Abstract
The preferentially expressed antigen in melanoma (PRAME) is expressed in several hematologic malignancies, but either is not expressed or is expressed at only low levels in normal hematopoietic cells, making it a target for cancer therapy. PRAME is a tumor-associated antigen and has been described as a corepressor of retinoic acid signaling in solid tumor cells, but its function in hematopoietic cells is unknown. PRAME mRNA expression increased with chronic myeloid leukemia (CML) disease progression and its detection in late chronic-phase CML patients before tyrosine kinase inhibitor therapy was associated with poorer therapeutic responses and ABL tyrosine kinase domain point mutations. In leukemia cell lines, PRAME protein expression inhibited granulocytic differentiation only in cell lines that differentiate along this lineage after all-trans retinoic acid (ATRA) exposure. Forced PRAME expression in normal hematopoietic progenitors, however, inhibited myeloid differentiation both in the presence and absence of ATRA, and this phenotype was reversed when PRAME was silenced in primary CML progenitors. These observations suggest that PRAME inhibits myeloid differentiation in certain myeloid leukemias, and that its function in these cells is lineage and phenotype dependent. Lastly, these observations suggest that PRAME is a target for both prognostic and therapeutic applications.
Introduction
PRAME, or the preferentially expressed antigen in melanoma, was originally described as an HLA-A24–restricted tumor-associated antigen in melanoma.1 PRAME is expressed in a number malignancies; however, its expression is low or absent in various normal tissues, including CD34+ hematopoietic progenitors.2-4 Until recently its function remained unknown. Epping et al have characterized PRAME as a ligand-dependent corepressor of retinoic acid receptor α (RARα), RARβ, and RARγ signaling.5 The authors demonstrated that PRAME protein expression in solid tumor cell lines inhibited differentiation in the presence of the RARα ligand all-trans retinoic acid (ATRA). The authors also hypothesized that the polycomb group protein EZH2 may act together with PRAME to mediate the block in differentiation.5
In hematologic malignancies PRAME is expressed in 22% to 62% of unsorted bone marrow (BM) or peripheral blood (PB) samples from chronic myeloid leukemia (CML) patients and in 25% to 62% of pediatric acute myeloid leukemia (AML) cases.2-4,6 In our analyses of gene expression that increased with CML progression and also discriminated leukemic blasts from normal CD34+ sorted BM, PRAME demonstrated the most statistically significantly increased expression with disease progression.2 PRAME hypomethylation may contribute to its increased expression in blast crisis (BC) CML and AML.7,8 Whereas increased PRAME expression is associated with poor outcomes in solid tumors,9-11 the data in hematologic malignancies appear contradictory. Increased PRAME expression discriminates acute megakaryoblastic leukemia from a transient myeloproliferative disorder in Down syndrome neonates and is associated with CML progression.2,12 However, in pediatric AML, acute promyelocytic leukemia (APL), and now most recently in adult AML with normal cytogenetics, increased PRAME expression is associated with better outcomes.4,13,14 These observations are not mutually exclusive. When present in de novo AML, PRAME expression appears to be associated with good risk cytogenetic abnormalities.4,13,14 However, when associated with BCR-ABL in CML PRAME expression is a marker of an acute leukemia where outcomes, in general, are very poor.2,12
Retinoic acid receptor signaling is important in both normal and malignant hematopoietic cell proliferation and differentiation.15-17 In APL, where PML-RARα inhibits retinoic acid–induced gene transcription and cell differentiation, supraphysiologic ATRA concentrations overcome this block and promote granulocytic differentiation. ATRA's effects on normal progenitor cells, however, are cell phenotype and concentration dependent.16 Whereas supraphysiologic concentrations of ATRA in culture shift hematopoiesis toward granulopoiesis, physiologic concentrations increase proliferation and promote colony formation of several cell lineages.18 As a consequence of these observations, we sought to determine how PRAME protein expression affects myeloid differentiation in hematopoietic cells and whether PRAME expression in chronic-phase (CP) CML patients is associated with outcomes on tyrosine kinase inhibitor (TKI) therapy.
Methods
Patient samples
Patient samples used for these investigations were obtained at the Fred Hutchinson Cancer Research Center (FHCRC) from Institutional Review Board–approved protocols with written informed consent, in accordance with the Declaration of Helsinki. We have previously examined gene expression profiles in several normal and leukemic patient samples using microarrays.2,19 These studies examined bone marrow (BM) and peripheral blood (PB) samples from 42 CP, 17 accelerated phase (AP), and 31 BC CML patients2 ; 29 myelodysplastic syndrome (MDS) patients; 26 AML patients; 32 B-acute lymphoblastic leukemia (ALL), 7 T-ALL, and an additional 17 CP CML patients.2,19 CD34+ sorted cells from normal BM (n = 8) and peripheral blood stem cell (PBSC) products (n = 10), in addition to unselected BM (n = 10), PB (n = 10), and sorted B (n = 4) and T (n = 3) lymphocytes were also examined.2,19 For functional studies, additional normal CD34+ PBSC (n = 3) and CML BC leukopheresis (n = 3) samples were obtained from volunteer donors at the FHCRC and from the FHCRC's Leukemia Repository. Acute leukemia was defined as more than 30% peripheral blood blasts.
Samples for independent quantitative reverse-transcription–polymerase chain reaction (QPCR) validation studies included the following: 3 normal CD34+ and 4 unsorted BM samples, 35 APL cases, 31 BC CML cases, 58 diagnostic samples from newly diagnosed CP CML cases enrolled on the Novartis RIGHT study who received imatinib mesylate (IM) at 800 mg/day, and 25 late CP patients who previously failed IM therapy who subsequently received nilotinib at 800 mg/day on the Novartis AMN107 study. CP RIGHT study patients were within 6 months of diagnosis and were untreated except for hydrea or anagrelide. Seven patients received IM for less than 1 month before enrollment. With regard to last IM response before nilotinib therapy, among the 25 AMN study patients, 7 patients had evidence of a complete hematologic response (CHR). In addition, only 2 had partial cytogenetic responses, whereas the remaining patients had minor, minimal, or no cytogenetic responses. Before therapy, the median white blood cell and peripheral blood blast counts were as follows: 52.6 thousands per μL (range, 3.6 to 60.1 thousands per μL) and 0% (range, 0%-9%), respectively, for the RIGHT study patients and 12.6 thousands per μL (range, 1.8 to 70.7 thousands per μL) and 0% (range, 0%-3%), respectively, for the AMN study patients. Median follow up for the RIGHT study patients was 18 months (range, 9-20 months). Median follow up for the Novartis AMN107 study patients was 12 months (range, 3-18 months).
Lentiviral and retroviral vectors
The PRAME expression vector was modified from an existing published lentiviral vector (gift from Dr C. A. Blau, University of Washington, Seattle).20 PRAME was expressed from an MSCV promoter and GFP, from a phosphoglycerate kinase (hPGK) promoter. The shPRAME and shGFP (control) pRETRO-SUPER vectors were received from M.T.E. and Dr Rene Bernards (Netherlands Cancer Institute, Amsterdam) and are published.5 The shPRAME and shGFP hairpins were removed from the pRETRO-SUPER vectors and cloned into a lentiviral shRNA vector with an H1 Pol III promoter that also expressed YFP from an hPGK promoter (gift from Dr Hans Peter Kiem, FHCRC). Complete details are given in supplemental methods (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell line culture, progenitor cell culture, and transduction
Human leukemia cell lines and normal and CML CD34+ hematopoietic progenitor cells were transduced and cultured as described in supplemental methods. Transduction efficiencies for cell lines ranged from 50% to 85%, for normal CD34+ hematopoietic progenitors from 35% to 45%, and for CML CD34+ hematopoietic progenitors from 15% to 25%. GFP or YFP expression in these cells was greater than 99% for the duration of the experiments.
Cell line experiments were performed in duplicate or triplicate. Cells were treated with vehicle control or ATRA (Sigma-Aldrich) at 0.001, 0.01, 0.1, 1, and 10 μM. Cell proliferation was examined using either a CellTiter 96Aqueous One proliferation Assay (Promega) or an ATPlite 1-step Single Addition Luminescence ATP Detection Assay System (PerkinElmer). Proliferation was normalized to signal at time 0 to account for any discrepancies in cell numbers between conditions at the initiation of the experiment. Cell morphology was examined by Wright-Giemsa (WG) staining.
An experimental overview of the primary cell experiments is given in supplemental Figure 1. PRAME was consistently overexpressed for the duration of the experiment (supplemental Figure 2). Serum-free media with 1% lot-tested BSA (Biocell Laboratories Inc) was used in all primary progenitor cell experiments after transduction. To examine the effects of PRAME protein expression on in vitro myeloid differentiation, after GFP (or YFP) selection cells were cultured in 100 ng/mL stem cell factor (SCF), 30 ng/mL IL3, 20 ng/mL IL6, 20 ng/mL granulocyte colony-stimulating factor (G-CSF), and 20 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF; all cytokines from Peprotech).21 All patient samples were examined without ATRA. For CML progenitors, cells were also compared after treatment with ATRA at 0.001 μM (∼ physiologic concentrations, 1 patient), as well as 0.01 μM and 0.1 μM (2 patients). For normal hematopoietic progenitors, cells were compared at 0.001 μM (n = 2) and 0.01 μM (n = 1). On day 0, 100 000 cells were set up in independent culture dishes for each experimental day and condition to ensure that the data were not influenced by previous cell culture manipulation. Primary cells in culture were counted using trypan blue exclusion and were examined by WG staining on each experimental day.
Western blotting
Thirty micrograms of nuclear and cytoplasmic protein were run on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels. The following primary antibodies were used: rabbit polyclonal to PRAME (gift from Dr P. Coulie), rabbit polyclonal to PRAME (AbCam), rabbit polyclonal to lamin A/C (Cell Signaling), and mouse monoclonal to β-actin (Sigma-Aldrich) and GAPDH (AbCam). The following secondary antibodies were used: Amersham ECL anti–mouse IgG, HRP-linked species-specific whole antibody (GE Healthcare) or Amersham ECL anti–rabbit IgG, HRP-linked species-specific whole antibody (GE Healthcare).
Flow cytometry
For cell line experiments, flow cytometry was performed on a FACSCalibur system (Becton Dickinson Immunocytometry Systems). FlowJo (TreeStar Inc) was used for analysis. Cells were analyzed using CD11b/Mac-1-APC antibody (Becton Dickinson [BD]), CD11b-PE antibody (Caltag/Invitrogen), and 7-AAD (BD). Other antibodies included CD15-FITC (BD), CD64-FITC (Beckman-Coulter [BC]), CD41-PE (BC) and CD235a (glycophorin A)–PE (DAKO), CD14-PerCP-Cy5.5 (BD), IgG2-FITC (BD), and IgG1-PE (BD). Flow cytometry for human progenitor cells was performed using an LSRII (Becton Dickinson Immunocytometry Systems). Data were analyzed using software developed by Dr Brent Wood. Positivity for DAPI was used to exclude nonviable cells, and thresholds for positivity were determined using unstained cells and isotype control antibodies, as appropriate. Antibodies used included: CD11b PE (BD), CD117 PE-Cy5 (BC), CD33 PE-Cy7 (BD), and CD34 APC (BD). Data for surface marker expression are shown as either percentage expression or as absolute numbers of cells in culture expressing each marker on each experimental day.
Colony-forming assays
Colony-forming assays for cell lines were performed in triplicate using Methylcellulose Medium for Colony Assays of Human Cells without cytokines (StemCell Technologies) with or without the addition of ATRA at the concentrations listed under “Cell line culture, progenitor cell culture, and transduction.”
Normal hematopoietic progenitor cells were plated in triplicate on days 0, 2, 4, and 7 and CML progenitor cells were plated in triplicate on days 0 and 2 (and on day 4 for 2 patients) in 0.7% lot-tested agarose containing 30% lot-tested FBS, 1% BSA, 100 ng/mL SCF, 30 ng/mL IL3, 20 ng/mL IL6, 20 ng/mL G-CSF, and 20 ng/mL GM-CSF.21-23 Granulocyte-macrophage colony-forming unit (CFU-GM), granulocyte CFU (CFU-G), and monocyte CFU (CFU-M) colonies were counted 12 to 18 days after plating. Colony size was scored as +1 to +3 (50-199, 200-1000, and > 1000 cells, respectively). In addition to colony morphology, individual colonies were plucked and stained with WG. The numbers of progenitor cells in culture on each given experimental day with the ability to give rise to a colony were calculated as follows: (number of colonies counted)*(number of cells in culture on day of plating)/(number of cells plated). These numbers are reported on the Y-axis of all colony-forming assay figures. Micrographs were obtained using a Nikon E800 microscope (Nikon) and either a 10 ×/0.45 or 20 ×/0.75 PlanApo objective. Color images were acquired using a Coolsnap cf (Photometrics) CCD camera and Metamorph software (Molecular Devices).
Quantitative RT-PCR
After cDNA synthesis, PRAME expression was quantitated and normalized to GUSB expression. Relative expression is reported (ΔCt and ΔΔCt calculations). In addition, a more sensitive QPCR assay was used in which PRAME mRNA copy numbers in samples were extrapolated from known copy numbers of serially diluted PRAME plasmid cDNA and normalized to B2M expression. The assay reproducibly detected as few as 1 to 10 copies of PRAME mRNA. All samples were run in duplicate.
Microarray studies
Two published data sets were reanalyzed to identify gene expression correlating with PRAME expression in patient samples and to examine PRAME expression in normal and leukemia patient samples (“Patient samples”).2,19 In addition, PRAME and control vector–transduced normal progenitor cells were compared (2 independent experiments, each performed in duplicate) on days 4, 7, and 14. The procedures for RNA extraction, amplification, labeling, and hybridization, as well as statistical analysis methods for both the Rosetta platform and the Affymetrix HU-133A platform, are as previously published2,19,24 or as described in supplemental methods. For the hematopoietic progenitor cells, due to limited cell numbers, 1 μg of high-quality RNA was amplified using a single-stranded linear amplification protocol.25 CEL files and normalized data are available at http://www.fhcrc.org/science/labs/radich.26 Microarray data have also been deposited in the GEO public database under accession number GSE17100.27 Ingenuity Pathways Analyses (Ingenuity Systems, http://www.ingenuity.com) were used to determine biologically enriched pathways and functions, and the DAVID (National Institute of Allergy and Infectious Diseases [NIAID]) algorithm28 was used to identify enriched Gene Ontology categories29 (supplemental methods).30
Statistical analysis
Cell line experiments were performed in duplicate or triplicate. Proliferation and cell surface marker expression were compared across experimental conditions via linear regression models with adjustment for ATRA concentration and day in culture.
In the primary progenitor cell experiments we compared cell surface marker expression between experimental and control conditions over time using a least squares line fit to the average of the treatment group differences over independent experiments.
Mixed linear models were used to compare colony numbers in experimental versus control vector–transduced cells. These models included a random subject effect to account for correlation between repeated measurements among subjects, and fixed effects to adjust for day in culture and ATRA concentration. An interaction effect was included to test for a difference in colony type comparisons.
PRAME expression was compared between disease status groups using Wilcoxon rank-sum tests. The Fisher exact test was used to compare PRAME expression (defined as PRAME = 0 or PRAME > 0) across treatment response groups and by mutation status (present or absent), and t tests were used to test the associations between diagnostic blood counts and IM response before enrollment on the AMN107 trial and nilotinib outcomes or mutation status on the trial.
Results
PRAME expression in normal hematopoietic and leukemia cells
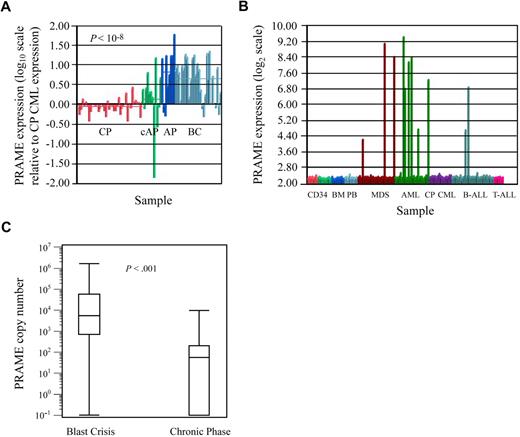
Microarray-based gene expression analyses revealed that PRAME was most highly expressed in BC CML patients (Figure 1A), was heterogeneously expressed in AML and MDS, and was not expressed in CP CML and most cases of ALL (Figure 1B). PRAME was also not expressed in normal hematopoietic cells including CD34+ BM, CD34+ PBSC, unsorted BM, unsorted PB (Figure 1B), or sorted B and T lymphocytes (data not shown).
PRAME expression in leukemia cell lines and patient samples. (A) PRAME expression increases with CML disease progression. PRAME expression is shown as the log10 ratio of the normalized expression of PRAME in each patient sample compared with PRAME expression in a pool of CML CP samples. Red bars represent CP patients (n = 42); green bars represent patients with either AP disease by virtue of additional cytogenetic abnormalities (cAP; n = 7) or patients who returned to CP after treatment for BC disease (last 3); dark blue bars represent patients with AP disease by both cytogenetic and blast count criteria (n = 10); and light blue represents BC patients (n = 28). The difference between CP and BC is statistically significant (P << .001). (B) PRAME expression in normal and malignant hematopoietic cells. Normalized PRAME expression is shown on a log2 scale. PRAME was heterogeneously expressed in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) patients with either refractory anemia with excess blasts or refractory anemia with excess blasts in transition. From left to right: CD34+ selected bone marrow (BM; red, n = 8), CD34+ selected peripheral blood stem cell products (light green, n = 10), unsorted BM (medium blue, n = 10), unsorted peripheral blood (light blue, n = 10), MDS (brown, n = 29), AML (green, n = 26), CP CML (purple, n = 17 independent cases from those shown in A), B-ALL (medium blue, n = 32), and T-ALL (pink, n = 7). (C) Quantitative PCR validation of PRAME expression in independent diagnostic BM samples from BC (n = 31) and early CP (n = 58) CML patients demonstrates increased expression in BC. Data are shown on a logarithmic (log10) scale. The line within each box represents the median; the upper and lower lines defining the box represent the 75th and 25th percentiles, respectively; and the lines outside the box extend to the maximum and minimum values (10−1 indicates undetectable PRAME expression). PRAME was expressed in all but 1 BC patient and in 50% of the CP patients. Mean expression was 9.0 × 104 copies (median, 5.5 × 104; range, 0 to 1.6 × 106 copies) in BC patients versus 560 copies (median, 200; range, 0 to 9.8 × 103 copies) in CP patients (P < .001).
PRAME expression in leukemia cell lines and patient samples. (A) PRAME expression increases with CML disease progression. PRAME expression is shown as the log10 ratio of the normalized expression of PRAME in each patient sample compared with PRAME expression in a pool of CML CP samples. Red bars represent CP patients (n = 42); green bars represent patients with either AP disease by virtue of additional cytogenetic abnormalities (cAP; n = 7) or patients who returned to CP after treatment for BC disease (last 3); dark blue bars represent patients with AP disease by both cytogenetic and blast count criteria (n = 10); and light blue represents BC patients (n = 28). The difference between CP and BC is statistically significant (P << .001). (B) PRAME expression in normal and malignant hematopoietic cells. Normalized PRAME expression is shown on a log2 scale. PRAME was heterogeneously expressed in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) patients with either refractory anemia with excess blasts or refractory anemia with excess blasts in transition. From left to right: CD34+ selected bone marrow (BM; red, n = 8), CD34+ selected peripheral blood stem cell products (light green, n = 10), unsorted BM (medium blue, n = 10), unsorted peripheral blood (light blue, n = 10), MDS (brown, n = 29), AML (green, n = 26), CP CML (purple, n = 17 independent cases from those shown in A), B-ALL (medium blue, n = 32), and T-ALL (pink, n = 7). (C) Quantitative PCR validation of PRAME expression in independent diagnostic BM samples from BC (n = 31) and early CP (n = 58) CML patients demonstrates increased expression in BC. Data are shown on a logarithmic (log10) scale. The line within each box represents the median; the upper and lower lines defining the box represent the 75th and 25th percentiles, respectively; and the lines outside the box extend to the maximum and minimum values (10−1 indicates undetectable PRAME expression). PRAME was expressed in all but 1 BC patient and in 50% of the CP patients. Mean expression was 9.0 × 104 copies (median, 5.5 × 104; range, 0 to 1.6 × 106 copies) in BC patients versus 560 copies (median, 200; range, 0 to 9.8 × 103 copies) in CP patients (P < .001).
PRAME expression was validated in independent patient samples by QPCR, where sensitivity and range of expression are greater. PRAME was highly expressed in all but 1 of 31 BC patients (median expression, 5500 copies), and could be detected in 50% of 58 early CP patients (Figure 1C). Median expression, however, was significantly lower, at 200 copies among CP patients who expressed PRAME. PRAME was expressed in all but 6 of 35 APL patients and expression varied over 3 orders of magnitude. It was either undetectable or detectable at only very low levels in 3 normal CD34+ and 4 unsorted BM samples.
PRAME expression in leukemia cell lines inhibits granulocytic differentiation
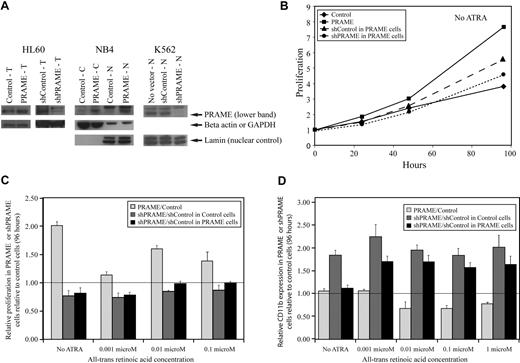
A lentiviral GFP-selectable PRAME expression vector and YFP-selectable vectors containing shRNA hairpins silencing PRAME (supplemental methods) were used to examine the effects of PRAME protein expression in HL60, NB4, and K562 cells (Figure 2A). The AML cell lines NB4 and HL60 express some PRAME protein and differentiate along the granulocytic lineage, with increasing CD11b expression, after treatment with supraphysiologic concentrations of ATRA. HL60 and NB4 cells overexpressing PRAME protein proliferated more rapidly than control vector–transduced cells both in the absence of ATRA and after treatment with increasing concentrations of ATRA (Figure 2B-C). PRAME-overexpressing HL60 cells also exhibited decreased CD11b expression relative to control cells after treatment with various ATRA concentrations over 96 hours (Figure 2D). Although NB4 cells transduced with PRAME exhibited similar differences, these were smaller and mostly evident at lower ATRA concentrations (≤ 0.01 μM). PRAME was then silenced in both the PRAME-overexpressing and control vector–transduced HL60 cell lines. The shPRAME silenced cells exhibited decreased proliferation and increased CD11b expression relative to shContol (shGFP) cells in both cell lines (Figure 2B-D). These data suggest that PRAME protein expression in these cell lines promotes proliferation and inhibits granulocytic differentiation.
PRAME expression in all-trans retinoic acid–responsive leukemia cell lines increases proliferation and inhibits granulocytic differentiation. (A) PRAME protein expression is shown in HL60, NB4, and K562 cells by Western blotting. PRAME protein overexpression by lentiviral PRAME expression vector transduction is shown in total cell lysates (T) from HL60 cells and in both cytoplasmic (C) and nuclear (N) lysates from NB4 cells. PRAME silencing using lentiviral vectors containing short-hairpins targeting PRAME is shown in T-cell lysates in HL60 cells and N lysates in K562 cells. In the HL60 cells shown here, PRAME was silenced in HL60 cells overexpressing PRAME protein. Lamin served as the nuclear control; beta actin and GAPDH served as the cytoplasmic or total lysate control. (B) PRAME-overexpressing HL60 cells (PRAME) proliferated more rapidly than control cells (Control) over 96 hours. When PRAME was silenced in these PRAME-overexpressing cells (shPRAME), proliferation decreased compared with control cells (shControl). PRAME cells are PRAME-overexpressing cells; control cells are the empty vector control. PRAME was silenced (shPRAME) in both PRAME-overexpressing cells and the corresponding empty vector–transduced cells. shControl is a hairpin that targets GFP. (C) The increase in proliferation in HL60 PRAME cells relative to control cells (P < .001) as well as the decrease in proliferation of shPRAME HL60 cells relative to shControl HL60 cells (P = .06 in PRAME-overexpressing cells and P < .001 in control HL60 cells) were seen both in the absence and presence of increasing concentrations of all-trans retinoic acid (ATRA). For simplicity, proliferation in PRAME or shPRAME cells is shown relative to the corresponding control cell line (experimental condition/control condition). Thus, the line at 1.00 indicates the expected ratio if there is no difference. Data at 96 hours are used and show the mean of 3 independent experiments. The P values, however, were calculated using all data (ATRA and no ATRA) at 0, 24, 48, 72, and 96 hours. (D) Granulocytic differentiation as measured by CD11b expression over time was inhibited in PRAME-overexpressing HL60 cells. PRAME cells expressed significantly less CD11b over time than control cells at various ATRA concentrations (P = .003). When PRAME was silenced in these PRAME-overexpressing cells as well as the empty vector control cells, CD11b expression was greater in shPRAME cells compared with shControl cells in both the PRAME-overexpressing cell line and the control vector cell line (P < .001 for both). The ratio of CD11b expression in PRAME or shPRAME cells is shown relative to CD11b expression in control or shControl cells (experimental percentages CD11b/control percentage CD11b) after 96 hours. The line at 1.00 indicates the expected ratio if there is no difference.
PRAME expression in all-trans retinoic acid–responsive leukemia cell lines increases proliferation and inhibits granulocytic differentiation. (A) PRAME protein expression is shown in HL60, NB4, and K562 cells by Western blotting. PRAME protein overexpression by lentiviral PRAME expression vector transduction is shown in total cell lysates (T) from HL60 cells and in both cytoplasmic (C) and nuclear (N) lysates from NB4 cells. PRAME silencing using lentiviral vectors containing short-hairpins targeting PRAME is shown in T-cell lysates in HL60 cells and N lysates in K562 cells. In the HL60 cells shown here, PRAME was silenced in HL60 cells overexpressing PRAME protein. Lamin served as the nuclear control; beta actin and GAPDH served as the cytoplasmic or total lysate control. (B) PRAME-overexpressing HL60 cells (PRAME) proliferated more rapidly than control cells (Control) over 96 hours. When PRAME was silenced in these PRAME-overexpressing cells (shPRAME), proliferation decreased compared with control cells (shControl). PRAME cells are PRAME-overexpressing cells; control cells are the empty vector control. PRAME was silenced (shPRAME) in both PRAME-overexpressing cells and the corresponding empty vector–transduced cells. shControl is a hairpin that targets GFP. (C) The increase in proliferation in HL60 PRAME cells relative to control cells (P < .001) as well as the decrease in proliferation of shPRAME HL60 cells relative to shControl HL60 cells (P = .06 in PRAME-overexpressing cells and P < .001 in control HL60 cells) were seen both in the absence and presence of increasing concentrations of all-trans retinoic acid (ATRA). For simplicity, proliferation in PRAME or shPRAME cells is shown relative to the corresponding control cell line (experimental condition/control condition). Thus, the line at 1.00 indicates the expected ratio if there is no difference. Data at 96 hours are used and show the mean of 3 independent experiments. The P values, however, were calculated using all data (ATRA and no ATRA) at 0, 24, 48, 72, and 96 hours. (D) Granulocytic differentiation as measured by CD11b expression over time was inhibited in PRAME-overexpressing HL60 cells. PRAME cells expressed significantly less CD11b over time than control cells at various ATRA concentrations (P = .003). When PRAME was silenced in these PRAME-overexpressing cells as well as the empty vector control cells, CD11b expression was greater in shPRAME cells compared with shControl cells in both the PRAME-overexpressing cell line and the control vector cell line (P < .001 for both). The ratio of CD11b expression in PRAME or shPRAME cells is shown relative to CD11b expression in control or shControl cells (experimental percentages CD11b/control percentage CD11b) after 96 hours. The line at 1.00 indicates the expected ratio if there is no difference.
K562 cells express PRAME protein highly, but do not differentiate into granulocytes after ATRA treatment. It has previously been shown that increasing RARα numbers, such that K562 cells express the same numbers as HL60 cells, do not promote granulocytic differentiation after ATRA exposure.31 We examined whether PRAME silencing in these K562 cells with increased numbers of RARα, compared with the corresponding control with an unchanged complement of RARα, promoted granulocytic differentiation. PRAME silencing in these cell lines did not promote granulocytic differentiation after ATRA treatment. Granulocytic differentiation was not observed by WG staining and CD11b expression remained low, suggesting that even with increased numbers of RARα, PRAME silencing was not sufficient to induce granulocytic differentiation. Other lineage markers including CD33 (moderate), CD15 (moderate), CD64 (moderate), CD41 (low), CD34 (low), and glycophorin A (CD235a, high) were no different between shControl and shPRAME K562 cell lines. Similar to published data, however, proliferation was increased in shPRAME silenced K562 cells (supplemental Figure 3).32 These data suggest that PRAME's inhibition of ATRA-induced granulocytic differentiation requires other factors in some cell types.
PRAME expression in normal and CML CD34+ hematopoietic progenitor cells inhibits myeloid differentiation
To examine the consequences of PRAME protein expression on cell proliferation and myeloid differentiation in multipotent progenitor cells, PRAME was aberrantly expressed in normal CD34+ progenitor cells from healthy donors (who do not express PRAME) and silenced in CD34+ CML myeloid BC primary cells (supplemental Figures 1-2).
In normal hematopoietic progenitors, PRAME was expressed using a GFP-selectable lentiviral vector (supplemental methods). Myeloid differentiation was examined in the absence of ATRA and after exposure to various concentrations of ATRA. Cells were cultured in SCF, IL3, IL6, GM-CSF, and G-CSF as discussed in “Cell line culture, progenitor cell culture, and transduction.” Forced PRAME expression did not alter proliferation, but inhibited myeloid differentiation, similar to the effect observed in NB4 and HL60 cells (Figure 3). PRAME cells, compared with control cells, demonstrated increased expression of early precursor cell markers (CD34 and CD117, P = .001 and P = .002, respectively) and decreased expression of terminal myeloid markers (CD11b, P = .04) in culture over time in the absence of ATRA. As shown in Figure 3D-I, these differences persisted in PRAME cells exposed to physiologic concentrations of ATRA (0.001 μM; P = .02 for CD117, P = .15 for CD11b), but were smaller than those observed in the absence of ATRA. After exposure to supraphysiologic concentrations of ATRA (0.01 μM), myeloid differentiation was primarily granulocytic by WG staining and occurred more rapidly. A similar difference in CD117 expression between PRAME and control cells, however, was observed earlier in culture (between days 0 to 7).
Aberrant PRAME expression in CD34+ normal hematopoietic progenitors inhibits myeloid differentiation. (A-C) The numbers of cells in culture expressing CD34 (A), CD117 (B), and CD11b (C) are shown over time in PRAME vector–transduced cells compared with control vector–transduced cells in the absence of ATRA. The data shown represent the mean of 3 independent experiments. On day 0 there were 100 000 cells in culture for each condition. The early precursor cell markers, CD34 and CD117 (P = .001 and P = .002, respectively), were more highly expressed in PRAME cells and the terminal myeloid marker, CD11b (P = .04), demonstrated lower expression in PRAME cells compared with control vector–transduced cells over time. (D-I) The consequences of ATRA exposure on percentage of CD117 and CD11b expression over time are shown. In the absence of ATRA, PRAME-expressing cells, compared with control cells, exhibited up to 41% greater expression of CD117 and 35% lower expression of CD11b (D,G). These differences persisted in PRAME cells exposed to low (physiologic) concentrations of ATRA (0.001 μM; E,H), but were overcome with increasing concentrations of ATRA of 0.01 μM (F,I; CD117 [P = .02] and CD11b [P = .15]). However, as shown in panel F, PRAME cells expressed increased CD117 early in culture between days 0 and 7, compared with later in culture for cells exposed to no ATRA or 0.001 μM ATRA concentrations (D-E).
Aberrant PRAME expression in CD34+ normal hematopoietic progenitors inhibits myeloid differentiation. (A-C) The numbers of cells in culture expressing CD34 (A), CD117 (B), and CD11b (C) are shown over time in PRAME vector–transduced cells compared with control vector–transduced cells in the absence of ATRA. The data shown represent the mean of 3 independent experiments. On day 0 there were 100 000 cells in culture for each condition. The early precursor cell markers, CD34 and CD117 (P = .001 and P = .002, respectively), were more highly expressed in PRAME cells and the terminal myeloid marker, CD11b (P = .04), demonstrated lower expression in PRAME cells compared with control vector–transduced cells over time. (D-I) The consequences of ATRA exposure on percentage of CD117 and CD11b expression over time are shown. In the absence of ATRA, PRAME-expressing cells, compared with control cells, exhibited up to 41% greater expression of CD117 and 35% lower expression of CD11b (D,G). These differences persisted in PRAME cells exposed to low (physiologic) concentrations of ATRA (0.001 μM; E,H), but were overcome with increasing concentrations of ATRA of 0.01 μM (F,I; CD117 [P = .02] and CD11b [P = .15]). However, as shown in panel F, PRAME cells expressed increased CD117 early in culture between days 0 and 7, compared with later in culture for cells exposed to no ATRA or 0.001 μM ATRA concentrations (D-E).
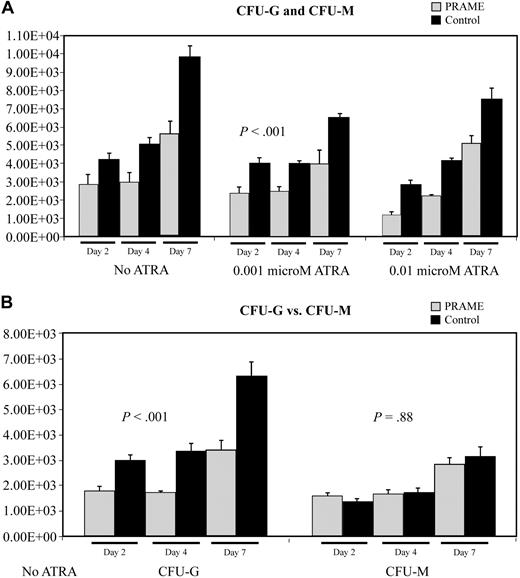
Colony-forming assay data also suggested that myeloid differentiation was inhibited (Figure 4). PRAME-expressing cells consistently formed fewer colonies then control cells (P < .001) on agarose containing SCF, IL3, IL6, GM-CSF, and G-CSF. Comparing granulocytic to monocytic/macrophage colony formation, PRAME-expressing cells formed relatively fewer granulocytic colonies than monocytic/macrophage colonies (difference in CFU-G, P < .001; difference in CFU-M, P = .88). These data suggest that granulocytic differentiation was preferentially impaired.
Aberrant PRAME expression in CD34+ hematopoietic progenitors inhibits colony formation due to impaired myeloid differentiation. (A) The numbers of CFU-G and CFU-M in PRAME-transduced cells were compared with control vector–transduced cells. The mean colony numbers from 3 independent experiments performed in triplicate are shown. Colony numbers in PRAME-expressing cells compared with control cells were decreased on days 2, 4, and 7 both in the absence and presence of increasing concentrations of ATRA (P < .001). The numbers on the y-axis represent the numbers of progenitor cells in culture on each experimental day that gave rise to a colony. These numbers were calculated from the colony counts, the numbers of cells in culture on the day of plating, and the numbers of cells plated. (B) To more fully characterize the impact of aberrant PRAME expression on myeloid differentiation, individual colonies were plucked and stained to discriminate CFU-G from CFU-M. There were significantly fewer CFU-G colonies in PRAME cells compared with control cells on days 2, 4, and 7 (P < .001). These differences were not observed for CFU-Ms (P = .88).
Aberrant PRAME expression in CD34+ hematopoietic progenitors inhibits colony formation due to impaired myeloid differentiation. (A) The numbers of CFU-G and CFU-M in PRAME-transduced cells were compared with control vector–transduced cells. The mean colony numbers from 3 independent experiments performed in triplicate are shown. Colony numbers in PRAME-expressing cells compared with control cells were decreased on days 2, 4, and 7 both in the absence and presence of increasing concentrations of ATRA (P < .001). The numbers on the y-axis represent the numbers of progenitor cells in culture on each experimental day that gave rise to a colony. These numbers were calculated from the colony counts, the numbers of cells in culture on the day of plating, and the numbers of cells plated. (B) To more fully characterize the impact of aberrant PRAME expression on myeloid differentiation, individual colonies were plucked and stained to discriminate CFU-G from CFU-M. There were significantly fewer CFU-G colonies in PRAME cells compared with control cells on days 2, 4, and 7 (P < .001). These differences were not observed for CFU-Ms (P = .88).
PRAME was then silenced in 3 PRAME-expressing myeloid BC CML patients using YFP-selectable shRNA lentiviral constructs (supplemental methods). In 1 patient, PRAME silenced cells proliferated more rapidly in culture; however, no difference in proliferation was observed in the remaining patients. Decreased CD34 and CD117 expression was observed in shPRAME cells, compared with shControl cells, in 2 patients. In 1 patient, decrease in CD34 and CD117 expression was apparent in the absence of ATRA (14% and 7%, respectively); whereas in the second patient the effect was greater after exposure to ATRA at 0.01 or 0.1 μM (10% and 12% decrease in CD34, respectively and 9% and 5% decrease in CD117, respectively). Cell numbers were limited for the third patient, but did not demonstrate a difference.
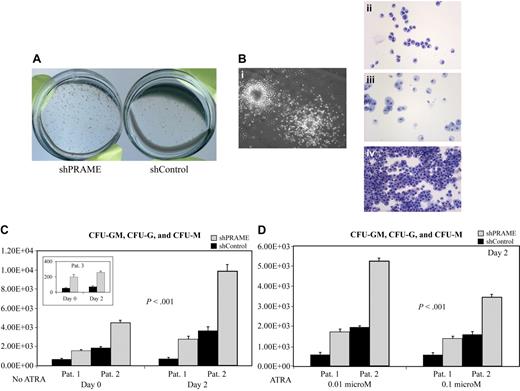
In contrast to the CD34+ normal progenitors, which formed fewer CFU-G and CFU-M colonies with forced PRAME expression, all PRAME silenced patient samples consistently demonstrated the reverse phenotype with increased numbers of CFU-GM, CFU-G, and CFU-M colonies (Figure 5A-C). This difference was also observed in 2 patient samples after cells were treated with ATRA (Figure 5D). Both CFU-G and CFU-M colony numbers were increased in shPRAME cells. In summary, these data suggest that PRAME expression inhibits myeloid differentiation.
PRAME silencing in primary CML progenitor cells increases myeloid colony formation. (A) shPRAME-transduced cells (left) compared with shControl cells (shGFP, right) from 3 CML blast crisis patients exhibited increased colony formation on agarose both in the absence and presence of ATRA at 0.01 and 0.1 μM. Colonies arising from shPRAME and shControl cells exposed to 0.01 μM ATRA from 1 patient are shown. (B) To confirm colony morphology after exposure to GM-CSF and G-CSF individual colonies (i) were plucked and stained with Wright-Giemsa. Colony types were predominately granulocytic (CFU-G, ii) or monocytic (CFU-M, iii), but CFU-GMs (iv) were also seen. (C) shPRAME silenced CML cells compared with shControl cells from 3 patients demonstrated increased numbers of CFU-GMs, CFU-Gs, and CFU-Ms on days 0 and 2. The mean colony numbers from independent experiments performed in triplicate are shown. The numbers represented on the y-axis indicate the numbers of progenitor cells in culture on each experimental day that gave rise to a colony. (D) For 2 patients, sufficient cells were available to assess CFU formation after exposure to ATRA at 0.01 μM and 0.1 μM. Similar to the phenotype seen in the absence of ATRA, shPRAME cells compared with shControl cells formed increased numbers of CFU-GM, CFU-G, and CFU-M colonies. Day 2 is shown; day 4 shows the same increase in CFUs in shPRAME cells compared with shControl cells (included in calculations of statistical significance). Furthermore, the same differences were observed in 1 patient whose cells were treated with 0.001 μM ATRA. The mean numbers of colonies are shown from independent experiments performed in triplicate. The numbers represented on the y-axis indicate the numbers of cells in culture on a particular experimental day that gave rise to a colony.
PRAME silencing in primary CML progenitor cells increases myeloid colony formation. (A) shPRAME-transduced cells (left) compared with shControl cells (shGFP, right) from 3 CML blast crisis patients exhibited increased colony formation on agarose both in the absence and presence of ATRA at 0.01 and 0.1 μM. Colonies arising from shPRAME and shControl cells exposed to 0.01 μM ATRA from 1 patient are shown. (B) To confirm colony morphology after exposure to GM-CSF and G-CSF individual colonies (i) were plucked and stained with Wright-Giemsa. Colony types were predominately granulocytic (CFU-G, ii) or monocytic (CFU-M, iii), but CFU-GMs (iv) were also seen. (C) shPRAME silenced CML cells compared with shControl cells from 3 patients demonstrated increased numbers of CFU-GMs, CFU-Gs, and CFU-Ms on days 0 and 2. The mean colony numbers from independent experiments performed in triplicate are shown. The numbers represented on the y-axis indicate the numbers of progenitor cells in culture on each experimental day that gave rise to a colony. (D) For 2 patients, sufficient cells were available to assess CFU formation after exposure to ATRA at 0.01 μM and 0.1 μM. Similar to the phenotype seen in the absence of ATRA, shPRAME cells compared with shControl cells formed increased numbers of CFU-GM, CFU-G, and CFU-M colonies. Day 2 is shown; day 4 shows the same increase in CFUs in shPRAME cells compared with shControl cells (included in calculations of statistical significance). Furthermore, the same differences were observed in 1 patient whose cells were treated with 0.001 μM ATRA. The mean numbers of colonies are shown from independent experiments performed in triplicate. The numbers represented on the y-axis indicate the numbers of cells in culture on a particular experimental day that gave rise to a colony.
PRAME expression is associated with differential gene expression
Little is known regarding gene expression associated with PRAME expression. We first identified a gene signature correlating with PRAME expression in our CML microarray data set.2 Using a correlation coefficient greater than 0.6, 142 probe sets were negatively correlated and 191 were positively correlated with PRAME expression (supplemental Table 1). The most highly negatively correlated gene was INPP5D (or Ship-1) and the most highly positively correlated genes were GARP, ALDH7A1, FKBP10, MTA3, RAB3GAP, and FHL1. Within the gene expression network around PRAME (Ingenuity Pathways Analysis), decreased expression of differentiation-associated genes such as CEBPA, CEBPE, and EP300 was identified.
To more directly identify genes, biologic and molecular functions, and pathways that distinguish cells that express PRAME from those that do not, normal CD34+ cells with forced PRAME expression were compared with control vector–transduced cells in culture over time (days 4, 7, 14) using gene expression microarrays (Affymetrix HU-133). Linear time covariate analysis (GenePlus; Enodar Biologic Corporation) identified fewer genes changing over time in PRAME cells compared with control cells (supplemental Tables 2-4), possibly consistent with a block in differentiation. By day 14, PRAME-expressing cells were skewed toward differentially decreased gene expression. Genes with decreased expression in PRAME cells included granule-associated genes (PRTN3 and MPO), caspases (1, 2, and 8), and several tumor suppressors (PTEN, RASA1, and CDKN1A). Top statistically significantly enriched molecular and cellular functions included apoptosis; cell development and differentiation; DNA replication, recombination, and repair; and immune response (supplemental Tables 5-6, DAVID [NIAID] and Ingenuity Pathways Analysis). In these analyses, the gene expression network around PRAME demonstrated a small but significant increase in EZH2 and YY1 expression (supplemental Figure 4).
PRAME expression before TKI therapy is associated with response and mutation status in late CP patients
The association between PRAME expression and outcomes has not been characterized in CML. Because PRAME expression is associated with advanced disease where therapeutic outcomes are much poorer, we hypothesized that PRAME expression in CP cases may be associated with poorer outcomes on TKI therapy. We examined 25 late CP CML patients who previously failed IM using samples before second TKI therapy with nilotinib (AMN107 study). Among these patients, 12 were considered nilotinib failures. Failure on nilotinib was defined as no or loss of a hematologic response. In addition, patients who lost a cytogenetic response or who did not achieve a cytogenetic response or achieved only minor or minimal cytogenetic responses on nilotinib were also considered failures. The remaining 13 patients maintained ongoing hematologic or cytogenetic responses for the duration of follow up. As shown in Tables 1–2, PRAME expression before nilotinib therapy was statistically significantly associated with nilotinib failure (P = .04) and the presence of ABL tyrosine kinase domain (TKD) point mutations developing either before or on therapy (P = .03). The associations between PRAME expression before nilotinib therapy and nilotinib response and mutation status were independent of prior IM response at AMN study entry and white blood cell and peripheral blast count at study entry. There was no association between cytogenetic or hematologic response on IM before nilotinib therapy and treatment outcomes (P = .76 and P = .81, respectively) or mutation status (P = .10 and P = .69, respectively). Furthermore, there was also no association between white blood cell count or peripheral blast count at AMN study entry and treatment outcomes (P = .76 and P = .86, respectively) or mutation status (P = .87 and P = .61, respectively).
PRAME expression is associated with poorer response to nilotinib in late CP patients
| . | PRAME detected, no. (%) . | PRAME not detected, no. (%) . |
|---|---|---|
| Responded to nilotinib | 5 (38) | 8 (62) |
| Failed nilotinib | 10 (83) | 2 (17) |
| . | PRAME detected, no. (%) . | PRAME not detected, no. (%) . |
|---|---|---|
| Responded to nilotinib | 5 (38) | 8 (62) |
| Failed nilotinib | 10 (83) | 2 (17) |
Twenty-five late CP patients received nilotinib as second-line therapy after imatinib failure. PRAME expression was measured prior to beginning nilotinib therapy using QPCR. PRAME expression was detected in 10 (83%) of 12 patients who failed nilotinib therapy compared with 5 (38%) of 13 patients who responded (P = .04).
PRAME expression is associated with an increased incidence of ABL tyrosine kinase domain point mutations in late CP patients
| . | PRAME detected, no. (%) . | PRAME not detected, no. (%) . |
|---|---|---|
| Mutation at entry | 8 (89) | 1 (11) |
| Mutation at any time | 11 (79) | 3 (21) |
| No mutation at entry | 5 (42) | 7 (58) |
| No mutation at any time | 2 (25) | 6 (75) |
| . | PRAME detected, no. (%) . | PRAME not detected, no. (%) . |
|---|---|---|
| Mutation at entry | 8 (89) | 1 (11) |
| Mutation at any time | 11 (79) | 3 (21) |
| No mutation at entry | 5 (42) | 7 (58) |
| No mutation at any time | 2 (25) | 6 (75) |
ABL tyrosine kinase domain (TKD) point mutation data were available for 21 patients at study entry and for 22 patients during nilotinib treatment. PRAME expression was detected in 11 (79%) of 14 patients who developed ABL TKD point mutations either prior to therapy or during therapy compared with 2 (25%) of 8 patients who never had evidence of a point mutation (P=.03). When restricted to patients with mutations at study entry only, P=.07.
We also examined PRAME expression in 58 newly diagnosed, untreated CP CML patients within 6 months of diagnosis (RIGHT study). Seven patients did not achieve a major cytogenetic response (≤ 35% Philadelphia chromosomes) by 12 months or progressed on IM. As shown in Table 3, although the proportions of patients expressing PRAME in the responder and nonresponder groups were similar to those reported for the late CP patients, this observation did not reach statistical significance (possibly due to the overall low failure rate in these patients).
PRAME expression and response in early CP patients
| . | PRAME detected, no. (%) . | PRAME not detected, no. (%) . |
|---|---|---|
| Responded to imatinib | 23 (45) | 28 (55) |
| Failed imatinib | 5 (71) | 2 (29) |
| . | PRAME detected, no. (%) . | PRAME not detected, no. (%) . |
|---|---|---|
| Responded to imatinib | 23 (45) | 28 (55) |
| Failed imatinib | 5 (71) | 2 (29) |
A similar difference was observed in 58 newly diagnosed and untreated CP CML patients. The proportions of patients expressing PRAME in the imatinib mesylate (IM) responder and IM nonresponder groups were similar to the late CP patients receiving nilotinib, but did not reach statistical significance (P=.25). PRAME expression was detected in 5 (71%) of 7 patients who failed IM compared with 23 (45%) of 51 patients who responded to IM.
Discussion
Although PRAME is frequently expressed in myeloid malignancies, its function and clinical relevance remain unclear.2,3,6 Given that retinoic acid receptor–mediated signaling is important in myeloid differentiation and is perturbed in leukemia,15-17 and that PRAME expression is primarily associated with acute leukemia where there is a block in differentiation, we examined whether PRAME protein expression inhibits myeloid differentiation. When normal hematopoietic progenitors aberrantly expressed PRAME, myeloid differentiation was inhibited not only in the presence of ATRA, but also in its absence. This block, however, did not increase the numbers of progenitor cells capable of forming differentiated myeloid cells, but rather resulted in impaired differentiation with fewer progenitors giving rise to fully differentiated myeloid cells. Correspondingly, when PRAME was silenced in primary myeloid blast crisis CML cells, more progenitors were capable of forming differentiated myeloid cells.
Our findings also suggest that PRAME may require additional factors to inhibit differentiation in certain cell types, and thus support the apparently conflicting observations of several groups.5,10,32,33 Similar to the report by Epping et al, in which PRAME inhibited differentiation in melanoma cell lines in the presence of ATRA,5 we found that PRAME protein expression in HL60 and NB4 cells inhibited, although did not completely block, granulocytic differentiation after exposure to ATRA. In contrast, in K562 cells, even in K562 cells with increased numbers of RARα receptors, PRAME silencing did not promote granulocytic differentiation after ATRA exposure. Rather, similar to published data, PRAME silencing promoted proliferation.32,33 An unexpected finding was that myeloid differentiation in both normal and CML hematopoietic progenitors was inhibited by PRAME not only in the presence, but also the absence, of ATRA. Although, it is possible that residual retinoic acid bound to albumin in serum or BSA may contribute to the ATRA-independent effects observed, this possibility seems less likely. In addition, primary cell experiments were performed primarily in serum-free media where ATRA levels were likely well below physiologic levels, if not absent. This observation also suggests that investigations of PRAME in primary cells are particularly important as they may elucidate and clarify differences reported in cell line studies.
It remains unclear how PRAME mediates this observed block in myeloid differentiation in the absence of retinoic acid, although it may be mediated by RARα or by another nuclear receptor. Our gene expression studies in primary cells identified possible effects of PRAME expression on other genes involved in the regulation of myeloid differentiation. CEBPA and CEBPE, which promote myeloid differentiation, decreased with increasing PRAME expression, whereas YY1, which inhibits granulocytic differentiation, increased with increasing PRAME expression.34-36 SOX9, however, which has recently been reported to regulate PRAME expression and ATRA sensitivity in melanoma cells,37 was not differentially expressed in BC CML cases that express or do not express PRAME. SOX9 expression was low or absent in all cases.
The suggestion that PRAME may be involved in CML progression led us to investigate if PRAME expression correlated with response to TKI therapy, as phase of disease is the most important factor determining TKI therapy efficacy and the likelihood of developing ABL TKD point mutations.2,38 Similar to advanced disease patients, PRAME-expressing late CP patients were more likely to fail therapy and to develop or have baseline ABL TKD point mutations. This association between PRAME expression and poor outcome contrasts, but does not conflict, with the observation that PRAME expression is associated with better outcomes and good risk cytogenetic abnormalities in AML patients.4,13,14 Our studies of CML progression2 together with our studies in late CP patients suggest that PRAME expression is a molecular marker for more advanced disease where outcomes, overall, are significantly poorer.
Because of its limited expression in normal tissues, PRAME is an attractive target for therapy. Promoting differentiation in leukemia cells can make these cells more susceptible to other therapies including targeted therapies and chemotherapy. In advanced CML, PRAME mRNA is expressed heterogeneously in the hematopoietic stem cell compartment, but is more consistently expressed in the common myeloid and granulocyte/macrophage progenitor compartments.39 The latter has been described as a potential leukemia stem cell compartment in myeloid BC.40 That PRAME impairs the efficacy of histone deacetylase inhibitors in solid tumor cells has also recently been described.41 Thus, PRAME overexpression may contribute to drug resistance in CML. As the polycomb group protein EZH2 may mediate transcriptional repression by PRAME,5 histone deacetylase inhibitors that deplete EZH2 may be an effective strategy in treating PRAME-expressing CML.42 Lastly, as a leukemia-associated antigen, PRAME has recently become a target for immunologic interventions. HLA-A2–restricted cytotoxic T lymphocyte clones induced against PRAME can lyse solid tumor cells43 and recognize and respond to primary CML cells.44,45
In conclusion, these data suggest that PRAME expression plays a role in the block in differentiation seen in progression from CP to myeloid BC, although it likely requires other genetic events, such as BCR-ABL, to drive proliferation. PRAME inhibits myeloid differentiation in a retinoic acid–dependent manner, but also appears to act independently of retinoic acid. In hematopoietic cells its function depends upon cell phenotype, suggesting other factors are required to inhibit differentiation in some cell types. Lastly, PRAME expression is associated with ABL TKD point mutations and poor treatment response and as such may be a marker of considerable clinical utility.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to the following people: Drs Pierre Coulie and Rene Bernards for providing the original PRAME antibody and retroviral shRNA vectors used in these experiments; Dr C. A. Blau for providing the lentiviral expression vectors; Dr Stephen Collins for the kind loan of the K562 RARA and control cell lines; and Dr Hans P. Kiem and his laboratory for lentiviral shRNA vectors and assistance with the preparation of viral supernatants (Core Grant no. DK56465). We also thank Karen McDougall and August J. Salvado of Novartis Pharmaceuticals Corporation for providing outcomes data for the RIGHT study patients and Hongyue Dai of Rosetta Inpharmatics, Merck and Co for assistance with microarray analysis of the CML progression data.
This work was supported by National Cancer Institute (NCI) grants CA18029 (J.P.R.) and CA106796, a Leukemia & Lymphoma Society Translational Research Program grant, and a V Foundation for Cancer Research V Scholar grant (V.G.O.).
National Institutes of Health
Authorship
Contribution: V.G.O. initiated the study, performed experiments, collected and analyzed all data, created all figures, and wrote the paper; K.A.G. and T.G. performed the statistical analyses; C.L.C. and T.Y. assisted with PRAME quantitative PCR, Western blotting, flow cytometry, and cell transduction; K.S. assisted with progenitor cell experiments; B.L.W. performed flow cytometry for progenitor cell studies; M.T.E. provided vectors and shRNA hairpins for silencing studies; Y.S. provided outcomes for the AMN107 study patients; E.P.-A. and P.L. prepared microarrays; D.L.S. analyzed AML and MDS array data; J.L.A. provided experimental advice for progenitor cell experiments and analysis; and J.P.R. helped organize the studies and the paper.
Conflict-of-interest disclosure: Y.S. is employed at Novartis Pharmaceuticals Corporation. J.P.R. receives laboratory support and honoraria from Novartis Pharmaceuticals Corporation. The remaining authors declare no competing financial interests.
Correspondence: Vivian G. Oehler, 1100 Fairview Ave N, D4-100, PO Box 19024, Seattle, WA 98109-1024; e-mail: voehler@u.washington.edu.

![Figure 3. Aberrant PRAME expression in CD34+ normal hematopoietic progenitors inhibits myeloid differentiation. (A-C) The numbers of cells in culture expressing CD34 (A), CD117 (B), and CD11b (C) are shown over time in PRAME vector–transduced cells compared with control vector–transduced cells in the absence of ATRA. The data shown represent the mean of 3 independent experiments. On day 0 there were 100 000 cells in culture for each condition. The early precursor cell markers, CD34 and CD117 (P = .001 and P = .002, respectively), were more highly expressed in PRAME cells and the terminal myeloid marker, CD11b (P = .04), demonstrated lower expression in PRAME cells compared with control vector–transduced cells over time. (D-I) The consequences of ATRA exposure on percentage of CD117 and CD11b expression over time are shown. In the absence of ATRA, PRAME-expressing cells, compared with control cells, exhibited up to 41% greater expression of CD117 and 35% lower expression of CD11b (D,G). These differences persisted in PRAME cells exposed to low (physiologic) concentrations of ATRA (0.001 μM; E,H), but were overcome with increasing concentrations of ATRA of 0.01 μM (F,I; CD117 [P = .02] and CD11b [P = .15]). However, as shown in panel F, PRAME cells expressed increased CD117 early in culture between days 0 and 7, compared with later in culture for cells exposed to no ATRA or 0.001 μM ATRA concentrations (D-E).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/15/10.1182_blood-2008-07-170282/4/m_zh89990941870003.jpeg?Expires=1765891109&Signature=Tdce2huqwzWQd0CwznMLqnolkVFojynL7cp9vvxRhBStxu1YqQtpid3TwnoAet6jhvYrQlRJEr9hu569J74dlzxGZYm20OWzHAADcDJylURvXzG1HNmqImf1zlzVkX4wWRyNa0QGgYW9Q7ms5-SsjdDzQagN0OXPdbR~l6YsXCQ5EovxacqLZA4S75xVxWGQN0CFvlN8pozxRUbFWxBUQtQpsDxgH7ciw0ZqJV1J1oS5sgOuDUrPqjsb6e86TiXvo1EeG8Inep1GTvy~BnkD~O770hGnmtvqfPXteQn0b7UBSdWJfiJSl42BDwh7Y9yBkpZFVkEcZgziCoIMHURduA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)