Abstract
The capacity of human cytomegalovirus (HCMV) to establish and maintain a latent infection from which it can later reactivate ensures its widespread distribution in the population, but the mechanisms enabling maintenance of latency in the face of a robust immune system are poorly understood. We examined the role of the HCMV UL111A gene, which encodes homologs of the immunosuppressive cytokine interleukin-10 in the context of latent infection of myeloid progenitor cells. A UL111A deletion virus was able to establish, maintain, and reactivate from experimental latency in a manner comparable with parental virus, but major histocompatibility complex class II levels increased significantly on the surfaces of cells infected with the deletion virus. Importantly, there was an increase in both allogeneic and autologous peripheral blood mononuclear cells and CD4+ T-cell responses to UL111A deletion virus-infected myeloid progenitors, indicating that loss of the capacity to express viral interleukin-10 during latency results in latently infected cells becoming more readily recognizable by a critical arm of the immune response. The detection of a viral gene that suppresses CD4+ T-cell recognition of latently infected cells identifies an immune evasion strategy that probably enhances the capacity of HCMV to persist in a latent state within the human host.
Introduction
Human cytomegalovirus (HCMV) is a species-specific β-herpesvirus that infects a majority of the world's population.1 During primary productive infection, both innate and adaptive immune responses are activated, and replicating virus is eventually cleared from the host. However, the virus is able to evade complete immune clearance by establishing and maintaining a life-long latent infection in hematopoietic cells, specifically those of the myeloid lineage.2-7 During latency, detectable infectious virus production ceases, the viral genome is maintained as an extrachromosomal plasmid8 at a low viral genome copy number,7 and only a subset of viral genes remain transcriptionally active.9-14 Reactivation from latency results in reinitiation of the full replicative cycle, with production of new infectious virus.
Reactivation of HCMV from latency during states of reduced immune surveillance has profound implications for the management of patients with hematologic and immune deficiency disorders. In these clinical contexts, HCMV reactivation can lead to tissue infection causing major morbidity and mortality. As a result, the presence of HCMV seropositivity and thus of latent virus in donors or recipients of unrelated stem cell transplantations is an adverse prognostic feature despite major advances in HCMV monitoring and management.15,16 Reactivation of HCMV is also of importance in seropositive patients with nontransplantation cellular immunodeficiencies, such as those resulting from human immunodeficiency virus infection,17 or in seropositive patients with iatrogenically induced immunodeficiency, such as that resulting from treatment with the anti-CD52 antibody Campath.18,19
There is no known therapy for the elimination of virus that persists during latency. Instead, where necessary, medical treatment has concentrated on the preemptive treatment or the prophylaxis of viral reactivation. Even here, no single strategy prevents reactivation while universally avoiding unnecessary therapy. There is an urgent need to better understand the mechanisms of viral latency to provide insights into methods that will eliminate this pool of residual virus.
Both CD4+ and CD8+ T cells play critical roles in the cell-mediated control of HCMV infection and clinical disease,20,21 with a remarkably large proportion of the host immune system devoted to controlling HCMV. Approximately 5% of all circulating CD4+ and CD8+ T cells are HCMV-specific in seropositive persons,22 and this proportion increases significantly in elderly persons, a process that may impair responses to other pathogens.23 CD4+ T cells act as antiviral effectors and provide helper functions for maintaining HCMV-specific CD8+ T-cell responses,24 and impaired HCMV-specific CD4+ T-cell response correlates with long-term virus secretion in young children.25 During latent infection, dominant HCMV-specific CD4+ T-cell clones emerge that are poorly represented in the primary phase of infection, reinforcing a role for the CD4+ T cells in the control of both productive and latent infections.26 Interestingly, a substantial portion of these HCMV-specific CD4+ T cells that emerge after resolution of primary infection acquire immediate cytotoxic capacity, which is manifested in a major histocompatibility complex (MHC) class II–restricted manner.26,27
A wide range of viral genes expressed during the productive phase of infection have been shown to encode immunomodulatory functions, which probably provide at least a transient advantage to the virus during the initial stages of virus replication and dissemination in the human host.28 One of these genes, UL111A, expresses a doubly spliced transcript encoding a homolog of the potent immunosuppressive cytokine human interleukin-10 (IL-10). This virally encoded IL-10 homolog (denoted cmvIL-10) is a 175-amino acid protein that shares only 27% identity with human IL-10 but exhibits a broad range of comparable functions. Like human IL-10, recombinant cmvIL-10 has been shown to bind to and signal through the human IL-10 receptor29-32 as well as inhibit the proliferation of peripheral blood mononuclear cells (PBMCs) and production of proinflammatory cytokines, down-regulate MHC class I and MHC class II expression, inhibit maturation of monocyte-derived dendritic cells (MDDCs), and decrease matrix metalloproteinase activity by cytotrophoblasts.30,31,33-35 In addition, expression of CD1 transcripts coding for antigen presentation proteins is down-regulated in MDDCs treated with recombinant cmvIL-10, and treatment of MDDCs with supernatant from cells productively infected with the UL111A deletion virus RVAdIL10C supports these findings.36 Thus, cmvIL-10 exhibits a broad range of immunosuppressive properties, although these are cell type dependent as both human IL-10 and cmvIL-10 exert a stimulatory effect on B cells.37,38
During latent infection, the UL111A gene expresses an alternate transcript termed latency associated (LA) cmvIL-10. This transcript is colinear with the cmvIL-10 transcript at the amino terminus but, because of the lack of splicing of a second intron, codes for an in-frame stop codon, which results in a truncated protein of 139 amino acids.14 Transcripts predicted to encode LAcmvIL-10 have also been detected during productive infection of human foreskin fibroblasts (HFs),39 as have several additional viral IL-10 variants.40 The detection of LAcmvIL-10 transcription during latent infection raised the possibility that LAcmvIL-10 may encode similar immunosuppressive properties to cmvIL-10. However, analysis of cells treated with recombinant LAcmvIL-10 protein demonstrated that its immunosuppressive properties are more restricted than those of cmvIL-10 as the only function reported to date has been the ability to down-regulate MHC class II on myeloid cells.31 Interestingly, this function appeared to occur independently of the human IL-10 receptor, suggesting a mechanism of receptor interaction, which differs from that used by cmvIL-10.31
In this study, we examined the function of the viral IL-10–encoding UL111A gene in the context of latent infection of primary human myeloid progenitors, with a focus on the control of immune recognition. We used a combination of quantitative polymerase chain reaction (PCR) and infectious virus assays, together with immunostaining and flow cytometry to show that a UL111A deletion virus was able to establish, maintain, and reactivate from experimental latency as efficiently as parent virus, but that cells latently infected with the UL111A deletion virus expressed significantly higher levels of cell surface MHC class II. We applied cell proliferation and intracellular interferon-γ (IFN-γ) staining assays to examine the capacity of PBMC samples and purified CD4+ T cells to respond to latently infected myeloid progenitors in an allogeneic setting, and showed that UL111A can modulate this immune response. Finally, we examined the response of CD4+ T cells toward autologous latently infected myeloid progenitors and show that UL111A can inhibit the recognition of latently infected cells by CD4+ T cells derived from HCMV-seropositive but not HCMV-seronegative donors.
Methods
Cells
HFs were propagated in Dulbecco modified Eagle media (Invitrogen) supplemented with 10% fetal calf serum (FCS; CSL). PBMCs were derived from healthy human whole blood or granulocyte-colony stimulating factor mobilized peripheral blood from HCMV-seropositive or HCMV-seronegative donors by Ficoll gradient centrifugation and made available by the Bone Marrow Transplant Unit at Westmead Hospital. CD34+ myeloid progenitors were derived by magnetic bead separation from human fetal liver or mobilized PBMCs as described previously.11 CD34+ myeloid progenitors were cultured in granulocyte macrophage progenitor (GM-P) media consisting of Iscove modified Dulbecco media (Invitrogen) supplemented with 5% FCS (CSL) and 5% bladder cell carcinoma (ATCC #HTB-9) conditioned media as previously described.4 The CD34− cell fraction from each donor sample was resuspended in 90% human AB serum and 10% dimethyl sulfoxide and stored at −80°C for later isolation of CD4+ T cells. CD4+ cells from PBMCs or CD34− cell fraction from each mobilized peripheral blood sample was isolated using CD4 microbeads according to the manufacturer's instructions (Miltenyi Biotec). This study had the approval of the Sydney West Area Human Research Ethics Committee.
Viruses
RVAdIL10C is a previously constructed virus containing a deletion of the UL111A gene generated in HCMV strain AD169.41 Infection of myeloid progenitors with HCMV was performed by washing the cells in Hanks Buffered Salt Solution (Invitrogen), followed by resuspending the cells in DMEM plus 10% FCS at a density of 106 cells/mL, and adding virus at a multiplicity of infection (MOI) = 3. After incubation with virus inoculum for 3 hours, cells were washed 3 times in Hanks Buffered Salt Solution and cultured in GM-P media.
Flow cytometry for cell surface antigens
A total of 105 myeloid progenitors were washed in fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline/1% FCS/2 mM sodium azide) and incubated for 30 minutes at 4°C with anti–CD34-phycoerythrin (PE; Miltenyi Biotec) and isotype control IgG2a-PE (BD Biosciences) to assess purity after isolation, or anti–HLA-DR-allophycocyanin (APC; BD Biosciences) and anti–IgG2a-APC (BD Biosciences) to assess surface MHC class II expression. CD34+ cells were assessed for CD33, CD38, and c-kit expression by flow cytometry using anti–CD33-PE, anti–CD38-APC, and anti–CD117-APC conjugated antibodies, respectively (BD Biosciences). CD4+ T cells were surface stained with anti–CD3-APC (BD Biosciences) and anti–CD4-PE (BD Biosciences) conjugated antibodies. Flow cytometry data were acquired using FACSCalibur flow cytometer (BD Biosciences) and analyzed by CellQuest (BD Biosciences).
CFSE proliferation assay
PBMCs or CD4+ cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) according to the manufacturer's instructions (CFSE CellTrace proliferation kit; Invitrogen). A total of 105 myeloid progenitors were mixed with 106 CSFE-labeled PBMCs or CD4+ cells in a total volume of 1 mL RPMI media/10% FCS (Invitrogen), and incubated for 5 days before analysis by flow cytometry.
Intracellular IFN-γ assay
A total of 105 myeloid progenitors were mixed with 106 PBMCs or CD4+ cells in 1 mL RPMI/10% FCS media, and incubated for 16 hours at 37°C/5% CO2. A total of 4 μL GolgiStop (BD Biosciences) was added and cells incubated for a further 5 hours. Cells were washed in FACS buffer and stained for surface CD4 and CD3, before being fixed and permeabilized according to the manufacturer's instructions (BD Cytofix/Cytoperm kit; BD Biosciences), followed by staining with anti–IgG1-fluorescein isothiocyanate isotype (BD Biosciences) or anti–IFN-γ–fluorescein isothiocyanate (BD Biosciences) in 100 μL BD Perm/Wash solution for 30 minutes at 4°C and analysis by flow cytometry.
Results
Establishment of latent infection with UL111A deletion virus RVAdIL10C
To investigate the role of the viral IL-10 coding region in the context of latent infection, we used a UL111A deletion virus (RVAdIL10C) that has previously been described and used in several studies.36,41 This virus deletes amino acids 49 to 174 of cmvIL-10 and therefore removes the capacity to express both cmvIL-10 and LAcmvIL-10. Multiple cycle growth analyses demonstrated that this virus replicated with the same efficiency as the parental strain AD169,41 confirming the nonessential nature of this gene in productive infection.42 However, this virus had not previously been assessed in the context of latent infection. Thus, we first sought to assess the ability of the UL111A deletion virus RVAdIL10C to establish latency in primitive hematopoietic progenitor cells. CD34+ cells were enriched from human fetal liver using magnetic bead separation. Immunostaining and flow cytometry analysis for CD34 and for the myeloid lineage-committed cell-surface marker CD33 revealed that more than 95% of these cells expressed both CD33 and CD34, consistent with our previous analyses11 and demonstrating that these cells were myeloid progenitors (data not shown). Cells were mock-infected or infected with either AD169 (parent virus) or RVAdIL10C at a MOI = 3. Mock and infected suspension cells were harvested at various time points (days 1, 3, 5, 8, 11, and 14 after infection), and the distribution and quantity of viral genomes were determined using a combination of cell-dilution PCR and quantitative competitive (QC)–PCR as we have previously described11 (Figure 1A-B). By cell dilution PCR, viral DNA was detected down to 1 cell equivalent at each time point after infection in cells infected with either virus, and QC-PCR enabled the number of viral genomes per cell to be determined.
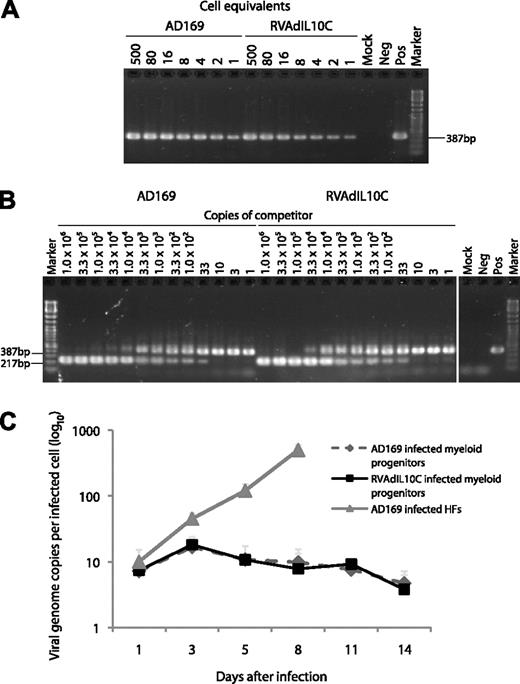
Level of viral DNA in myeloid progenitor cells infected with parental or viral IL-10 deletion viruses. Lysates of myeloid progenitors at day 8 after infection with parental virus (AD169) or viral interleukin-10 (IL-10) deletion virus (RVAdIL10C) were analyzed by (A) cell-dilution PCR using primers IEP3C and IEP4BII4 to detect a 387-bp product derived from HCMV genomic DNA, and (B) QC-PCR in the presence of between 1.0 and 1.0 × 106 copies of competitor template, an HCMV ie1/ie2 cDNA plasmid,4 to determine the amount of viral genomes present in latently infected myeloid progenitors. The number of copies of competitor template added to each reaction is indicated on top of each lane. Arrows represent the position of the 387-bp product derived from HCMV genomic DNA and the 217-bp product derived from the cDNA competitor template. Controls located in another section of the same gel were samples from either mock-infected myeloid progenitors (Mock), productively infected HFs (Pos), or samples without any added DNA template (Neg). (C) At the indicated time points after infection, cells from HCMV strain AD169- and RVAdIL10C-infected myeloid progenitor cell or HF cultures (MOI = 3) were analyzed by cell-dilution PCR and QC-PCR to determine the number of viral genome copies per infected cell. The average from 6 independent replicate experiments is shown as a line graph with SEM.
Level of viral DNA in myeloid progenitor cells infected with parental or viral IL-10 deletion viruses. Lysates of myeloid progenitors at day 8 after infection with parental virus (AD169) or viral interleukin-10 (IL-10) deletion virus (RVAdIL10C) were analyzed by (A) cell-dilution PCR using primers IEP3C and IEP4BII4 to detect a 387-bp product derived from HCMV genomic DNA, and (B) QC-PCR in the presence of between 1.0 and 1.0 × 106 copies of competitor template, an HCMV ie1/ie2 cDNA plasmid,4 to determine the amount of viral genomes present in latently infected myeloid progenitors. The number of copies of competitor template added to each reaction is indicated on top of each lane. Arrows represent the position of the 387-bp product derived from HCMV genomic DNA and the 217-bp product derived from the cDNA competitor template. Controls located in another section of the same gel were samples from either mock-infected myeloid progenitors (Mock), productively infected HFs (Pos), or samples without any added DNA template (Neg). (C) At the indicated time points after infection, cells from HCMV strain AD169- and RVAdIL10C-infected myeloid progenitor cell or HF cultures (MOI = 3) were analyzed by cell-dilution PCR and QC-PCR to determine the number of viral genome copies per infected cell. The average from 6 independent replicate experiments is shown as a line graph with SEM.
Analysis of 6 replicates obtained from 6 independent CD34+ myeloid progenitor cell samples revealed that the level of infection by AD169 and RVAdIL10C was similar over the time course of infection (Figure 1C). The distribution of viral DNA genome copies across the time course was comparable between AD169- and RVAdIL10C-infected cultures, ranging between 4.7 to 16.7 copies per cell and 3.8 to 18.1 copies per cell, respectively. In contrast to productively infected HFs analyzed in parallel (increasing from 10 copies per cell on day 1 to 500 copies per cell by day 8), the number of viral genome copies per infected myeloid progenitor cell remained low and relatively constant throughout the time course, with little change from the initial 7.4 RVAdIL10C and 7.2 AD169 genome copies per cell detected on day 1 after infection. These data are consistent with the UL111A deletion virus RVAdIL10C, establishing a nonproductive latent infection in myeloid progenitor cells in a manner comparable with parental virus.
We next sought to determine whether both viruses had the capacity to reactivate from latency. Aliquots of 5 × 104 cells harvested from AD169- and RVAdIL10C-infected CD34+ myeloid progenitor cell cultures at days 3, 5, 8, and 11 after infection were subjected to a reactivation assay whereby cells were cocultured with uninfected permissive HF monolayers and examined for cytopathic effect as previously described.4,11,12 Cytopathic effect in HF was detected between days 9 and 13 of coculture, but not when HFs were cocultured with mock-infected myeloid progenitors, lysates of infected or mock-infected myeloid progenitor cells, or supernatants from infected myeloid progenitors (Table 1; and data not shown). The ability to induce reactivation from myeloid progenitor cells infected with AD169 and RVAdIL10C after extended coculture with HFs demonstrated that both the parent and UL111A deletion virus established a comparable reactivatable latent infection in myeloid progenitor cells.
Virus reactivation from myeloid progenitor cells infected with AD169 and RVAdIL10C by coculture with permissive HFs
| Day after coculture at which reactivation determined by the appearance of cytopathic effect (CPE) in HFs was first detected . | ||||||
|---|---|---|---|---|---|---|
| Day after infection . | AD169 infected myeloid progenitor cells . | RVAdIL10C infected myeloid progenitor cells . | AD169 infected myeloid progenitor cell lysate . | RVAdIL10C infected myeloid progenitor cell lysate . | Supernatant from AD169 infected myeloid progenitor cell culture . | Supernatant from RVAdIL10C infected myeloid progenitor cell culture . |
| 3 | 13.7 (2.7 × 10−4) | 14.3 (2.9 × 10−4) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) |
| 5 | 12.3 (2.5 × 10−4) | 12.3 (2.5 × 10−4) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) |
| 8 | 9.3 (1.6 × 10−4) | 10.0 (2.0 × 10−4) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) |
| Day after coculture at which reactivation determined by the appearance of cytopathic effect (CPE) in HFs was first detected . | ||||||
|---|---|---|---|---|---|---|
| Day after infection . | AD169 infected myeloid progenitor cells . | RVAdIL10C infected myeloid progenitor cells . | AD169 infected myeloid progenitor cell lysate . | RVAdIL10C infected myeloid progenitor cell lysate . | Supernatant from AD169 infected myeloid progenitor cell culture . | Supernatant from RVAdIL10C infected myeloid progenitor cell culture . |
| 3 | 13.7 (2.7 × 10−4) | 14.3 (2.9 × 10−4) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) |
| 5 | 12.3 (2.5 × 10−4) | 12.3 (2.5 × 10−4) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) |
| 8 | 9.3 (1.6 × 10−4) | 10.0 (2.0 × 10−4) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) | No CPE (< 1.0 × 10−5) |
Values represent the average from 3 independent replicate experiments. Values in parentheses indicate the average frequency of myeloid progenitors that reactivated to produce infectious virus determined by infectious center assay.
CPE indicates cytopathic effect; and HFs, human foreskin fibroblasts.
No CPE indicates no cytopathic effect observed after 30 days of coculture with permissive HFs.
Modulation of MHC class II surface expression by UL111A during latent HCMV infection
To determine whether UL111A alters expression of cell surface MHC class II during latent infection, CD34+ myeloid progenitor cells were mock-, AD169-, or RVAdIL10C-infected at an MOI = 3. Cells were harvested at multiple times after infection (days 1, 3, 5, 8, 11, and 14), and immunostained with anti–HLA-DR-APC conjugated antibody or its isotype before being examined for cell surface MHC class II (HLA-DR) expression by flow cytometry (Figure 2A). The mean fluorescence intensity values of surface MHC class II by AD169- and RVAdIL10C-infected myeloid progenitor cells were normalized to those of mock-infected cells from the same replicate and time point (Figure 2B).
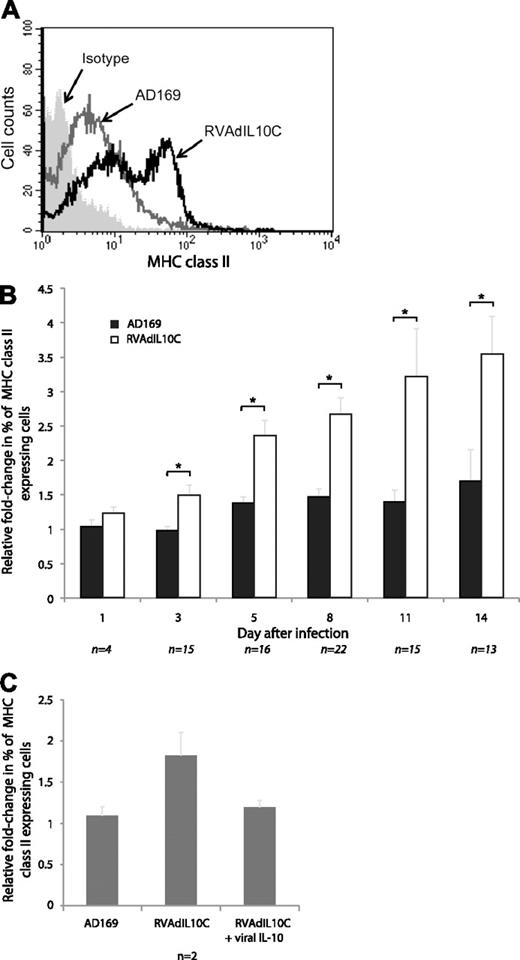
MHC class II (HLA-DR) surface expression by latently infected myeloid progenitors. Mock-, AD169-, and RVAdIL10C-infected myeloid progenitors harvested at days 1, 3, 5, 8, 11, and 14 after infection were immunostained for surface MHC class II (HLA-DR). (A) Flow cytometry histogram of cells analyzed at day 8 after infection. (B) The relative fold change in the percentage of MHC class II–positive AD169 or RVAdIL10C-infected myeloid progenitors determined by flow cytometry is shown normalized to the mock-infected value for each individual time point. (C) The relative fold change in the percentage of MHC class II–positive myeloid progenitors on day 8 after infection with either AD169 or RVAdIL10C with or without addition to cultures of 100 ng/mL recombinant viral IL-10 proteins (cmvIL-10 with LAcmvIL-10) on days 6 and 7. The number of independent replicates (n) for each time point is shown. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test: *P < .05.
MHC class II (HLA-DR) surface expression by latently infected myeloid progenitors. Mock-, AD169-, and RVAdIL10C-infected myeloid progenitors harvested at days 1, 3, 5, 8, 11, and 14 after infection were immunostained for surface MHC class II (HLA-DR). (A) Flow cytometry histogram of cells analyzed at day 8 after infection. (B) The relative fold change in the percentage of MHC class II–positive AD169 or RVAdIL10C-infected myeloid progenitors determined by flow cytometry is shown normalized to the mock-infected value for each individual time point. (C) The relative fold change in the percentage of MHC class II–positive myeloid progenitors on day 8 after infection with either AD169 or RVAdIL10C with or without addition to cultures of 100 ng/mL recombinant viral IL-10 proteins (cmvIL-10 with LAcmvIL-10) on days 6 and 7. The number of independent replicates (n) for each time point is shown. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test: *P < .05.
MHC class II levels were comparable on day 1 after infection; but from day 3, there was a statistically significant (P < .05) increase in the amount of MHC class II expressed by RVAdIL10C-infected myeloid progenitors compared with AD169-infected counterparts (Figure 2B). This increase in MHC class II was maintained for the remainder of the 14-day time course of infection. To confirm that this phenotype was the result of a lack of capacity of RVAdIL10C to express viral IL-10, on day 6 and 7 after infection, cultures of RVAdIL10C-infected myeloid progenitors were supplemented with 100 ng/mL recombinant viral IL-10 proteins (cmvIL-10 and LAcmvIL-10); and on day 8, MHC class II levels were measured by flow cytometry. In contrast to the increased MHC class II expression by RVAdIL10C-infected myeloid progenitors that were not supplemented with viral IL-10 proteins, addition of viral IL-10 proteins resulted in a lack of increased MHC class II levels, which remained comparable with parent virus-infected cells (Figure 2C). It was concluded that RVAdIL10C latently infected myeloid progenitor cells expressed a higher level of surface MHC class II molecules compared with the parent AD169 virus-infected cells as a consequence of a lack of capacity to express viral IL-10, implicating the HCMV UL111A region in modulating MHC class II during the latent phase of infection.
Allogeneic PBMC responses to myeloid progenitor cells latently infected with parental and UL111A deletion viruses
Our finding that the UL111A gene functioned to modulate MHC class II during latency suggested that the capacity of immune cells to respond to latently infected myeloid progenitors may be controlled by this viral gene. To examine the impact of the UL111A gene on PBMC response to latently infected cells, a CFSE cell proliferation assay was performed. Allogeneic CFSE-labeled PBMCs from normal blood donors were mixed with mock-, AD169-, or RVAdIL10C-infected myeloid progenitor cells harvested at various time points after infection (days 3, 5, 8, 11, and 14). After 5 days of culture, cells were harvested and analyzed by flow cytometry to determine the extent of PBMC proliferation in response to cells latently infected with either AD169 or RVAdIL10C (Figure 3A). The percentage of PBMC proliferation was determined from up to 15 replicates, each from an independent donor sample, after normalizing to the level of PBMC proliferation induced by mock-infected myeloid progenitors (Figure 3B). Compared with PBMCs treated with AD169-infected myeloid progenitors, PBMCs treated with RVAdIL10C-infected myeloid progenitors exhibited a statistically significant increase in proliferation for all time points investigated except for day 3 after infection, where the level of PBMC proliferation of parent and mutant-infected myeloid progenitors remained at a similar level.
CFSE proliferation assay of PBMC incubated with allogeneic latently infected myeloid progenitor cells. CFSE-labeled PBMCs were incubated for 5 days with mock-, AD169-, and RVAdIL10C-infected myeloid progenitors (harvested at days 3, 5, 8, 11, and 14 after infection) and assayed for proliferation using flow cytometry. The mitogen phytohemagglutinin (PHA) was added to PBMC as a positive control (PBMC + PHA), with CFSE-labeled PBMCs (PBMC only) as negative control. (A) Representative histogram plots of live-gated PBMCs undergoing proliferation measured by CFSE are shown. (B) Mean fold change in the percentage of PBMC proliferation in response to AD169- or RVAdIL10C-infected myeloid progenitors normalized to mock-infected myeloid progenitor cells. The number of independent replicates performed for each time point is indicated by n. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test: *P < .05, ***P < .001.
CFSE proliferation assay of PBMC incubated with allogeneic latently infected myeloid progenitor cells. CFSE-labeled PBMCs were incubated for 5 days with mock-, AD169-, and RVAdIL10C-infected myeloid progenitors (harvested at days 3, 5, 8, 11, and 14 after infection) and assayed for proliferation using flow cytometry. The mitogen phytohemagglutinin (PHA) was added to PBMC as a positive control (PBMC + PHA), with CFSE-labeled PBMCs (PBMC only) as negative control. (A) Representative histogram plots of live-gated PBMCs undergoing proliferation measured by CFSE are shown. (B) Mean fold change in the percentage of PBMC proliferation in response to AD169- or RVAdIL10C-infected myeloid progenitors normalized to mock-infected myeloid progenitor cells. The number of independent replicates performed for each time point is indicated by n. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test: *P < .05, ***P < .001.
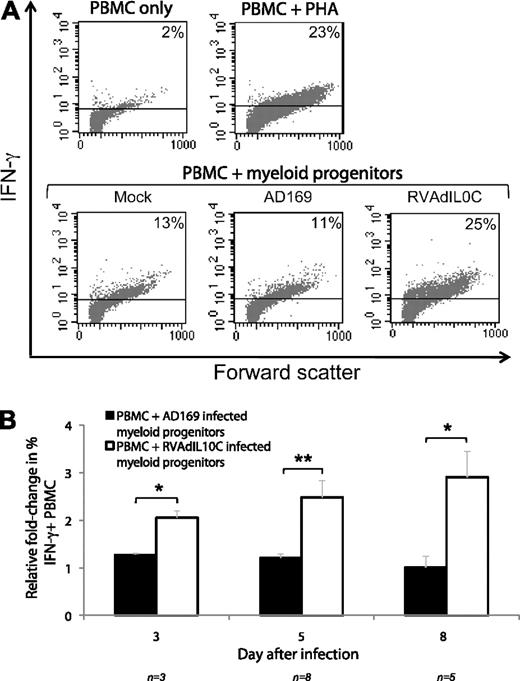
To further assess the impact of the UL111A region on allogeneic PBMC responses to RVAdIL10C latently infected myeloid progenitor cells, an intracellular IFN-γ staining assay was performed as an additional indicator of PBMC stimulation. PBMCs were incubated for 16 hours with mock-, AD169-, and RVAdIL10C-infected myeloid progenitors harvested at days 5, 8, and 11 after infection and assessed for IFN-γ production using intracellular immunostaining and flow cytometry (Figure 4A). From multiple replicate experiments from independent donor samples, PBMCs incubated with AD169-infected myeloid progenitors showed a similar level of IFN-γ–positive cells compared with those incubated with mock-infected myeloid progenitors. However, at all 3 time points tested, incubation of PBMCs with myeloid progenitors infected with RVAdIL10C resulted in a statistically significant increase in the percentage of IFN-γ–positive cells compared with PBMCs incubated with myeloid progenitor cells infected with AD169 (Figure 4B). These analyses demonstrate that deletion of UL111A in latently infected myeloid progenitor cells results in increased PBMC activation, suggesting that this viral gene functions during latency to limit the allogeneic immune response.
Intracellular IFN-γ staining of PBMCs incubated with allogeneic latently infected myeloid progenitor cells. Mock-, AD169-, and RVAdIL10C-infected myeloid progenitors harvested at days 3, 5, and 8 after infection were mixed (1:10) with PBMCs for 16 hours and assayed for intracellular IFN-γ using flow cytometry. The mitogen PHA was added to PBMCs as a positive control (PBMC + PHA), with PBMCs alone as a negative control. (A) Representative flow cytometry scatter plots of IFN-γ+ live-gated PBMCs for different treatments. (B) Column graphs showing mean fold change in the percentage of IFN-γ+ PBMCs after coculture with AD169- or RVAdIL10C-infected myeloid progenitors, normalized to mock-infected myeloid progenitor cells. The number of independent replicates (n) is shown. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test: *P < .05, **P < .005.
Intracellular IFN-γ staining of PBMCs incubated with allogeneic latently infected myeloid progenitor cells. Mock-, AD169-, and RVAdIL10C-infected myeloid progenitors harvested at days 3, 5, and 8 after infection were mixed (1:10) with PBMCs for 16 hours and assayed for intracellular IFN-γ using flow cytometry. The mitogen PHA was added to PBMCs as a positive control (PBMC + PHA), with PBMCs alone as a negative control. (A) Representative flow cytometry scatter plots of IFN-γ+ live-gated PBMCs for different treatments. (B) Column graphs showing mean fold change in the percentage of IFN-γ+ PBMCs after coculture with AD169- or RVAdIL10C-infected myeloid progenitors, normalized to mock-infected myeloid progenitor cells. The number of independent replicates (n) is shown. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test: *P < .05, **P < .005.
Modulation of allogeneic CD4+ T-cell response to myeloid progenitor cells latently infected with a UL111A deletion virus
The demonstration that RVAdIL10C-infected myeloid progenitor cells expressed higher levels of MHC class II compared with AD169-infected myeloid progenitors, and also induced PBMC activation/proliferation, implicated control of the CD4+ T-cell response by UL111A during latency. We therefore determined whether purified CD4+ T cells responded in the same manner by assessing their response to myeloid progenitors latently infected with either RVAdIL10C or AD169.
CD4+ lymphocytes were freshly isolated from PBMCs using anti-CD4 microbeads. The purity of CD4+ cells isolated by this procedure was consistently more than 95% as determined by immunostaining with anti–CD4-PE–conjugated antibody and flow cytometry (data not shown). CD4+ T cells were incubated at a ratio of 10:1 with mock-, AD169-, or RVAdIL10C-infected myeloid progenitor cells that had been harvested on day 8 after infection. Cells were immunostained for T-cell markers CD4 and CD3, followed by intracellular IFN-γ staining. Flow cytometry was used to determine the proportion of IFN-γ–positive cells in the gated CD3+/CD4+ population from 4 independent experiments, each using cells from different donors, and the results shown relative to the effect of mock-infected myeloid progenitor cells (Figure 5A). A comparable proportion of CD3+/CD4+ T cells were IFN-γ–positive when incubated with mock- or AD169-infected myeloid progenitors. In contrast, compared with incubation with AD169-infected myeloid progenitors, incubation with RVAdIL10C-infected myeloid progenitors resulted in a statistically significant (P < .05) increase in the percentage of IFN-γ–producing CD4+ T cells.
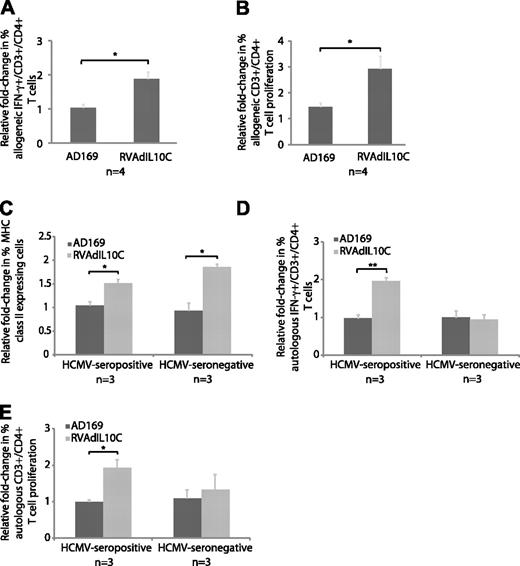
Impact of viral IL-10 on CD4+ T-cell response to latently infected myeloid progenitor cells. (A-B) Allogeneic settings. (C-E) Autologous settings. Mock-, AD169-, and RVAdIL10C-infected myeloid progenitors harvested at day 8 after infection were mixed (1:10) with allogeneic CD4+ T cells and intracellular IFN-γ expression or cell proliferation determined by flow cytometry on CD3+/CD4+ cells from 4 independent replicate experiments (n). Column graphs show mean fold-change of the percentage of (A) IFN-γ+ CD3+/CD4+ T cells and (B) proliferative response of CFSE-labeled CD3+/CD4+ T cells to allogeneic AD169- and RVAdIL10C-infected myeloid progenitors. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test. (C) Surface expression of MHC class II (HLA-DR) on mobilized peripheral blood-derived myeloid progenitor cells. Mock-, AD169-, and RVAdIL10C-infected CD34+ myeloid progenitors from 3 HCMV-seropositive and 3 HCMV-seronegative stem cell donors were harvested on day 8 after infection and assessed for surface MHC class II expression by flow cytometry. The mean fold change in the percentage of MHC class II–positive myeloid progenitors infected with AD169 or RVAdIL10C is shown relative to mock-infected counterparts. (D) Column graphs represent mean fold change (relative to mock infection) of the percentage of IFN-γ+ CD3+/CD4+ T cells and (E) proliferative response of CFSE-labeled CD3+/CD4+ T cells after incubation with autologous mock-, AD169-, and RVAdIL10C-infected myeloid progenitors from HCMV-seropositive and HCMV-seronegative stem cell donors. Significant differences between AD169 and RVAdIL10C infections using 3 HCMV-seropositive and 3 HCMV-seronegative donors were determined using 1-tailed, paired Student t test: *P < .05, **P < .005.
Impact of viral IL-10 on CD4+ T-cell response to latently infected myeloid progenitor cells. (A-B) Allogeneic settings. (C-E) Autologous settings. Mock-, AD169-, and RVAdIL10C-infected myeloid progenitors harvested at day 8 after infection were mixed (1:10) with allogeneic CD4+ T cells and intracellular IFN-γ expression or cell proliferation determined by flow cytometry on CD3+/CD4+ cells from 4 independent replicate experiments (n). Column graphs show mean fold-change of the percentage of (A) IFN-γ+ CD3+/CD4+ T cells and (B) proliferative response of CFSE-labeled CD3+/CD4+ T cells to allogeneic AD169- and RVAdIL10C-infected myeloid progenitors. Significant differences between AD169 and RVAdIL10C treatments were determined using 1-tailed, paired Student t test. (C) Surface expression of MHC class II (HLA-DR) on mobilized peripheral blood-derived myeloid progenitor cells. Mock-, AD169-, and RVAdIL10C-infected CD34+ myeloid progenitors from 3 HCMV-seropositive and 3 HCMV-seronegative stem cell donors were harvested on day 8 after infection and assessed for surface MHC class II expression by flow cytometry. The mean fold change in the percentage of MHC class II–positive myeloid progenitors infected with AD169 or RVAdIL10C is shown relative to mock-infected counterparts. (D) Column graphs represent mean fold change (relative to mock infection) of the percentage of IFN-γ+ CD3+/CD4+ T cells and (E) proliferative response of CFSE-labeled CD3+/CD4+ T cells after incubation with autologous mock-, AD169-, and RVAdIL10C-infected myeloid progenitors from HCMV-seropositive and HCMV-seronegative stem cell donors. Significant differences between AD169 and RVAdIL10C infections using 3 HCMV-seropositive and 3 HCMV-seronegative donors were determined using 1-tailed, paired Student t test: *P < .05, **P < .005.
The extent of allogeneic CD4+ T-cell proliferation in response to myeloid progenitor cells latently infected with either AD169 or RVAdIL10C was determined by CFSE T-cell proliferation assay. PBMC-derived CD4+ T cells labeled with CFSE were cocultured with mock-, AD169-, or RVAdIL10C-infected myeloid progenitors harvested on day 8 after infection. After 5 days of coculture, cells were immunostained for CD4 and CD3, and the extent of proliferation determined by flow cytometry. The proliferation profile of CD3+/CD4+ T cells was very similar to that obtained using total PBMCs. That is, CD3+/CD4+ cells incubated with RVAdIL10C-infected myeloid progenitors displayed an increased level of proliferation compared with CD3+/CD4+ cells incubated with AD169- or mock-infected myeloid progenitors (P < .05; Figure 5B). In summary, these data provide evidence that the UL111A gene region impacts the allogeneic CD4+ T-cell response to latently infected myeloid progenitor cells. These findings are consistent with this increased response being a consequence of higher cell surface MHC class II expression on myeloid progenitor cells latently infected with the UL111A region deletion virus.
Modulation of autologous CD4+ T-cell response to myeloid progenitor cells latently infected with a UL111A deletion virus
In an extension of our analysis of the UL111A-mediated control of an allogeneic CD4+ T-cell response to latently infected cells, the experimental approach was modified so as to examine the CD4+ T-cell response in an autologous setting. For these experiments, myeloid progenitors and CD4+ T cells were sourced from the same donor. Granulocyte-colony stimulating factor mobilized peripheral blood stem cell donors were selected for this study because of the higher numbers of circulating myeloid progenitor cells available for enrichment and experimental latent infection. Donors were clinically healthy and HCMV IgG seropositive. Three separate stem cell donor samples that met these criteria and were no longer required for transplantation in the clinical setting were made available.
From 70 to 80 mL of mobilized peripheral blood leukocytes, 0.7% (± 0.1%) of cells were CD34+ and the purity of CD34+ after CD34 magnetic bead enrichment was 81.9% (± 4.8%) for the 3 donors. With respect to additional cell surface proteins, which mark primitive progenitors, the majority of these cells (73.4% ± 1.3%) were CD34+/CD38− and 15.6% (± 10.6%) were CD34+/c-kit+, which is similar to the proportions we detected previously in fetal liver derived CD34+ cells (86.6% ± 1.8% and 38.4% ± 12.1%, respectively).11 At the time of CD34+ cell enrichment, the CD34− cell fraction from each donor sample was stored at −80°C for later isolation of CD4+ T cells.
To determine whether CD34+ myeloid progenitor cells isolated from mobilized peripheral blood samples could be latently infected in vitro with HCMV, CD34+ enriched cells were mock-infected or infected with AD169 or RVAdIL10C at a MOI = 3, and cell dilution and QC-PCR were performed to assess infectivity and distribution of viral DNA at day 3 and day 8 after infection. In all 3 samples, viral DNA was detected down to 1 cell equivalent in both AD169- and RVAdIL10C-infected cultures at approximately 15 viral genome copies at both time points (data not shown). This level of infection was comparable with that which we observed in fetal liver-derived CD34+ myeloid progenitors from this study and our previous work.11
To determine whether cell surface MHC class II expression was modulated by UL111A during latent infection of CD34+ myeloid progenitors from healthy HCMV-seropositive stem cell donors, on day 8 after infection (MOI = 3) nonadherent AD169-, RVAdIL10C-, and mock-infected myeloid progenitors were immunostained for cell surface MHC class II (HLA-DR) and analyzed by flow cytometry (Figure 5C). There was a statistically significant increase in the percentage of MHC class II expressing myeloid progenitors infected with RVAdIL10C compared with those infected with the parent AD169 (P < .05), consistent with that observed using fetal liver-derived myeloid progenitors. In addition to HCMV-seropositive donors, we performed the same experiment using CD34+ myeloid progenitors from healthy HCMV-seronegative stem cell donors. In this instance, we also observed a statistically significant increase (P < .05) in MHC class II by RVAdIL10C-infected cells compared with AD169-infected counterparts (Figure 5C).
To compare the response of autologous CD4+ T cells to myeloid progenitors latently infected with either parent or UL111A deletion viruses, the frozen CD34− cell fractions from either HCMV-seropositive or HCMV-seronegative donors were resuscitated and CD4+ cells isolated using magnetic bead separation. These CD4+ T cells were then cocultured with their respective autologous myeloid progenitors that had been mock-, AD169-, or RVAdIL10C-infected (day 8 after infection) and the extent of CD4+/CD3+ T-cell activation measured by intracellular IFN-γ staining and CFSE-labeled proliferation assay.
In the context of cells from HCMV-seropositive donors, RVAdIL10C-infected myeloid progenitors induced a statistically significant increase (P < .005) in the percentage of autologous IFN-γ+ CD4+/CD3+ T cells compared with mock- or AD169-infected myeloid progenitors, which remained at similar levels (Figure 5D). In contrast, when this experiment was performed using cells from HCMV-seronegative donors, RVAdIL10C-infected myeloid progenitors did not induce a CD4+/CD3+ T-cell response as determined by IFN-γ staining (Figure 5D). The CFSE proliferation assay correlated with this result, whereby CD4+/CD3+ T cells from HCMV-seropositive donors, but not HCMV-seronegative donors were induced to proliferate when incubated with autologous RVAdIL10C-infected myeloid progenitors (Figure 5E). These results demonstrated that in an autologous environment, in contrast to parental virus (AD169) or mock infection, myeloid progenitor cells latently infected with RVAdIL10C-induced CD4+ T-cell activation. Furthermore, this increased CD4+ T-cell response was observed only when HCMV-seropositive donor cells were used, as no response was observed when cells were obtained from HCMV-seronegative donors were used. Collectively, these findings using autologous CD4+ T cells are consistent with UL111A functioning to inhibit the CD4+ T-cell response to latently infected myeloid progenitors by suppressing the capacity of latently infected cells to present antigens via MHC class II to CD4+ T cells.
Discussion
The elimination of latent HCMV with replicative potential within the myeloid reservoir would abolish one of the primary concerns of physicians managing patients with severe cellular immunodeficiency states: the reactivation of HCMV in seropositive patients and the resultant potential for organ infections, including pneumonitis, hepatitis, retinitis, and colitis. In normal persons, eradication of a myeloid HCMV reservoir would eliminate viral transmission from seropositive donors to stem cell or solid organ transplantation recipients and minimize any adverse effects induced by persistence of small viral loads over long periods. Thus, the study of antigen expression and immune modulation during HCMV latency is critically important in advancing the management of HCMV latency. In this study, we identified a viral gene UL111A expressed during the latent phase of infection that renders latently infected cells refractory to CD4+ T-cell recognition, thus contributing to viral persistence during latency. The capacity of HCMV to maintain a life-long latent infection in healthy persons conflicts with several fundamental aspects of viral latency. First, unlike α-herpesviruses, herpes simplex virus and varicella zoster virus, which target immune-privileged neurons, HCMV confronts the challenge of latently infecting myeloid cells that are themselves components of the immune system. Second, the virus remains at least partially active during latency, expressing several viral genes.9-14 Third, a remarkably large proportion of the circulating CD4+ and CD8+ T-cell population in healthy, latently infected persons are HCMV-specific.22 Thus, it might be expected that, like replicating virus, latent virus would also be cleared by the immune response. That latent virus is never cleared indicates the presence of mechanisms promoting viral latency through interference with normal antiviral immune mechanisms, and our study identifies a mechanistic basis for latent HCMV-encoded modulation of a critical arm of the immune response.
We show that, in the context of latent infection of primary human myeloid progenitor cells, a UL111A gene deletion virus was able to establish, maintain, and reactivate from latency as efficiently as parental virus, but cells infected with this virus expressed higher levels of MHC class II. The UL111A gene encodes homologs of human IL-10, one of which, LAcmvIL-10, has been shown to be expressed during the latent phase of infection.14 Recombinant LAcmvIL-10 protein modulates MHC class II expression by myeloid lineage cells, leading to the hypothesis that this protein expressed by the UL111A gene may function during the latent phase of infection.31 Endogenously expressed HCMV-encoded proteins have been shown to be efficiently presented to CD4+ T cells via the MHC class II pathway,43,44 suggesting that any viral proteins similarly presented on the surfaces of latently infected cells in an MHC class II–restricted manner would render these cells susceptible to CD4+ T-cell recognition. Our finding that UL111A deletion virus (RVAdIL10C)–infected myeloid progenitors became recognizable by allogeneic and autologous CD4+ T cells supports the notion that viral IL-10 expressed by UL111A modulates the presentation of endogenously derived viral peptides by latently infected cells via the MHC class II pathway.
The demonstration of increased MHC class II on RVAdIL10C-infected myeloid progenitors together with elevated allogeneic PBMC and CD4+ T-cell proliferation and intracellular IFN-γ staining demonstrated a role for UL111A in controlling the immune response during latency. In an autologous setting using cells from healthy, HCMV-seropositive donors, an increased percentage of IFN-γ–producing CD4+ T cells and increased CD4+ T-cell proliferative responses were observed to RVAdIL10C-infected myeloid progenitors compared with those mock infected or infected with parent virus. Comparable results were obtained in experiments repeated using culture media supplemented with inactivated human AB serum rather than FCS, ruling out the possibility that bovine serum antigens in FCS may have been presented by myeloid progenitors as foreign peptides to stimulate CD4+ T cells (data not shown). In addition, in the autologous setting, we did not observe any increased CD4+ T-cell response to RVAdIL10C-infected myeloid progenitors setting when healthy, HCMV-seronegative donor cells were used. These experiments do not provide definitive evidence that UL111A modulates the response of HCMV-specific CD4+ T cells, but they do demonstrate that CD4+ T cells derived from HCMV-seropositive donors cells were required for this response. Whether these CD4+ T cells are HCMV-specific therefore remains to be established, and this will be an important component of studies aimed at identifying both HCMV proteins expressed during the latent phase of infection as well as CD4+ T-cell clones, which are reactive to peptides derived from these proteins.
A rare subset of CD4+ T cells with cytotoxic capability has been described in human peripheral blood.45 Significantly, studies by van Leeuwen et al identified a subpopulation of HCMV-specific CD4+CD28− granzyme B+ cytotoxic T cells, which emerges in the circulation only after cessation of viral replication and is detectable at much higher frequencies in HCMV-seropositive persons during latency.27 Additional studies by this group demonstrated that these T cells could lyse HCMV antigen-expressing target cells in an MHC class II–dependent manner and that the dominant HCMV-specific CD4+ T-cell clones present during latency were poorly represented during the acute phase, indicating that selection of CD4+ T-cell clones continues once the virus has become latent.26 Thus, determining whether UL111A represses the capacity of HCMV-specific CD4+CD28− granzyme B+ cytotoxic T cells to recognize latently infected myeloid progenitors will be an important focus of future studies to define the extent of immunoevasion encoded by this viral gene, as will determining the impact of UL111A on regulation of the CD8+ T-cell response via modulation of MHC class I during latency. During the productive phase of infection, viral IL-10 expressed from the UL111A gene does not impair MHC class I–restricted peptide presentation on bystanding antigen-presenting cells,41 but it remains to be determined whether the same occurs in the context of latent infection, or if directly infected cells are resistant to CD8+ T-cell recognition as a consequence of UL111A expression. Analysis of CD4+ recognition of bystander myeloid progenitor cells will also be important to determine whether viral IL-10 acts as an autocrine and paracrine factor.
Latent infection of magnetic bead-enriched primitive CD34+ myeloid progenitors with the parent strain AD169 had little effect on cell surface MHC class II levels compared with mock-infected cells. This finding differed from that observed previously after latent infection of more heterogeneous GM-P cells with HCMV strain Towne, where MHC class II levels decreased on CD14+ GM-Ps compared with mock infection.46 The reason for this difference is not clear but may reflect fundamental differences in the response of latently infected myeloid progenitor cells at different stages of differentiation and/or virus strain-specific differences. Profiling changes in both cellular and viral gene expression as latently infected primitive CD34+ myeloid progenitors differentiate along divergent pathways to become monocytes/macrophages or myeloid dendritic cells will ultimately be required to define the complex changes, which probably occur at different stages of differentiation. The results we obtained were derived from a single laboratory strain of HCMV (AD169). This strain lacks 19 open reading frames found in lower passage clinical isolates, such as the Toledo strain,47 and it remains to be confirmed experimentally whether a more natural isolate of HCMV will replicate the same phenomenon that we observed in the current study.
In addition to primate cytomegaloviruses,29,48 the γ-herpesviruses Epstein-Barr virus (EBV) and ovine herpesvirus 2, the α-herpesvirus equine herpesvirus type 2, and the Orf poxvirus encode homologs of IL-10.49-53 The EBV IL-10 homolog (ebvIL-10) has been subjected to considerable analysis. This homolog encoded by the BCRF1 gene shares approximately 83% amino acid identity with human IL-1053 and exhibits similar functions to human IL-10, including the inhibition of cytokine synthesis in monocytes and deactivation of macrophage activity.54 Like cmvIL-10, LAcmvIL-10, and human IL-10, ebvIL-10 has been shown to modulate the expression of MHC class II molecules on human monocytes. In doing so, ebvIL-10 has been shown to decrease the proliferation of EBV-specific human T cells.55 However, unlike LAcmvIL-10, ebvIL-10 is not expressed during latent infection, with its activity restricted to the productive phase of infection.56 So although both viruses use homologs of IL-10 to directly modulate immune recognition, EBV differs from HCMV as it does not appear to require this function for maintenance of latency.
In conclusion, we have identified a role for UL111A in the modulation of MHC class II expression to limit CD4+ T-cell recognition of latently infected cells. This is the first viral gene that has been shown to function in an immunomodulatory capacity during the latent phase of infection. Our results have a practical significance beyond the understanding of HCMV immunobiology and latency. The use of antibody or antisense therapy to inhibit mechanisms that down-regulate expression of histocompatibility antigens presenting viral peptides carries the promise of completely eradicating the reservoir of myeloid cells latently infected with HCMV. Furthermore, recent studies indicate that adoptive cellular immunotherapy directed at matrix and structural proteins expressed during productive infection effectively control active infection.57,58 Targeting latency-associated antigens with adoptive immunotherapy while simultaneously inhibiting down-regulation of antigen expression is a logical next step on the road to the elimination of HCMV persistence in those with latent infection.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Vicki Antonenas and Kenneth Bradstock of the Department of Hematology, Westmead Hospital, for provision of mobilized peripheral blood samples, and Winnie Garcia for assistance with virus stock preparation.
This work was supported by Australian National Health and Medical Research Council (Program Grant 358399; B.S., A.L.C.) and Deutsche Forschungsgemeinschaft (KFO 183; B.P., S.P.-K.).
Authorship
Contribution: A.K.L.C. designed and performed research, wrote the paper, and analyzed data; D.J.G., B.P., and S.P.-K. contributed vital new reagents; S.A. analyzed data and wrote the paper; A.L.C. analyzed data; and A.A. and B.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barry Slobedman, Centre for Virus Research, Westmead Millennium Institute and University of Sydney, PO Box 412, Westmead, NSW 2145, Australia; e-mail: barry_slobedman@wmi.usyd.edu.au.
References
Author notes
*A.A. and B.S. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal