To the editor:
AML1-ETO is the chimeric protein generated as a result of the t(8;21) in acute myeloid leukemia. Understanding which of the proteins that interact with AML1-ETO are essential for its activity is vital for developing targeted small-molecule AML1-ETO inhibitors. Two papers recently assessed the importance of CBFβ, the non–DNA-binding subunit of the core binding factors, for AML1-ETO's activity.1,2 In both studies, the authors introduced amino acid substitutions at the CBFβ-binding interface of the Runt domain of AML1-ETO, and assessed the effects of these mutations on AML1-ETO's ability to confer serial replating ability to primary mouse bone marrow cells. Our group (Roudaia et al)1 and that of Kwok et al2 reached opposite conclusions. Roudaia et al1 reported that CBFβ is essential for AML1-ETO's activity, whereas Kwok et al2 claimed it is dispensable.
Roudaia et al1 compared the effect of a single T161A mutation, which decreases the affinity of CBFβ for the Runt domain of AML1-ETO by 63-fold, with a combined Y113A/T161A mutation that decreases binding by 430 fold, on AML1-ETO's activity (Figure 1A). The side chains of both T161 and Y113 make direct contacts to CBFβ.3-5 The extent to which binding was impaired was determined by isothermal titration calorimetry using purified proteins.1 We showed that the T161A mutation alone only slightly impaired AML1-ETO's activity in serial replating assays, whereas the combined Y113A/T161A mutations severely compromised AML1-ETO's serial replating activity, as well as its ability to cooperate with TEL-PDGFβR to cause acute myeloid leukemia in mice.1
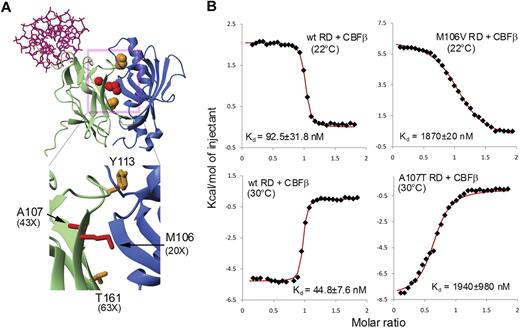
Location of mutations and results of binding measurements. (A) Structure of the CBFβ–Runt domain–DNA complex (PDB code 1h9d) with Runt domain amino acids mutated in the Roudaia et al (orange) and Kwok et al (red) studies indicated.1,2 CBFβ is indicated in blue; Runt domain in green; and DNA in pink. (B) Results of isothermal titration calorimetry (ITC) measurements of the binding of CBFβ to wild-type Runt domain, M106V Runt domain, and A107T Runt domain. Measurements for the M106V mutant were carried out at 22°C, as done in the Roudaia et al study. Measurements for the A107T mutant were carried out at 30°C as a result of very low signals at 22°C for this mutant, precluding an accurate measurement. The wild-type Runt domain has also been measured at this temperature to provide an accurate comparison between the 2. The enthalpy of the reaction changes from exothermic to endothermic upon going from 22°C to 30°C, resulting in the mirror image appearance of the data for the wild-type Runt domain at the 2 different temperatures.
Location of mutations and results of binding measurements. (A) Structure of the CBFβ–Runt domain–DNA complex (PDB code 1h9d) with Runt domain amino acids mutated in the Roudaia et al (orange) and Kwok et al (red) studies indicated.1,2 CBFβ is indicated in blue; Runt domain in green; and DNA in pink. (B) Results of isothermal titration calorimetry (ITC) measurements of the binding of CBFβ to wild-type Runt domain, M106V Runt domain, and A107T Runt domain. Measurements for the M106V mutant were carried out at 22°C, as done in the Roudaia et al study. Measurements for the A107T mutant were carried out at 30°C as a result of very low signals at 22°C for this mutant, precluding an accurate measurement. The wild-type Runt domain has also been measured at this temperature to provide an accurate comparison between the 2. The enthalpy of the reaction changes from exothermic to endothermic upon going from 22°C to 30°C, resulting in the mirror image appearance of the data for the wild-type Runt domain at the 2 different temperatures.
Kwok et al2 introduced M106V and A107T mutations into AML1-ETO. The M106 side chain directly contacts CBFβ, although we previously showed that a very similar M106A mutation seriously perturbed the structure of the Runt domain.3 A107 is buried in the hydrophobic core of the Runt domain,3-5 making the rationale for this mutation unclear. Kwok et al reported that neither mutation impaired AML1-ETO's serial replating activity, and concluded that the interaction of AML1-ETO with CBFβ was not essential.
We purified AML1-ETO Runt domains containing the M106V and A107T mutations used by Kwok et al2 and measured their affinity for CBFβ by isothermal titration calorimetry (Figure 1B). We found that the M106V and A107T mutations impaired binding to CBFβ by 20- and 43-fold, respectively. Neither mutation impaired CBFβ binding as much as the T161A mutation, which by itself had only very modest effects on AML1-ETO's activity. Therefore, the contradictory conclusions reached by the 2 groups result from differences in the extent to which CBFβ binding was disrupted by the amino acid substitutions used in each study. Twenty- to 60-fold decreases in CBFβ-binding affinity in both studies were insufficient to cripple AML1-ETO, whereas a 400-fold decrease severely impaired its activity. This is likely a result of the relatively high concentration of CBFβ in cells,6 necessitating a more robust disruption of the interaction to shift the ratio of free to bound protein. We assert that CBFβ is, indeed, essential for AML1-ETO's activity.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr John H. Bushweller, University of Virginia, PO Box 800736, Charlottesville, VA 22908; e-mail: jhb4v@virginia.edu

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal