some patients with acute myeloid leukemia (AML).12 We now show that the Ser585 survival-only “arm” of the binary switch is deregulated in 20 (87%) of 23 AML samples, and bioinformatics analysis identifies a Ser585/PI3-kinase gene network. Although the expression level of one Ser585-regulated gene, osteopontin (OPN; aka SPP1, secreted phosphoprotein 1) correlated with overall survival (OS) in a heterogeneous cohort of 52 AML samples, it was not deemed significant by multivariate analysis. Importantly, however, high OPN expression was an independent predictor of poor OS in a 60-patient cohort of cytogenetically normal patients. Furthermore, blockade of OPN expression induces cell death in AML blasts as well as leukemic stem and progenitor cells (LSPCs). Our findings define a novel cytokine receptor survival-only pathway that is deregulated in AML and highlight OPN as a valuable drug target and prognostic indicator in AML.
Methods
Microarray analysis, pathway analysis, mutational analysis, quantitative RT-PCR, and statistical analysis
Detailed descriptions of the methods for the microarray analysis, pathway analysis, mutational analysis, quantitative reverse transcription–polymerase chain reaction (RT-PCR), and statistical analysis are given in the supplemental Materials (available on the Blood website; see the Supplemental Materials link at the top of the online article). All microarray data have been deposited in the Gene Expression Omnibus public database under accession no. GSE18222.
Patient material
Apheresis product and bone marrow or peripheral blood samples were obtained from patients with AML. For the OPN expression studies, bone marrow aspirates of 95 consecutive patients diagnosed with AML between 1998 and 2008 at Royal Adelaide Hospital (RAH) Australia were obtained after informed consent according to institutional guidelines in keeping with the Declaration of Helsinki, and studies were approved by the RAH Human Ethics Committee (patient data in supplemental Table 1). Diagnosis was confirmed by using cytomorphology, cytogenetics, and leukocyte antigen expression and was evaluated according to the French-American-British (FAB) classification. Cytogenetic risk classification categories were defined according to the Medical Research Council schema.13 Patients were treated with standard induction chemotherapy (combination of cytarabine, idarubicin, etoposide) according to the Australian Leukemia Lymphoma Group M7 protocol.14 For OPN expression studies of normal karyotype AML, an additional collection of 35 normal karyotype AML patient bone marrow aspirates were collected from the Kumamoto University School of Medicine, Japan, between 1987 and 2003 after informed consent based on the revised Helsinki protocol and approval by the institutional review board of Kumamoto University School of Medicine, Japan. Samples were analyzed by RT-PCR at the National University of Singapore. All evaluable patients underwent induction and consolidation chemotherapy according to the Japan Adult Leukemia Study Group protocols AML8715 and AML9716 and had confirmed normal karyotype by the G-banding method examining 20 mitoses. In cases showing normal karyotype, the absence of fusion mRNAs for RUNX1-ETO or PEBP2β-MYH11 were further confirmed with the RT-PCR method.
Cytokine signaling
Mononuclear cells (MNCs) from healthy donors and patients with AML were isolated by Ficoll-Hypaque density-gradient centrifugation, washed, and resuspended in PBS containing 0.1% human albumin (CSL), and stimulated for 5 minutes with GM-CSF. MNCs were lysed, and the βc subunit was immunoprecipitated with 1C1 and 8E4 anti-βc mAb and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis.12 OPN was isolated from human milk, and thrombin was cleaved as previously described.17,18 Anti–active-ERK pAb (Promega) was used at 50 ng/mL. Anti–phosphorylated Ser473 Akt pAb (Cell Signaling Technology Inc) and anti-p85 pAb (UBI) were used at 1:1000. Affinity-purified phospho-specific anti-βc phospho-585 pAb, anti-βc phospho-577 pAb, and anti-14-3-3 pAb were used at 1:1000.12 PI3-kinase assays were performed as previously described.10
Cell survival assays
CD34+ cells were purified from AML MNCs using CD34 MicroBeads according to the manufacturer's instructions (Miltenyi Inc). CD34+ cells were transfected with 50nM BLOCK-iT fluorescent oligo and 50 to 150nM of either GC-control siRNA or scrambled OPN siRNA or OPN siRNA (Invitrogen; siRNA oligonucleotide sequences are presented in supplemental Table 2). Survival was determined by annexin V–Alexa 568 staining (Roche). Viable cell number was assessed with the use of Flow-Count Fluorospheres (BD Biosciences).
Patient characteristics
All patient characteristics and OPN expression levels are presented in supplemental Table 1.
Results
Phospho-Ser585 survival signaling regulates a gene transcriptional program
Our previous studies that used the factor-dependent CTL-EN hemopoietic cell line have shown that the phosphorylation of Ser585 in the cytoplasmic tail of the GM-CSF and IL-3 receptor βc subunit is essential for the binding of the 14-3-3 scaffolding proteins, the recruitment and activation of PI3-kinase, and the specific regulation of hematopoietic cell survival.10-12 We have now used this CTL-EN model system to interrogate transcriptional targets of Ser585 signaling by microarray screening (Figure 1A).
Microarray analysis of the Ser585-survival pathway. CTL-EN cells expressing either wild-type βc or the βcSer585Gly mutant were factor deprived for 18 hours and then stimulated with GM-CSF for 0, 12, and 18 hours. Total RNA was isolated, reverse transcribed, and labeled; cDNA was used to probe either NIA 15k cDNA or AMF 21K microarrays. Pairwise comparisons were performed as in panel A whereby each solid arrow represents a separate 15k cDNA slide (n = 14) and each dotted arrow represents a 21k oligonucleotide slide (n = 8) in which differential gene expression was examined in either (1) cells expressing the wild-type βc or the βcSer585Gly mutant at a single time point or (2) a single cell line at different time points. Microarray data were subjected to Bayesian analysis to identify genes that were differentially expressed in CTL-EN cells expressing the wild-type βc and βcSer585Gly mutant in response to GM-CSF at 12 hours (B) and 18 hours (C). Bayesian analysis is shown whereby the probability that a particular gene is differentially expressed (t statistic) is plotted against the magnitude (log2 scale) of the differential expression (interaction coefficient) for each gene represented on the cDNA array. Negative interaction coefficients correspond to genes that are more strongly expressed in cells expressing the wild-type βc than the βcSer585Gly mutant (eg, genes that are induced by GM-CSF in cells expressing the wild-type βc and not in the βcSer585Gly mutant). Positive interaction coefficients correspond to genes that are more strongly expressed in cells expressing the βcSer585Gly mutant than the wild-type βc (eg, genes that are repressed by GM-CSF in cells expressing the wild-type βc and not in the βcSer585Gly mutant). Selected genes identified by GO analysis in Figure 3 as having a role in cell survival or cytokine signaling and that also showed differential expression are highlighted. (D-E) Microarray data presented as the response in wild-type βc cells (M = log2 [fold change in response to GM-CSF in CTL-EN cells expressing the wild-type βc]) plotted against the response in the βcSer585Gly mutant cells (M = log2 [fold change in response to GM-CSF in CTL-EN cells expressing the βcSer585Gly mutant]). Although most genes are either not GM-CSF regulated (M values close to 0 for both wild-type βc and βcSer585Gly mutant) or are GM-CSF regulated in an equivalent manner in cells expressing the wild-type βc and βcSer585Gly mutant (M values lie close to the diagonal line), a restricted subset lies off the diagonal. The M values for the genes highlighted in panels B to E at 0, 12, and 18 hours were plotted for cells expressing either the wild-type βc (○) or the βcSer585Gly mutant (Δ) (F). (G) A clustered heat map of the 138 genes identified as being differentially expressed between the wild-type βc and βcSer585Gly mutant CTL-EN cells in response to GM-CSF was generated with the use of a Euclidean matrix. The M value (log2 fold change) for both CTL-EN cells expressing the wild-type βc or the βcSer585Gly mutant was converted to a color scale with red indicating that the gene is induced by GM-CSF, green indicating that the gene is repressed, and the color intensity indicating the magnitude of regulation.
Microarray analysis of the Ser585-survival pathway. CTL-EN cells expressing either wild-type βc or the βcSer585Gly mutant were factor deprived for 18 hours and then stimulated with GM-CSF for 0, 12, and 18 hours. Total RNA was isolated, reverse transcribed, and labeled; cDNA was used to probe either NIA 15k cDNA or AMF 21K microarrays. Pairwise comparisons were performed as in panel A whereby each solid arrow represents a separate 15k cDNA slide (n = 14) and each dotted arrow represents a 21k oligonucleotide slide (n = 8) in which differential gene expression was examined in either (1) cells expressing the wild-type βc or the βcSer585Gly mutant at a single time point or (2) a single cell line at different time points. Microarray data were subjected to Bayesian analysis to identify genes that were differentially expressed in CTL-EN cells expressing the wild-type βc and βcSer585Gly mutant in response to GM-CSF at 12 hours (B) and 18 hours (C). Bayesian analysis is shown whereby the probability that a particular gene is differentially expressed (t statistic) is plotted against the magnitude (log2 scale) of the differential expression (interaction coefficient) for each gene represented on the cDNA array. Negative interaction coefficients correspond to genes that are more strongly expressed in cells expressing the wild-type βc than the βcSer585Gly mutant (eg, genes that are induced by GM-CSF in cells expressing the wild-type βc and not in the βcSer585Gly mutant). Positive interaction coefficients correspond to genes that are more strongly expressed in cells expressing the βcSer585Gly mutant than the wild-type βc (eg, genes that are repressed by GM-CSF in cells expressing the wild-type βc and not in the βcSer585Gly mutant). Selected genes identified by GO analysis in Figure 3 as having a role in cell survival or cytokine signaling and that also showed differential expression are highlighted. (D-E) Microarray data presented as the response in wild-type βc cells (M = log2 [fold change in response to GM-CSF in CTL-EN cells expressing the wild-type βc]) plotted against the response in the βcSer585Gly mutant cells (M = log2 [fold change in response to GM-CSF in CTL-EN cells expressing the βcSer585Gly mutant]). Although most genes are either not GM-CSF regulated (M values close to 0 for both wild-type βc and βcSer585Gly mutant) or are GM-CSF regulated in an equivalent manner in cells expressing the wild-type βc and βcSer585Gly mutant (M values lie close to the diagonal line), a restricted subset lies off the diagonal. The M values for the genes highlighted in panels B to E at 0, 12, and 18 hours were plotted for cells expressing either the wild-type βc (○) or the βcSer585Gly mutant (Δ) (F). (G) A clustered heat map of the 138 genes identified as being differentially expressed between the wild-type βc and βcSer585Gly mutant CTL-EN cells in response to GM-CSF was generated with the use of a Euclidean matrix. The M value (log2 fold change) for both CTL-EN cells expressing the wild-type βc or the βcSer585Gly mutant was converted to a color scale with red indicating that the gene is induced by GM-CSF, green indicating that the gene is repressed, and the color intensity indicating the magnitude of regulation.
Data from the National Institute of Aging (NIA) 15K cDNA and Adelaide Microarray Facility (AMF) 21K oligonucleotide microarrays identified 138 genes of which 76 were common to both arrays (supplemental Table 3). Bayesian analysis for the identification of differentially regulated genes was performed on both datasets, and the results from the NIA 15K cDNA arrays are shown (Figure 1B-C). The fold change (M) in response to GM-CSF shows a restricted subset of genes lie off the diagonal, indicating differential response (Figure 1D-E), which included osteopontin (Opn), cyclin-dependent kinase 8 (Cdk8), vaccinia-related kinase 1 (Vrk1), and protein tyrosine phosphatase receptor type S (Ptprs). High-ranked genes selected by Gene Ontogeny (GO) analysis (see Figure 2) as having potential roles in cell survival or cytokine signaling are indicated in Figure 1B through E, and the microarray expression profiles for these genes are shown in Figure 1F. A heat map was generated for the 138 genes and shows that Ser585 signaling regulates a complex gene program consisting of multiple clusters (Figure 1G).
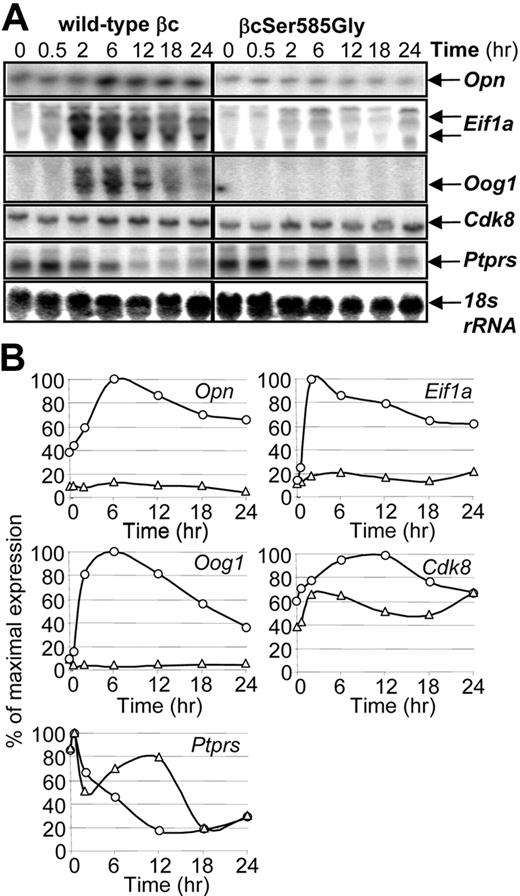
Ser585 regulates the expression of specific genes. CTL-EN cells expressing either the wild-type βc or the βcSer585Gly mutant were stimulated with GM-CSF for the indicated times after which time total RNA was purified and subjected to Northern analysis by using 32P-labeled cDNA probes (A). The Northern blot signals for each gene in CTL-EN cells expressing the wild-type βc (○) and βcSer585Gly mutant (Δ) were quantified by ImageQuant analysis from PhosphorImager screens and plotted as a percentage of the maximum signal observed (B).
Ser585 regulates the expression of specific genes. CTL-EN cells expressing either the wild-type βc or the βcSer585Gly mutant were stimulated with GM-CSF for the indicated times after which time total RNA was purified and subjected to Northern analysis by using 32P-labeled cDNA probes (A). The Northern blot signals for each gene in CTL-EN cells expressing the wild-type βc (○) and βcSer585Gly mutant (Δ) were quantified by ImageQuant analysis from PhosphorImager screens and plotted as a percentage of the maximum signal observed (B).
Genes were then selected for further validation by Northern analysis based on their Lod-odds ranking in microarray analysis (Figure 1; supplemental Table 3), GO analysis, Connectivity mapping, and Ingenuity Pathway Analysis (IPA; Figure 3). Although GM-CSF was able to induce the expression of Opn, eukaryotic translation initiation factor 1A (Eif1a), oogenesin 1 (Oog1), and Cdk8 in CTL-EN cells expressing the wild-type βc, this induction was reduced in cells expressing the βcSer585Gly mutant (Figure 2A-B). In contrast, although Ptprs expression was repressed by GM-CSF stimulation of CTL-EN cells expressing the wild-type βc, this repression was reduced in cells expressing the βcSer585Gly mutant (Figure 2A-B).
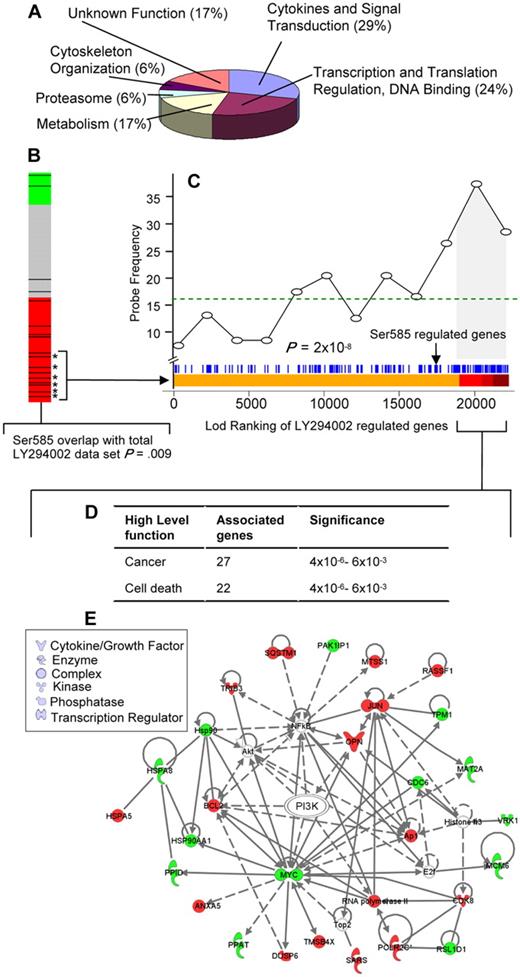
Identification of a Ser585/PI3-kinase transcriptional network. (A) Gene ontology (GO) (www.geneontology.org/) classifications are shown for the 138 Ser585-regulated genes identified by microarray screening. (B) Connectivity mapping19 of the 138 Ser585-regulated genes indicated negative connectivity with the expression change induced by the PI3-kinase inhibitor, LY294002. A negative connectivity (red) represents genes that are either induced by Ser585 and repressed by LY294002 or repressed by Ser585 and induced by LY294002. All 453 experiments in the Connectivity Map database were ranked by their connectivity score with the 138 Ser585-regulated genes identified in our studies. Each of the LY294002 comparisons (n = 17) is highlighted as a black bar on the heat map, and colors represent a negative (red), no connection (gray), or positive connection (green) between the 138 Ser585-regulated genes identified in our studies and individual LY294002-induced gene expression changes for each array in the Connectivity Map database. A cluster of LY294002 experiments (13 of 17) lie within the red region, indicating a statistically significant enrichment of our 138 Ser585-regulated genes and the LY294002 differentially expressed genes (P = .009). (C) Analysis of the overlap between the 138 Ser585-regulated gene set identified in our studies and the LY294002-sensitive genes in the Connectivity database. We pooled the data from the 6 top-ranked MCF7 cell LY294002 microarrays that showed significant overlap with our 138 gene set (bracketed asterisks) and performed a Wilcoxon rank sum test to compare a ranked list of LY294002 sensitive genes (Lod-ranking of LY294002-regulated genes) with our 138 gene set. Each blue line corresponds to an individual probe identified from our screen. Ser585-regulated genes (blue lines) cluster with top-ranked LY294002-sensitive genes (red region). Plotting this overlap shows a significant enrichment above a random distribution (dotted line) in Ser585-regulated genes (probe frequency) as the Lod-ranking of LY294002-regulated genes increases (P < .001). Of the 138 genes identified in our screen, 53 (38%) correspond to LY294002-sensitive genes encompassed by the gray portion of the graph (LY294002 cutoff; P = .01). (D) The 53 Ser585/PI3-kinase–regulated genes identified in panel C together with their fold change were uploaded into the IPA and overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of genes were then algorithmically generated based on their connectivity to known functional, biochemical, and disease interactions. A significant overlap with genes involved in cancer and cell death was observed (P < .001, right-tailed Fisher exact test). (E) The 53 Ser585/PI3-kinase–regulated genes identified in panel C were subjected to IPA mapping of network interactions. This analysis showed that 27 of 53 genes constitute a gene network with red denoting Ser585-induced genes and green denoting Ser585-repressed genes. Node shape denotes the indicated gene function or biologic process.
Identification of a Ser585/PI3-kinase transcriptional network. (A) Gene ontology (GO) (www.geneontology.org/) classifications are shown for the 138 Ser585-regulated genes identified by microarray screening. (B) Connectivity mapping19 of the 138 Ser585-regulated genes indicated negative connectivity with the expression change induced by the PI3-kinase inhibitor, LY294002. A negative connectivity (red) represents genes that are either induced by Ser585 and repressed by LY294002 or repressed by Ser585 and induced by LY294002. All 453 experiments in the Connectivity Map database were ranked by their connectivity score with the 138 Ser585-regulated genes identified in our studies. Each of the LY294002 comparisons (n = 17) is highlighted as a black bar on the heat map, and colors represent a negative (red), no connection (gray), or positive connection (green) between the 138 Ser585-regulated genes identified in our studies and individual LY294002-induced gene expression changes for each array in the Connectivity Map database. A cluster of LY294002 experiments (13 of 17) lie within the red region, indicating a statistically significant enrichment of our 138 Ser585-regulated genes and the LY294002 differentially expressed genes (P = .009). (C) Analysis of the overlap between the 138 Ser585-regulated gene set identified in our studies and the LY294002-sensitive genes in the Connectivity database. We pooled the data from the 6 top-ranked MCF7 cell LY294002 microarrays that showed significant overlap with our 138 gene set (bracketed asterisks) and performed a Wilcoxon rank sum test to compare a ranked list of LY294002 sensitive genes (Lod-ranking of LY294002-regulated genes) with our 138 gene set. Each blue line corresponds to an individual probe identified from our screen. Ser585-regulated genes (blue lines) cluster with top-ranked LY294002-sensitive genes (red region). Plotting this overlap shows a significant enrichment above a random distribution (dotted line) in Ser585-regulated genes (probe frequency) as the Lod-ranking of LY294002-regulated genes increases (P < .001). Of the 138 genes identified in our screen, 53 (38%) correspond to LY294002-sensitive genes encompassed by the gray portion of the graph (LY294002 cutoff; P = .01). (D) The 53 Ser585/PI3-kinase–regulated genes identified in panel C together with their fold change were uploaded into the IPA and overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of genes were then algorithmically generated based on their connectivity to known functional, biochemical, and disease interactions. A significant overlap with genes involved in cancer and cell death was observed (P < .001, right-tailed Fisher exact test). (E) The 53 Ser585/PI3-kinase–regulated genes identified in panel C were subjected to IPA mapping of network interactions. This analysis showed that 27 of 53 genes constitute a gene network with red denoting Ser585-induced genes and green denoting Ser585-repressed genes. Node shape denotes the indicated gene function or biologic process.
Phospho-Ser585 survival-only pathway regulates a PI3-kinase signaling network
GO analysis indicated that more than half of the 138 genes identified perform functions related to cytokines and signal transduction (29%) and transcription and translation (24%; Figure 3A; supplemental Table 3). The Ser585 gene program was examined in silico with the Connectivity Map to identify potential associations with transcriptional signatures from drug/compound treatments.19 This analysis identified a significant association between the 138 genes identified in our studies and the gene signatures produced by the PI3-kinase inhibitor, LY294002, in 13 of 17 microarray experiments across diverse cell types (Figure 3B red bar; connectivity score, −0.408; P = .009). As shown in Figure 3C, Ser585-regulated genes (blue lines) cocluster with top-ranked LY294002-sensitive genes (red region), and the frequency of Ser585-regulated genes (probe frequency) significantly increases over a random distribution (dotted line) as the Lod-ranking of LY294002-regulated genes increases (Figure 3C; P < .001, Wilcoxon rank sum test). Of the 138 genes identified in our screen, 53 (38%) correspond to LY294002-sensitive genes and are highlighted by the gray shading in Figure 3C as well as in supplemental Table 3. To further examine this Ser585/PI3-kinase network, we used IPA to identify biologic networks associated with known functional, biochemical, and disease interactions. This analysis showed a significant association with cancer (27 of 53, 51%) and cell death (24 of 53, 45%; Figure 3D) and identified a highly integrated Ser585/PI3-kinase signaling network (Figure 3E).
Phospho-Ser585 survival signaling is deregulated in AML
Previous preliminary studies raised the possibility that Ser585 phosphorylation could be deregulated in at least some patients with AML.12 Given that the IPA showed a significant association of Ser585-regulated genes with cancer and cell death (Figure 3D), we performed an investigation of the regulation of Ser585 phosphorylation in a panel of 23 AML patient samples. It was not possible to obtain sufficient numbers of normal CD34+ cells from bone marrow donors to examine the regulation of Ser585 phosphorylation with our phospho-specific antibodies. Furthermore, CD34+progenitors respond poorly to GM-CSF, most probably because of the low levels of GM-CSF receptor expression (data not shown). We therefore examined the Ser585 phosphorylation profile of primary MNCs because these cells have been previously shown to robustly respond to GM-CSF.12 Consistent with these previous findings, Ser585 and Tyr577 phosphorylation in MNCs derived from peripheral blood of healthy donors occurred in a biphasic manner with maximal Ser585 phosphorylation occurring at approximately 1pM and decreased Ser585 phosphorylation occurring at approximately 1000pM (Figure 4A). Quantification of Western blot analysis and unsupervised cluster analysis indicated that Ser585 was constitutively phosphorylated in 20 (87%) of 23 AML patient samples (P < .05, Mann-Whitney U test). AML38, AML96, and AML97 did not show statistically significant constitutive Ser585 phosphorylation (Figure 4C). In contrast, Tyr577 phosphorylation remained ligand dependent in 17 (94%) of 18 AML samples, occurring at approximately 1000pM (Figure 4C). Deregulated Ser585 phosphorylation in AML was evidenced as (1) increased basal Ser585 phosphorylation in the absence of GM-CSF (0pM) and (2) a failure to down-regulate Ser585 phosphorylation in response to 1000pM GM-CSF. Importantly, our results show that constitutive Ser585 phosphorylation was observed across diverse FAB and cytogenetic classifications (Figure 4C), indicating that constitutive Ser585 survival signaling is a common event in AML.
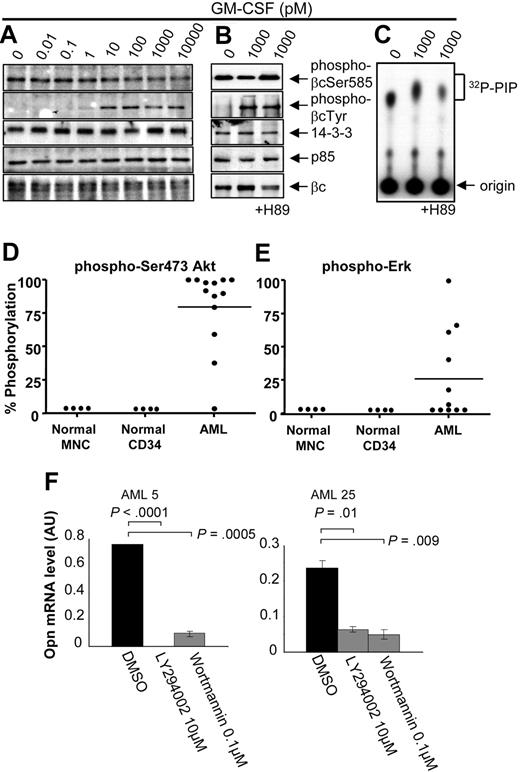
Ser585 phosphorylation is selectively deregulated in AML. MNCs from either healthy donors or patients with AML were stimulated with the indicated concentrations of GM-CSF for 5 minutes. The cells were then lysed, and βc was immunoprecipitated with βc-specific mAbs. Immunoprecipitates were subjected to SDS-PAGE and immunoblotted with affinity-purified phospho-specific anti–phospho-βcSer585 and anti–phospho-βcTyr577 antibodies. Typical results for a healthy donor and 2 AML samples are shown in panel A and panel B. The Western blot results from GM-CSF dose-response experiments on healthy donors (n = 4) and patients with AML (n = 23) were quantified by laser densitometry, and the quantified signals were converted to a heat map with the use of MeV (www.tm4.org/mev.html), where the intensity of red indicates the magnitude (%) of phosphorylation; gray, samples not done (ND); □, patients who did not show statistically significant constitutive Ser585 phosphorylation; †, samples derived from relapsed patients. Hierarchical cluster analysis was performed for Ser585 phosphorylation signals by using a Euclidean matrix with complete linkage, and the resultant heat maps are shown (C). FAB classifications, cytogenetics, blast count, and percentage of basal phosphorylation of βcSer585 for each AML are indicated alongside the heat maps. (D) Quantitative RT-PCR for the indicated genes was performed on total RNA extracted from purified bone marrow–derived CD34+ progenitor cells (n = 4; open boxes), mature CD14+ monocytes (n = 4; vertical striped boxes), and AML MNCs (n = 10; AML32-41, horizontal striped boxes). Boxes represent the interquartile range that contains 50% of the values, the horizontal lines mark the median, and the error bars indicate the SD. Data are normalized to β-actin expression and are presented as relative mRNA expression (log10). N/D denotes quantitative RT-PCR not done in bone marrow–derived CD34+ progenitor cells.
Ser585 phosphorylation is selectively deregulated in AML. MNCs from either healthy donors or patients with AML were stimulated with the indicated concentrations of GM-CSF for 5 minutes. The cells were then lysed, and βc was immunoprecipitated with βc-specific mAbs. Immunoprecipitates were subjected to SDS-PAGE and immunoblotted with affinity-purified phospho-specific anti–phospho-βcSer585 and anti–phospho-βcTyr577 antibodies. Typical results for a healthy donor and 2 AML samples are shown in panel A and panel B. The Western blot results from GM-CSF dose-response experiments on healthy donors (n = 4) and patients with AML (n = 23) were quantified by laser densitometry, and the quantified signals were converted to a heat map with the use of MeV (www.tm4.org/mev.html), where the intensity of red indicates the magnitude (%) of phosphorylation; gray, samples not done (ND); □, patients who did not show statistically significant constitutive Ser585 phosphorylation; †, samples derived from relapsed patients. Hierarchical cluster analysis was performed for Ser585 phosphorylation signals by using a Euclidean matrix with complete linkage, and the resultant heat maps are shown (C). FAB classifications, cytogenetics, blast count, and percentage of basal phosphorylation of βcSer585 for each AML are indicated alongside the heat maps. (D) Quantitative RT-PCR for the indicated genes was performed on total RNA extracted from purified bone marrow–derived CD34+ progenitor cells (n = 4; open boxes), mature CD14+ monocytes (n = 4; vertical striped boxes), and AML MNCs (n = 10; AML32-41, horizontal striped boxes). Boxes represent the interquartile range that contains 50% of the values, the horizontal lines mark the median, and the error bars indicate the SD. Data are normalized to β-actin expression and are presented as relative mRNA expression (log10). N/D denotes quantitative RT-PCR not done in bone marrow–derived CD34+ progenitor cells.
We then sought to examine the Ser585/PI3-kinase gene network identified in Figure 3 by determining the mRNA expression of specific genes in AML samples. For these experiments, we used both purified bone marrow–derived CD34+ progenitor cells and mature CD14+ monocytes as healthy donor controls. Our results show that BCL2, VRK1, POLR2C, CDK8, FNDC3, and PTPRS were more highly expressed in bone marrow CD34+ cells from healthy donors and became down-regulated in mature CD14+ monocytes (Figure 4D). Consistent with their immature phenotype, these genes (with the exception of PTPRS) were also highly expressed in AML blasts compared with CD14+ monocytes (Figure 4D). Thus, the genes identified in our screen for Ser585 targets are more strongly expressed in primitive CD34+ populations from both healthy donors and patients with AML. Attempts to disrupt the expression of Ser585-regulated genes by siRNA-mediated knockdown of βc expression were not successful despite obtaining high-transfection efficiencies (62%-74%) and using 7 different siRNAs obtained from 2 different companies (data not shown). The lack of significant βc knockdown may have been related to several factors, including the stability or transcriptional rates for βc mRNA. Interestingly, we noted that, although most Ser585-regulated genes in Figure 4D showed a narrow range of expression in AML, OPN was distinct in that it displayed a broad range of expression. Furthermore, pairs plots analysis (supplemental Figure 1) indicated that OPN expression was independently regulated with respect to all other genes analyzed in Figure 4D, suggesting that, although OPN was identified as a Ser585-regulated gene (Figures 1–2), other pathways also affect OPN expression in AML.
Deregulation of the phospho-Ser585/PI3-kinase signaling pathway and the regulation of OPN expression in AML
In line with the constitutive Ser585 phosphorylation observed in AML (Figure 4), we also found constitutive recruitment of both 14-3-3 and the p85 subunit of PI3-kinase to βc and deregulated PI3-kinase activity (Figure 5A-C). Tamburini et al20 have shown that the activation status of the PI3-kinase signaling pathway is a prognostic indicator of OS in AML. We therefore examined PI3-kinase and Ras-MAP-kinase signaling by examining the phosphorylation of Akt and ERK, respectively. We observed elevated basal Akt phosphorylation in 12 of 13 AML patient samples (Figure 5D), whereas elevated ERK phosphorylation was observed in 5 of 11 patient samples (Figure 5E). It is important to note that, although both Ser585 (Figure 4C) and Akt (Figure 5D) phosphorylation were both deregulated in most AML patient samples, a broad range of OPN expression (Figure 4D) was observed. Statistical analysis comparing OPN expression (high vs low) and basal Akt phosphorylation levels (high vs low) in 11 AML samples from Figure 5D was not significant (P = .3, χ2 test), indicating that elevated Akt phosphorylation does not directly correlate with increased expression of OPN. However, to examine the possibility that PI3-kinase signaling can promote OPN expression, we examined the effect of LY294002 and wortmannin treatment on OPN expression. Blockade of PI3-kinase signaling in 2 AML samples exhibiting high basal Akt phosphorylation resulted in a significant decrease in OPN mRNA expression (Figure 5F; P < .01). We next examined the possibility that OPN may also act upstream and activate PI3-kinase signaling and Akt phosphorylation. For these experiments we used the factor-dependent TF.1 erythroleukemia cell line because it exhibits low levels of basal Akt phosphorylation. Opn stimulation was not able to induce significant Akt phosphorylation, whereas clear Akt phosphorylation was detectable in response to GM-CSF (supplemental Figure 2). Together, these results suggest that, although PI3-kinase in AML can contribute to OPN expression, other pathways are also probably involved.
AML blasts exhibiting constitutive Ser585 phosphorylation also show deregulated PI3-kinase signaling. Mononuclear cells from patients with AML (A, AML37; B-C, AML101) were stimulated with the indicated concentrations of GM-CSF for 5 minutes. Where indicated, cells were preincubated for 1 hour with 10μM H89 or DMSO vehicle control before GM-CSF stimulation. Cells were then lysed and βc immunoprecipitated and subjected to SDS-PAGE and immunoblot analysis with the indicated antibodies. (C) In vitro kinase assay for PI3-kinase activity was performed on βc immunoprecipitates with phosphatidyl inositol 4,5 phosphate (PIP) and 32Pγ-ATP as described in “Cytokine signaling.” 32P-PIP and the origin are indicated (C). Whole-cell lysates were blotted with anti–phospho-Ser473Akt (D) or anti–phospho-ERK (E), and signals were quantified by laser densitometry. The level of phosphorylation in the absence of GM-CSF was plotted as a percentage of maximum signal observed after GM-CSF stimulation. (F) Purified CD34+ cells from the indicated AML patient samples were treated with 10μM LY294002 or 0.1μM Wortmannin or DMSO for 24 to 72 hours in IMDM medium containing 0.5% FCS, after which time viable cell number was assessed with Flow-Count Fluorospheres, and total RNA was isolated for OPN quantitative RT-PCR as described in Figure 4D.
AML blasts exhibiting constitutive Ser585 phosphorylation also show deregulated PI3-kinase signaling. Mononuclear cells from patients with AML (A, AML37; B-C, AML101) were stimulated with the indicated concentrations of GM-CSF for 5 minutes. Where indicated, cells were preincubated for 1 hour with 10μM H89 or DMSO vehicle control before GM-CSF stimulation. Cells were then lysed and βc immunoprecipitated and subjected to SDS-PAGE and immunoblot analysis with the indicated antibodies. (C) In vitro kinase assay for PI3-kinase activity was performed on βc immunoprecipitates with phosphatidyl inositol 4,5 phosphate (PIP) and 32Pγ-ATP as described in “Cytokine signaling.” 32P-PIP and the origin are indicated (C). Whole-cell lysates were blotted with anti–phospho-Ser473Akt (D) or anti–phospho-ERK (E), and signals were quantified by laser densitometry. The level of phosphorylation in the absence of GM-CSF was plotted as a percentage of maximum signal observed after GM-CSF stimulation. (F) Purified CD34+ cells from the indicated AML patient samples were treated with 10μM LY294002 or 0.1μM Wortmannin or DMSO for 24 to 72 hours in IMDM medium containing 0.5% FCS, after which time viable cell number was assessed with Flow-Count Fluorospheres, and total RNA was isolated for OPN quantitative RT-PCR as described in Figure 4D.
Targeting OPN blocks the survival of AML blasts and LSPCs
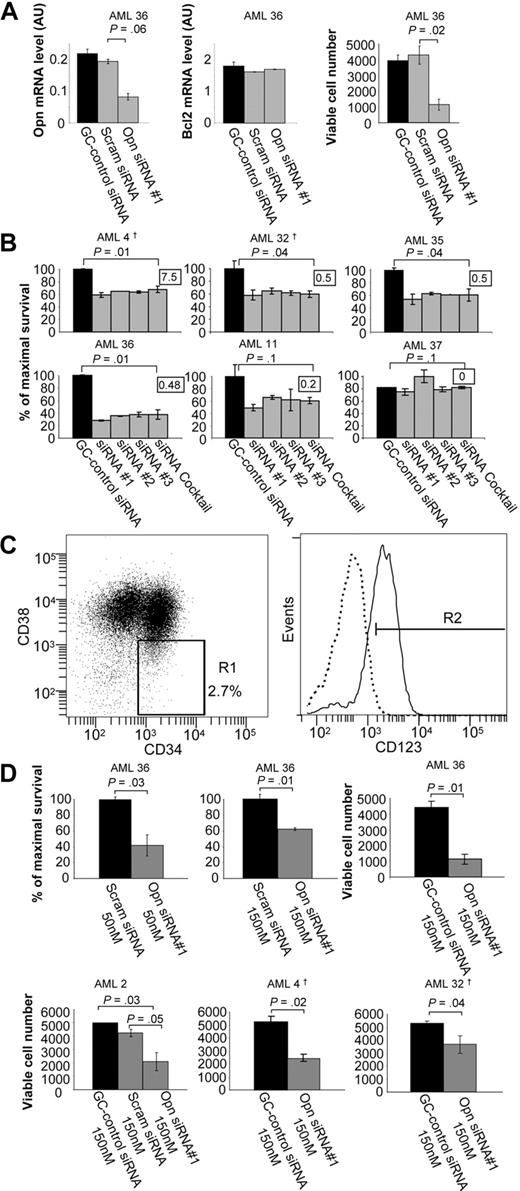
Of the genes identified in the Ser585/PI3-kinase gene network, several are known regulators of cell survival, including BCL2,21 CDK8,22 PTPRS,22 and OPN.23,24 However, OPN was unique in that it is a secreted protein and would be biologically accessible for therapeutic antagonists such as mAbs. In addition, OPN exhibited a high probability of differential regulation by Ser585 (supplemental Table 3), was validated by Northern blot (Figure 2), and was predicted by IPA to directly link with PI3-kinase, a known canonical survival-signaling hub (Figure 3E). We therefore addressed the possibility that OPN could function as a survival factor in AML by siRNA-mediated knockdown in CD34+ blasts. Compared with either a GC-control or scrambled control siRNAs, siRNA-mediated OPN knockdown resulted in a 58% decrease in OPN expression with no off-target effects observed for either BCL2 expression (Figure 6A) or actin (data not shown). Furthermore, siRNA-mediated OPN knockdown resulted in a significant reduction in cell survival (Figure 6A; P = .02). Similarly, 50nM of siRNA complexes also resulted in significant reduction in cell survival, suggesting minimal cytotoxicity (supplemental Figure 3). In addition, 4 of 5 AML patient samples exhibiting high OPN expression showed increased cell death after OPN siRNA knockdown (Figure 6B AML4, AML32, AML35, AML36; P < .05), whereas no significant induction of cell death was observed in an AML patient sample with undetectable levels of OPN expression (Figure 6B AML37). It is important to note that 100% of patient samples analyzed in Figure 6A and B coexpressed CD34 and CD33, with CD33+ cells in these samples ranging between 65% and 98%. These findings are consistent with previous reports showing that more than 85% of patients have AML CD34+ blast cells coexpressing significant levels of CD33.25 Because LSPCs are central to the long-term maintenance and growth of AML, we then sought to examine the effect of OPN expression knockdown on the survival of this clinically relevant target cell population. The CD34+/CD38−/CD123+ fraction has been shown to contain the self-renewing, tumor-initiating population of LSPCs.26,27 Opn knockdown in purified CD34+/CD38−/CD123+ LSPCs (Figure 6C) resulted in a significant decrease in cell survival in vitro in 4 of 4 patient samples compared with both GC-control siRNA and scrambled control siRNA (Figure 6D; P < .05). No evidence of cytotoxicity by either the scrambled siRNA control or siRNA concentration (50nM vs 150nM) was observed (Figure 6D). Quantitative RT-PCR confirmed knockdown of OPN expression in LSPCs (supplemental Figure 4). Our results would suggest that knockdown of OPN expression results in cell death of AML blasts and LSPCs, indicating that OPN can support cell survival. Although OPN has not previously been shown to act as a mitogen for hemopoietic cells, we do not exclude the possibility that OPN expression in AML may also promote deregulated proliferation.
siRNA-mediated knockdown of OPN expression reduces cell survival in AML blasts and LSPCs. Purified CD34+ cells from the indicated AML patient samples were transfected with 50nM BLOCK-iT fluorescent oligo and 50 to 150nM of (1) RNAi High GC negative control duplexes; (2) scrambled siRNA duplexes; (3) OPN siRNAs duplexes no. 1, no. 2, or no. 3; or (4) a cocktail consisting of all 3 OPN siRNAs duplexes (A-B,D). After transfection, cells were cultured for 2 to 3 days in IMDM medium containing 0.5% FCS, after which time total RNA was isolated for quantitative RT-PCR (A) and viable cell number was assessed by using Flow-Count Fluorospheres (A,D). (B) Cell survival was determined by annexin V–Alexa 568 staining and flow cytometry. The survival of purified CD34+ cells isolated from patients with AML in culture varied from 15% to 90%, and results are presented as the percentage of cell survival relative to the control siRNA-transfected cells. The inset indicates the level of OPN mRNA expression for each AML sample as determined in Figures 4 and 7. (C) AML CD34+/CD38−/CD123+ LSPCs were purified by fluorescence-activated cell sorting (FACS). Cells were stained with CD34-FITC, CD38-PE-Cy7, and CD123-PE antibodies after which time LSPCs were sorted with FACSAria cell sorter into CD34+CD38− subpopulation (1%-2% of total live cells) and gated for high CD123 expression (0.5%-1% of total). Approximately 5 × 105 to 1 × 106 cells were obtained from a single sort of 2 × 108 thawed cells. The purity of sorted populations was confirmed by secondary flow cytometry. (D) LSPCs were transfected and analyzed for cell survival as described in panel A. †Samples were derived from relapsed patients.
siRNA-mediated knockdown of OPN expression reduces cell survival in AML blasts and LSPCs. Purified CD34+ cells from the indicated AML patient samples were transfected with 50nM BLOCK-iT fluorescent oligo and 50 to 150nM of (1) RNAi High GC negative control duplexes; (2) scrambled siRNA duplexes; (3) OPN siRNAs duplexes no. 1, no. 2, or no. 3; or (4) a cocktail consisting of all 3 OPN siRNAs duplexes (A-B,D). After transfection, cells were cultured for 2 to 3 days in IMDM medium containing 0.5% FCS, after which time total RNA was isolated for quantitative RT-PCR (A) and viable cell number was assessed by using Flow-Count Fluorospheres (A,D). (B) Cell survival was determined by annexin V–Alexa 568 staining and flow cytometry. The survival of purified CD34+ cells isolated from patients with AML in culture varied from 15% to 90%, and results are presented as the percentage of cell survival relative to the control siRNA-transfected cells. The inset indicates the level of OPN mRNA expression for each AML sample as determined in Figures 4 and 7. (C) AML CD34+/CD38−/CD123+ LSPCs were purified by fluorescence-activated cell sorting (FACS). Cells were stained with CD34-FITC, CD38-PE-Cy7, and CD123-PE antibodies after which time LSPCs were sorted with FACSAria cell sorter into CD34+CD38− subpopulation (1%-2% of total live cells) and gated for high CD123 expression (0.5%-1% of total). Approximately 5 × 105 to 1 × 106 cells were obtained from a single sort of 2 × 108 thawed cells. The purity of sorted populations was confirmed by secondary flow cytometry. (D) LSPCs were transfected and analyzed for cell survival as described in panel A. †Samples were derived from relapsed patients.
OPN is a prognostic indicator of OS in AML
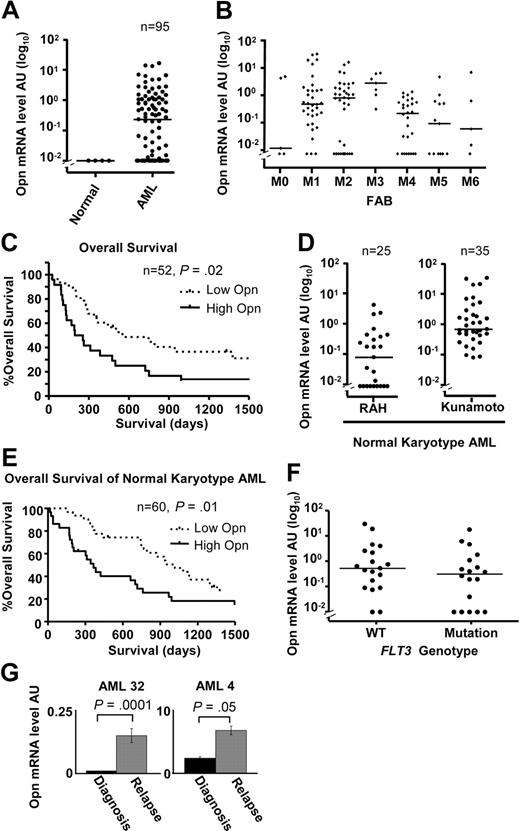
Given the role of OPN in promoting the survival of AML blasts and LSPCs, we considered the possibility that OPN expression levels may have prognostic significance. We quantified OPN mRNA expression in an expanded cohort of 95 consecutive diagnostic AML samples obtained at the RAH (supplemental Table 1). OPN expression ranged between 0 and 16 AU (median, 0.23 AU) with detectable expression in 67% of patients (64 of 95) (supplemental Table 1; Figure 7A). There was no striking association between OPN expression and FAB classification, although we noted that M3 had higher OPN levels compared with M4 (P < .05, Dunn post test) or M5 (P < .05; Figure 7B). Dividing patients into either low (< 0.23 AU, below median value) or high (> 0.23 AU, above median value) OPN expression, we examined OS by Kaplan-Meier estimates and the log-rank test. Of the 95 patients in our cohort, 52 (62%) had received standard induction chemotherapy and were evaluable for analysis. There was no significance difference in age, sex, white blood cell count (WCC), marrow blast percentage, FAB grouping, FLT3 mutation status, cytogenetic grouping, or allotransplantation numbers between the OPN-high and OPN-low groups (Table 1). Patients with high OPN (median OS, 225 days) showed a significantly reduced OS compared with patients with low OPN (median OS, 552 days; Figure 7C; n = 52; HR, 2.09; 95% CI, 1.16-4.22; P = .02), suggesting OPN expression may have prognostic significance in AML.
Increased OPN expression is associated with poor OS in AML. (A) Scatterplot showing OPN expression determined by quantitative RT-PCR in 4 normal CD14+ monocyte controls and 95 diagnostic samples from consecutive patients with AML collected at the RAH of which 60 were treated with induction chemotherapy. The horizontal lines indicate the median expression of OPN (0.23 AU). (B) Scatterplot of the range and median of OPN expression for various FAB classifications for all patients analyzed (95 RAH plus 35 Kunamoto normal karyotype patients). There is a significant difference in median OPN expression between FAB M3 and M4 or M5 subgroups (P = 0.01, Kruskal-Wallis test; P < .05, Dunn multiple comparisons test). (C) Kaplan-Meier log-rank analysis of all patients from RAH who underwent standard induction chemotherapy, excluding acute promyelocytic leukemia (APML; M3; n = 52). Taking a cutoff of 0.23 AU (median; split panel A), patients were divided into either high OPN expressers (> 0.23 AU) or low OPN expressers (< 0.23 AU). (D) Scatterplot showing the range and median of OPN expression for cytogenetically normal AML patient samples obtained at RAH (n = 25) and Kunamoto (n = 35), which were analyzed for OS (E). (E) Kaplan-Meier survival curves for the combined patients with normal cytogenetic AML shown in panel D after standard induction chemotherapy (n = 60), comparing high versus low OPN expression as determined by median split of each cohort as shown in panel D. High OPN is associated with poor OS (median, 384 days) compared with low OPN (median, 1017 days; P = .01). (F) Scatterplot comparing OPN expression in patients with cytogenetically normal AML analyzed in panel E for which FLT3 mutation status had been determined (ITD or D835 mutation; n = 37). There was no difference in median OPN expression between the 2 groups (n = 37; P = .2, Mann-Whitney U test). (G) Two patients for whom cryopreserved diagnosis and relapse samples were available were examined for OPN expression by quantitative RT-PCR. A significant increase in OPN expression was observed in both patients at relapse (P < .05, Mann-Whitney U test).
Increased OPN expression is associated with poor OS in AML. (A) Scatterplot showing OPN expression determined by quantitative RT-PCR in 4 normal CD14+ monocyte controls and 95 diagnostic samples from consecutive patients with AML collected at the RAH of which 60 were treated with induction chemotherapy. The horizontal lines indicate the median expression of OPN (0.23 AU). (B) Scatterplot of the range and median of OPN expression for various FAB classifications for all patients analyzed (95 RAH plus 35 Kunamoto normal karyotype patients). There is a significant difference in median OPN expression between FAB M3 and M4 or M5 subgroups (P = 0.01, Kruskal-Wallis test; P < .05, Dunn multiple comparisons test). (C) Kaplan-Meier log-rank analysis of all patients from RAH who underwent standard induction chemotherapy, excluding acute promyelocytic leukemia (APML; M3; n = 52). Taking a cutoff of 0.23 AU (median; split panel A), patients were divided into either high OPN expressers (> 0.23 AU) or low OPN expressers (< 0.23 AU). (D) Scatterplot showing the range and median of OPN expression for cytogenetically normal AML patient samples obtained at RAH (n = 25) and Kunamoto (n = 35), which were analyzed for OS (E). (E) Kaplan-Meier survival curves for the combined patients with normal cytogenetic AML shown in panel D after standard induction chemotherapy (n = 60), comparing high versus low OPN expression as determined by median split of each cohort as shown in panel D. High OPN is associated with poor OS (median, 384 days) compared with low OPN (median, 1017 days; P = .01). (F) Scatterplot comparing OPN expression in patients with cytogenetically normal AML analyzed in panel E for which FLT3 mutation status had been determined (ITD or D835 mutation; n = 37). There was no difference in median OPN expression between the 2 groups (n = 37; P = .2, Mann-Whitney U test). (G) Two patients for whom cryopreserved diagnosis and relapse samples were available were examined for OPN expression by quantitative RT-PCR. A significant increase in OPN expression was observed in both patients at relapse (P < .05, Mann-Whitney U test).
Characteristics of the 52 consecutive patients treated with induction chemotherapy according to OPN status
| . | Total patients, n = 52 . | OPN high, n = 25 (48%) . | OPN low, n = 27 (52%) . | P . |
|---|---|---|---|---|
| Median OPN level (range) | 0.20 (0.00-13.1) | 0.775 (0.23-13.7) | 0.00 (0.00-0.20) | N/A |
| Mean age, y, mean ± SD | 51 ± 17 | 55 ± 15 | 48 ± 18 | .2 |
| Male, n (%) | 29 (55) | 15 (60) | 14 (51) | .7 |
| Median WBC count, ×109/L (range) | 11.9 (1.3-300) | 15.9 (1.3-300) | 7 (1.68-171) | .7 |
| Median BM blasts, % (range) | 67 (20-99) | 55 (22-92) | 70 (20-99) | .8 |
| FAB, n (%) | ||||
| M0 | 2 (4) | 1 (4) | 1 (4) | N/A |
| M1 | 19 (36) | 10 (40) | 9 (33) | N/A |
| M2 | 14 (26) | 8 (32) | 6 (22) | N/A |
| M4 | 7 (13) | 2 (8) | 5 (18) | N/A |
| M5 | 7 (13) | 3 (12) | 4 (15) | N/A |
| M6 | 2 (7) | 1 (4) | 2 (7) | .9 |
| Cytogenetic subgroup, n (%) | ||||
| Unfavorable | 11 (21) | 7 (28) | 4 (14) | N/A |
| Intermediate | 37 (71) | 15 (60) | 22 (81) | N/A |
| Favorable | 3 (6) | 3 (12) | 0 (0) | .07 |
| FLT3 mutation, n (%) | 14/20 (70) | 5/8 (63) | 9/12 (75) | .6 |
| Induction chemotherapy, n (%) | 52 (100) | 25 (100) | 27 (100) | < .99 |
| Allotransplantation, n (%) | 11/52 (21) | 4/25 (16) | 7/27 (25) | .5 |
| . | Total patients, n = 52 . | OPN high, n = 25 (48%) . | OPN low, n = 27 (52%) . | P . |
|---|---|---|---|---|
| Median OPN level (range) | 0.20 (0.00-13.1) | 0.775 (0.23-13.7) | 0.00 (0.00-0.20) | N/A |
| Mean age, y, mean ± SD | 51 ± 17 | 55 ± 15 | 48 ± 18 | .2 |
| Male, n (%) | 29 (55) | 15 (60) | 14 (51) | .7 |
| Median WBC count, ×109/L (range) | 11.9 (1.3-300) | 15.9 (1.3-300) | 7 (1.68-171) | .7 |
| Median BM blasts, % (range) | 67 (20-99) | 55 (22-92) | 70 (20-99) | .8 |
| FAB, n (%) | ||||
| M0 | 2 (4) | 1 (4) | 1 (4) | N/A |
| M1 | 19 (36) | 10 (40) | 9 (33) | N/A |
| M2 | 14 (26) | 8 (32) | 6 (22) | N/A |
| M4 | 7 (13) | 2 (8) | 5 (18) | N/A |
| M5 | 7 (13) | 3 (12) | 4 (15) | N/A |
| M6 | 2 (7) | 1 (4) | 2 (7) | .9 |
| Cytogenetic subgroup, n (%) | ||||
| Unfavorable | 11 (21) | 7 (28) | 4 (14) | N/A |
| Intermediate | 37 (71) | 15 (60) | 22 (81) | N/A |
| Favorable | 3 (6) | 3 (12) | 0 (0) | .07 |
| FLT3 mutation, n (%) | 14/20 (70) | 5/8 (63) | 9/12 (75) | .6 |
| Induction chemotherapy, n (%) | 52 (100) | 25 (100) | 27 (100) | < .99 |
| Allotransplantation, n (%) | 11/52 (21) | 4/25 (16) | 7/27 (25) | .5 |
All patients were treated with standard AML induction therapy that included cytarabine, idarubicin, and etoposide followed by consolidation chemotherapy. Patients with APML(M3) treated with all-trans retinoic acid–based regimens were excluded. The mean age of our cohort was 51 years with a median follow-up of living patients at 5 years. The 5-year OS rate was 23%. FLT3 mutation status (internal tandem duplication of FLT3 receptor or D835 point mutation) was available for 20 patients (38%). Differences in the distribution of patients between FAB subgroups and cytogenetic risk groups were tested for categorical variables by χ2 test. Differences in continuous variables were tested with the Student t test or Mann-Whitney U test when appropriate.
WBC indicates white blood cell; N/A, not applicable; and cytogenetic subgroups, karyotype risk groups were defined according to the MRC10 criteria.
The effect of OPN expression on patient outcome was further examined by a multivariate Cox proportional hazards model. WCC and cytogenetics remained independent risk factors after multivariate analysis (Table 2; P < .05). Controlling for these factors, OPN expression was no longer a significant predictor of OS (Table 2; n = 52; P = .08), suggesting that variables such as cytogenetic risk classification may be influencing the effect of OPN on patient outcome.
Multivariate analysis of the prognostic effect of OPN and other prognostic variables on overall survival
| Prognostic marker . | Level . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|---|
| OPN | High | 1.79 | 0.94-3.39 | .08 |
| Age | Continuous | 1.02 | 0.99-1.05 | .06 |
| WCC | Continuous | 1.006 | 1.001-1.012 | .02 |
| Cytogenetics | .03* | |||
| High | 3.71 | 0.91-15.08 | .07 | |
| Intermediate | 1.26 | 0.37-4.32 | .7 | |
| Low | 1.0 | — | — |
| Prognostic marker . | Level . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|---|
| OPN | High | 1.79 | 0.94-3.39 | .08 |
| Age | Continuous | 1.02 | 0.99-1.05 | .06 |
| WCC | Continuous | 1.006 | 1.001-1.012 | .02 |
| Cytogenetics | .03* | |||
| High | 3.71 | 0.91-15.08 | .07 | |
| Intermediate | 1.26 | 0.37-4.32 | .7 | |
| Low | 1.0 | — | — |
The effect of OPN expression on overall survival for 52 patients with AML treated with chemotherapy in our institution was examined in a multivariate Cox proportional hazards model that included all variables that were deemed influential in survival: OPN, age, white cell count (WCC) at diagnosis, and cytogenetic risk group. The assumptions for proportional hazards was not violated.
— indicates not applicable.
Global P value for cytogenetics.
Patients with AML with normal cytogenetics falling within the intermediate-risk category remain a difficult group to treat because of their heterogeneous prognosis and response to therapies. We therefore examined the effect of OPN expression on OS within cytogenetically normal AML. Although there was a trend toward significance in normal karyotype AML in our RAH dataset (P = .1), our sample size was limited (n = 25). We therefore analyzed an expanded collection of 60 patients with normal-karyotype AML who had been previously treated with induction chemotherapy (25 patients from the RAH collection plus 35 patients from Kunamoto, Japan; supplemental Table 1). The range of expression for the RAH samples was 0 to 4.82 AU (median, 0.09 AU), whereas the range for the Kunamoto samples was 0.07 to 32.2 AU (median, 0.67 AU; Figure 7D). Taking an institutional-specific median split to divide patients into high versus low OPN expression, we performed Kaplan-Meier log-rank analysis for OS. We noted that, although there was no significance difference in sex, WCC, FLT3, NPM1 mutation status, or transplantation frequency between OPN-high and OPN-low groups, the OPN-high group was associated with higher mean age (52 vs 42 years; P = .02, Student t test) and increased M1 FAB classification (53% vs 23%; P = .01, χ2 test; supplemental Table 4). Importantly, our results show that patients with high OPN expression had a significantly shorter OS (median, 384 days) compared with patients with low OPN expression (median, 1017 days; Figure 7E; n = 60; HR, 2.05; 95% CI, 1.20-3.88; P = .01), suggesting that high OPN may be associated with inferior outcome in normal karyotype AML. In those patients for which FLT3 status had been determined, there was no significant association between OPN expression and the presence of a FLT3 mutation (P > .05, Mann-Whitney U test; Figure 7F). The prognostic effect of OPN in normal cytogenetic AML was further investigated by a multivariate Cox proportional hazards model that included age, WCC, and treatment center. Only age and OPN remained significant at a P value less than .05 (Table 3). When we controlled for these factors, OPN remained significantly associated with poor OS (n = 60; HR, 2.22; 95% CI, 1.23-4.02; P = .01), suggesting that high OPN expression is an independent prognostic indicator in normal karyotype AML (Table 3).
Multivariate analysis of the prognostic effect of OPN in normal karyotype AML on OS
| Prognostic marker . | Level . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|---|
| OPN | High | 2.226 | 1.231-4.027 | .01 |
| Age | Continuous | 1.028 | 1.007-1.050 | .01 |
| WCC | Continuous | 1.002 | 1.000-1.004 | .07 |
| Prognostic marker . | Level . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|---|
| OPN | High | 2.226 | 1.231-4.027 | .01 |
| Age | Continuous | 1.028 | 1.007-1.050 | .01 |
| WCC | Continuous | 1.002 | 1.000-1.004 | .07 |
The effect of OPN expression on OS of 60 consecutive patients with normal cytogenetics treated with induction chemotherapy was examined in a multivariate Cox proportional hazards model that included age, WCC, and treatment center as potentially influential variables. After controlling for age and WCC, high OPN expression (high vs low) remained a significant predictive factor for OS (P = .01). Treatment center was not influential on prognosis after multivariate analysis (P = .9; data not shown), and there was no significant interaction between OPN and other prognostic variables at each treatment center. The assumptions for proportional hazards was not violated.
Because OPN expression was associated with poor OS, we considered the possibility that OPN expression may be increased in relapsed patient samples. We therefore compared OPN expression in 2 patients for whom both diagnostic and relapsed cryopreserved AML samples were available. Analysis of both these samples showed that OPN mRNA expression levels were significantly increased after disease relapse (Figure 7G; P < .05), further supporting the findings (Figure 7A-E) that high OPN expression is a prognostic indicator linked to inferior patient outcome and early death after chemotherapy.
Discussion
Deregulated cell survival is a classic hallmark of cancer and therefore represents a key therapeutic target.28 We now show that Ser585-survival signaling regulates a transcriptional program comprising multiple gene networks (Figure 1), including a pathway that regulates OPN and a set of targets of the PI3-kinase pathway with established roles in cancer and cell death (Figure 3). We further show that Ser585 signaling is deregulated in the majority of AML patient samples (20 of 23) representing diverse FAB and cytogenetic classifications (Figure 4). In contrast, no deregulation of Tyr577 phosphorylation was observed in most of the AML patient samples, suggesting that only the Ser585 survival “arm” of the βc phospho-binary switch is subject to deregulation in AML.
Thus, we observe a clear distinction between the GM-CSF receptor Ser585 phosphorylation pathway (which is constitutive) and receptor tyrosine phosphorylation pathways (which remain cytokine dependent) in primary AML samples. Others have examined the prevalence of cytokine receptor tyrosine signaling pathways in AML samples. For example, constitutive STAT, p38, and ERK activation/phosphorylation have been observed in 25% to 70% of AML samples with many phospho-protein networks remaining cytokine dependent with low basal signals.29-31 Collectively, these studies show that, although cytokine signaling pathways are frequently subject to deregulation in AML, the extent of this deregulation varies. Given this variation, the prevalence of constitutive Ser585 phosphorylation observed in our studies (20 of 23) is particularly striking and represents a common theme observed across a spectrum of patients with AML.
The deregulation of the normal cellular machinery that controls the axis between cell survival and death is proposed to be a key event in cellular transformation. However, to our knowledge, the specific comparison of cell survival-only expression profiles between nontransformed and transformed cells has not been reported. Our results show that BCL2, VRK1, POLR2C, CDK8, FNDC3, and PTPRS were identified as being in the Ser585/PI3-kinase gene network and were highly expressed in both CD34+ cells derived from both healthy donors and AML patient samples (Figure 4). These results would suggest that the normal cell survival pathways that operate in nontransformed CD34+ cells also operate in AML blasts. The role of any of these genes in the pathogenesis of AML remains to be determined; however, BCL2 is a prosurvival gene that has been shown to cooperate with c-MYC in promoting lymphoma in animal models.21 Both POLR2C and CDK8 are part of the mediator complex,32 and CDK8 has recently been shown to act as an oncogene in colon cancer.33,34 Importantly, MacKeigan et al22 identified CDK8 and PTPRS in an siRNA screen for kinases and phosphatases regulating cell survival, suggesting that deregulation of these genes may promote autonomous cell survival in AML.
Another gene identified as being part of the Ser585/PI3-kinase survival pathway was OPN. We have analyzed OPN expression in 130 AML samples with diverse cytogenetic abnormalities and found a broad range of expression across all FAB subgroups (Figure 7B). Univariate analysis of OS in all patients treated with induction chemotherapy at our institution (n = 52) showed that high OPN expression was associated with poor OS (median, 225 days) compared with low OPN (median, 552 days; Figure 7C; P = .02), although there was no significance on multivariate analysis. However, multivariate analysis of an expanded cohort of 60 normal-karyotype AMLs from RAH and Kumamoto indicated that OPN expression was an independent prognostic indicator of OS (HR, 2.23; 95% CI, 1.23-4.03; Figure 7E; Table 3; P = .01) and did not correlate with FLT3 or NPM1 mutation status in these patients. Normal cytogenetic AML is the largest clinical subgroup in adult AML (50%-60%) and remains a major clinical challenge in terms of risk stratification, timing of allogeneic transplantation, and lack of efficacious targeted therapy. Only a few prognostic indicators such as FLT3, NPM1, and CEBPα are currently available. In addition to these markers, our study identifies OPN as a new prognostic indicator for OS in normal karyotype AML that may have utility in patient stratification and treatment selection. Expanded studies of OPN expression in multicenter trials in a range of AML subgroups are warranted.
In addition to acting as a prognostic factor, our results further indicate that high OPN expression may play a functional role in promoting deregulated cell survival in AML. Targeting OPN expression by siRNA in either AML blasts or LSPCs results in loss of cell viability (Figure 6) and therefore probably represents a key survival factor in leukemic stem cells. The precise mechanistic details by which OPN contributes to leukemogenesis remains unclear; however, studies in mice lacking OPN suggest that it promotes stem cell quiescence by inhibiting proliferation.17 Others have shown that OPN can act as a survival factor for activated T cells independently of their proliferation, leading to exacerbation of multiple sclerosis in animal models.23 Note that other studies using microarray screens of AML patient samples did not identify OPN overexpression as a prognostic indicator.35 However, microarray analysis is known to underestimate expression differences compared with quantitative RT-PCR. Nevertheless, high OPN protein levels in the plasma and blood of patients with a range of solid tumors as well as AML have been reported, and in some cases high OPN levels have been linked to poor prognosis.36-38 Thus, deregulated expression of OPN may play an important pathogenic role by deregulating survival programs in diverse cancers beyond leukemia.
OPN is a secreted phosphoprotein that acts as a cytokine and chemoattractant to regulate pleiotropic responses in diverse cell types, including hemopoietic cells.39 In part, the pleiotropic activities of OPN are probably due to its ability to bind different cell surface receptors that include CD44 and specific integrins in the αβ family.39 OPN expression in the bone marrow is tightly regulated and expressed at the endosteal interface of bone and hemopoietic tissue where it is secreted by osteoblasts.39 Interestingly, Lin et al24 have shown that IL-3 stimulation of Ba/F3 cells induces OPN expression, which in turn promotes hemopoietic cell survival via the CD44 receptor. It is not yet clear which receptor system OPN uses to promote cell survival in AML blasts and LSPCs in our studies. However, mAbs to CD44 not only reduce disease burden in animal models of AML but also abrogate AML in mice that serially received a transplant, indicating that the therapeutic response was most likely because of the targeting of CD44 in LSPCs.40 Similarly, a mAb to CD44 also blocks the homing and engraftment of chronic myeloid leukemia (CML) cells in murine models.41 Given that OPN-null mice are viable and fertile and have a mild phenotype,17 it is possible that targeting OPN may represent an important avenue for the development of therapeutics that block deregulated survival in leukemia.
Our identification of a phosphoserine pathway that is deregulated in AML has important implications for the treatment of leukemia. The early picture emerging from clinical trials that used tyrosine kinase inhibitors (TKIs) for the treatment of cancer is that they are often highly effective in providing an initial response by preventing cell proliferation, but they are less effective in blocking the survival of quiescent LSPCs. Even with the remarkable success of imatinib in treating CML, minimal residual disease is clearly detectable in more than 90% of patients in remission because of the survival of a population of CML progenitors that are refractory to TKI therapy.42-44 Furthermore, the results from clinical trials that used FLT3 TKIs indicate that, although they are very effective at blocking FLT3 tyrosine phosphorylation in patient samples and are able to block FLT3-mediated proliferation, they show modest anticancer activity that provides partial remissions of short duration.45-47 Thus, targeting phosphotyrosine-independent cell survival-only programs in AML such as OPN may provide a complementary therapeutic approach to eradicate the quiescent long-term surviving LSPCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Andrew Zannettino from the Department of Hematology, Center for Cancer Biology, for supplying reagents; Nancy Briggs and Tom Sullivan from the School of Medicine, University of Adelaide, for statistical support; and Kate Harrison and Monika Kutyna (Department of Hematology, Center for Cancer Biology) for patient mutational studies.
This work was supported by grants from the National Health and Medical Research Council of Australia and the Cancer Council of South Australia. M.A.G. was supported as a Peter Nelson Leukemia Research Fellow. D.T. is a recipient of a Haemtology and Oncology Targeted Therapy (HOTT) Fellowship administered by the Hematology Society of Australia and New Zealand (Clinical Oncology Society of Australia [COSA], Roche, Medical Oncology Group of Australia [MOGA]).
Authorship
Contribution: J.A.P., M.A.G. designed and performed research, analyzed and interpreted data, and wrote the paper; D.T. performed research, analyzed and interpreted data, and wrote the paper; E.F.B., B.J.M., A.B., and B.J. performed research; C.H.K., A.T., and T.P.S. performed statistical analysis; L.B.T., I.D.L., K.H., and N.A. provided vital AML patient samples; G.J.G., R.J.D., and A.F.L. analyzed and interpreted data; and M.O., D.N.H., and S.K.N. performed research and analyzed and interpreted data.
Conflict-of-interest disclosure: A.F.L. receives honoraria from CSL Limited, Australia. The remaining authors declare no competing financial interests.
Correspondence: Mark A. Guthridge, Cell Growth and Differentiation Laboratory, Division of Human Immunology, Centre for Cancer Biology, SA Pathology, Frome Rd, Adelaide, SA, Australia 5000; e-mail: mark.guthridge@imvs.sa.gov.au.

![Figure 1. Microarray analysis of the Ser585-survival pathway. CTL-EN cells expressing either wild-type βc or the βcSer585Gly mutant were factor deprived for 18 hours and then stimulated with GM-CSF for 0, 12, and 18 hours. Total RNA was isolated, reverse transcribed, and labeled; cDNA was used to probe either NIA 15k cDNA or AMF 21K microarrays. Pairwise comparisons were performed as in panel A whereby each solid arrow represents a separate 15k cDNA slide (n = 14) and each dotted arrow represents a 21k oligonucleotide slide (n = 8) in which differential gene expression was examined in either (1) cells expressing the wild-type βc or the βcSer585Gly mutant at a single time point or (2) a single cell line at different time points. Microarray data were subjected to Bayesian analysis to identify genes that were differentially expressed in CTL-EN cells expressing the wild-type βc and βcSer585Gly mutant in response to GM-CSF at 12 hours (B) and 18 hours (C). Bayesian analysis is shown whereby the probability that a particular gene is differentially expressed (t statistic) is plotted against the magnitude (log2 scale) of the differential expression (interaction coefficient) for each gene represented on the cDNA array. Negative interaction coefficients correspond to genes that are more strongly expressed in cells expressing the wild-type βc than the βcSer585Gly mutant (eg, genes that are induced by GM-CSF in cells expressing the wild-type βc and not in the βcSer585Gly mutant). Positive interaction coefficients correspond to genes that are more strongly expressed in cells expressing the βcSer585Gly mutant than the wild-type βc (eg, genes that are repressed by GM-CSF in cells expressing the wild-type βc and not in the βcSer585Gly mutant). Selected genes identified by GO analysis in Figure 3 as having a role in cell survival or cytokine signaling and that also showed differential expression are highlighted. (D-E) Microarray data presented as the response in wild-type βc cells (M = log2 [fold change in response to GM-CSF in CTL-EN cells expressing the wild-type βc]) plotted against the response in the βcSer585Gly mutant cells (M = log2 [fold change in response to GM-CSF in CTL-EN cells expressing the βcSer585Gly mutant]). Although most genes are either not GM-CSF regulated (M values close to 0 for both wild-type βc and βcSer585Gly mutant) or are GM-CSF regulated in an equivalent manner in cells expressing the wild-type βc and βcSer585Gly mutant (M values lie close to the diagonal line), a restricted subset lies off the diagonal. The M values for the genes highlighted in panels B to E at 0, 12, and 18 hours were plotted for cells expressing either the wild-type βc (○) or the βcSer585Gly mutant (Δ) (F). (G) A clustered heat map of the 138 genes identified as being differentially expressed between the wild-type βc and βcSer585Gly mutant CTL-EN cells in response to GM-CSF was generated with the use of a Euclidean matrix. The M value (log2 fold change) for both CTL-EN cells expressing the wild-type βc or the βcSer585Gly mutant was converted to a color scale with red indicating that the gene is induced by GM-CSF, green indicating that the gene is repressed, and the color intensity indicating the magnitude of regulation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/23/10.1182_blood-2009-02-204818/4/m_zh89990945230001.jpeg?Expires=1767717047&Signature=D55eXjk43vBocXQLb7gMl0jbENVmbb0saj00~Z3rQnHML6GqE4Mld5HdBmVieNPPf00Bu3~WCttWI1I6v3XmorT2DiOYCRBX7FPQLtZ5txaXTK~XC4oigIgXuN3~1GhEbQJ3BKEULp~jOQhnog-LhEm3ZL4XFq8wkh30Z9xpCMun6UV~mk4b9y~4xMqemxYi-DOltndZhRZhCiWce9CdWoguR6NIm9l8VcFk7orA1CLRhlIBmVlhB~3GMX36fzHNz5~bIbwUpaRT5wtLDd-w~Jk5nkew4P6hIFQclqN6507SA~YZd-BOBan09WMh-B-4107cXJTgy2JAgSP9V2JBQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal