Abstract
Impaired regulation of hepcidin expression in response to iron loading appears to be the pathogenic mechanism for hereditary hemochromatosis. Iron normally induces expression of the BMP6 ligand, which, in turn, activates the BMP/Smad signaling cascade directing hepcidin expression. The molecular function of the HFE protein, involved in the most common form of hereditary hemochromatosis, is still unknown. We have used Hfe-deficient mice of different genetic backgrounds to test whether HFE has a role in the signaling cascade induced by BMP6. At 7 weeks of age, these mice have accumulated iron in their liver and have increased Bmp6 mRNA and protein. However, in contrast to mice with secondary iron overload, levels of phosphorylated Smads 1/5/8 and of Id1 mRNA, both indicators of BMP signaling, are not significantly higher in the liver of these mice than in wild-type livers. As a consequence, hepcidin mRNA levels in Hfe-deficient mice are similar or marginally reduced, compared with 7-week-old wild-type mice. The inappropriately low levels of Id1 and hepcidin mRNA observed at weaning further suggest that Hfe deficiency triggers iron overload by impairing hepatic Bmp/Smad signaling. HFE therefore appears to facilitate signal transduction induced by the BMP6 ligand.
Introduction
Hereditary hemochromatosis (HH) is a genetic disorder characterized by increased absorption of iron from the gastrointestinal tract. Progressive accumulation of catalytically active iron in parenchymal tissues may lead to severe organ damage such as hepatic fibrosis, cirrhosis, and hepatocellular carcinoma. HH is efficiently treated by phlebotomy. In Northern Europe, most patients with HH are homozygous for a single mutation (C282Y) in the HFE gene encoding a nonclassical major histocompatibility complex class I molecule.1 This mutation disrupts a disulfide bond required for proper folding of the HFE molecule. Shortly after its discovery in 1996, the HFE protein was shown to physically interact with transferrin receptor 1 (TFR1) and impair the uptake of transferrin-bound iron in cells.2-4 However, these observations did not shed much light on how HFE controls systemic iron homeostasis.
In untreated patients with HH resulting from mutations in the HFE gene, ferritin is high but hepcidin is inappropriately low relative to body iron burden.5 Hepcidin, a small peptide secreted by the liver, has a key role in coordinating the use and storage of iron with iron acquisition.6 It acts by binding to ferroportin, an iron exporter present on the surface of enterocytes and macrophages, and induces its internalization and lysosomal degradation.7 The loss of ferroportin from the cell surface prevents iron efflux from intestinal enterocytes and recycling of iron from senescent erythrocytes by macrophages. Hepcidin expression is normally enhanced by dietary or parenteral iron loading,8 thus providing a feedback mechanism to limit intestinal iron absorption. Because this expected up-regulation of hepcidin in response to iron loading is impaired in patients with HH, HFE is thought to be involved in the regulation of hepcidin expression.
The role of the BMP-SMAD signaling pathway in directing hepcidin expression is now well established.9,10 BMP6, whose mRNA expression is regulated by iron in vivo,11 is critical to activate this signaling cascade.12 Like other members of the transforming growth factor beta superfamily of ligands, BMP6 binds 2 type I and 2 type II BMP receptors (BMPR-I and -II, respectively). This induces the phosphorylation of BMPR-I by BMPR-II and the activated complex, in turn, phosphorylates a subset of Smad proteins (Smads 1, 5, and 8). The receptor-activated Smads then form heteromeric complexes with the common mediator Smad4 and these translocate to the nucleus where they regulate the transcription of specific targets such as hepcidin.13 Hemojuvelin, a molecule involved in severe and early onset juvenile hemochromatosis, was shown to act as a BMP coreceptor9 and is as critical as BMP6 to hepcidin expression.14,15
In contrast to hemojuvelin, the exact molecular function of HFE remains uncertain. Definite clues as to the site of HFE regulatory function in the context of systemic iron homeostasis were recently provided by experiments with genetically engineered mice bearing a targeted, tissue-specific disruption of Hfe. Whereas ablation of Hfe in the intestine16 or in macrophages17 did not affect body iron metabolism, mice lacking Hfe expression in hepatocytes exhibited hyperabsorption of dietary iron, increased serum iron, transferrin saturation, and iron deposition in the liver.17 Hepatocyte HFE is therefore necessary to prevent iron overload. HFE forms protein complexes with transferrin receptor 1 (TFR1) and its liver-specific homolog, transferrin receptor 2 (TFR2).18 Results obtained with different TfR1 mutant mice suggest that TFR1 may normally sequester HFE and keep it inactive. When serum iron concentration is high, diferric transferrin and HFE compete for TFR1 binding. HFE then dissociates from TFR1,19 and Schmidt et al speculate that HFE released from TFR1 interacts with TFR2 to signal for production of hepcidin. Recently, the necessity of an interaction between HFE and TFR2 for signal transduction was clearly demonstrated.20 This interaction depends upon the alpha3 domain of HFE. However, whether HFE when free of TFR1 has a role in the signaling cascade induced by the BMP6 ligand is still unknown.
Our group has derived Hfe-deficient mice on different genetic backgrounds.21 These mice provide a unique opportunity to explore how Hfe deficiency affects the Bmp6/Smad signaling pathway in vivo. We show that these mice have elevated liver iron content and increased Bmp6 mRNA and protein, but inappropriately low levels of phosphorylated Smads 1, 5, and 8 and of hepcidin mRNA. Hfe deficiency thus triggers iron overload by impairing hepatic Smad1/5/8 phosphorylation, suggesting that the efficacy of the BMP6 signaling pathway is reduced when HFE is missing.
Methods
Mice
Hfe-deficient mice on the C57BL/6 (B6) and DBA/2 (D2) backgrounds were derived as previously described.22 They were maintained at the IFR30 animal facility, as well as wild-type controls of the same genetic backgrounds. All experiments were performed on males. Unless otherwise specified, mice received a standard rodent diet (200 mg iron/kg body weight; SAFE) and were killed at 7 weeks. Experimental iron overload was obtained by feeding 4-week-old B6 and D2 wild-type mice the same diet supplemented with 8.3 g/kg carbonyl iron (Sigma-Aldrich) for 3 weeks. Three-week-old Hfe-deficient mice and litter-matched wild-type controls were obtained from B6D2F1 heterozygous (Hfe+/−) parents. Experimental protocols were approved by the Midi-Pyrénées Animal Ethics Committee.
Tissue iron measurement
Quantitative measurement of hepatic nonheme iron was performed as described previously.21 Results are reported as micrograms of iron per gram dry weight of tissue.
RNA preparation and real-time quantitative PCR
Liver samples were dissected for RNA isolation, rapidly frozen, and stored in liquid nitrogen. Total RNA was extracted and purified using the RNeasy Lipid Tissue kit (QIAGEN). All primers11,12 were designed using the Primer Express 2.0 software (Applied Biosystems). Real-time quantitative polymerase chain reactions (Q-PCRs) were prepared with Moloney murine leukemia virus reverse transcriptase (Promega) and LightCycler 480 DNA SYBR Green I Master reaction mix (Roche Diagnostics) and run in duplicate on a LightCycler 480 Instrument (Roche Diagnostics).
Immunohistochemistry
Four-micrometer sections of paraffin-embedded tissues were mounted on glass slides. Antigen retrieval was performed by incubating tissue sections with trypsin (1 mg/mL) for 8 minutes at 37°C. Endogenous peroxidase activity was quenched by incubating specimens with Dako REAL Peroxidase Blocking Solution (Dako). Tissue sections were then blocked with normal horse blocking serum (Vector Laboratories) and incubated for 1 hour at room temperature with the primary anti-BMP6 (N-19) antibody (1/100; Santa Cruz Biotechnology) diluted in PBS–1% BSA and 1% FCS. Immunohistochemical staining was performed using the ImmPRESS Reagent (ImmPRESS Anti–Goat Ig peroxidase Kit; Vector Laboratories) according to the instructions of the manufacturer. Sections were counterstained with hematoxylin. Tissue sections from Bmp6-deficient mice were used to test antibody specificity.
Western blot analysis
Livers were homogenized in a FastPrep-24 Instrument (MP Biomedicals Europe) for 20 seconds at 4 m/s. The lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM EDTA, pH 8, 1% NP-40) included inhibitors of proteases (1 mM PMSF, 10 μg/mL leupeptin, 10 mg/mL pepstatin A, and 1 mg/mL antipain) and of phosphatases (10 μL/mL phosphatase inhibitor cocktail 2; Sigma-Aldrich). Proteins were quantified using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories) based on the method of Bradford. Protein extracts (30 μg for phospho-Smad and 60 μg for Smad5) were diluted in Laemmli buffer (Sigma-Aldrich), incubated for 5 minutes at 95°C, and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Proteins were then transferred to Hybond-C Extra nitrocellulose membranes (Amersham Biosciences). Membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences), incubated with a rabbit polyclonal antibody to phosphorylated Smad1/5/8 (1/500, lot 8; Cell Signaling Technology) or a goat polyclonal antibody to Smad5 (1/200; Santa Cruz Biotechnology) and a mouse monoclonal antibody to β-actin (1/20 000; Sigma-Aldrich) at 4°C overnight, and washed with PBS–0.1% Tween-20 buffer. After incubation with infrared IRDye 800 anti–rabbit or anti–goat and IRDye 680 anti–mouse secondary antibodies (1/15 000; LI-COR Biosciences), membranes were scanned on the Odyssey Infrared Imaging System (LI-COR Biosciences). Band sizing was performed using the Odyssey 3.0 software (LI-COR Biosciences) and quantification of phosphorylated Smads and of Smad5 was calculated by normalizing the specific probe band to β-actin.
Statistical analyses
Log-transformed values of liver iron contents were compared by Student t tests. The relative expression ratios (and standard errors) of liver transcripts between Hfe−/− mice and wild-type controls were calculated using the relative expression software tool (REST, http://rest.gene-quantification.info).23 The mathematic model is based on the mean crossing point deviation between sample and control groups of target genes, normalized by the mean crossing point deviation of the reference gene Hprt.24 An efficiency correction was performed and randomization tests, which have the advantage of making no distributional assumptions about the data, were used to determine statistical significance.
Results
Hfe deficiency promotes liver expression of Bmp6
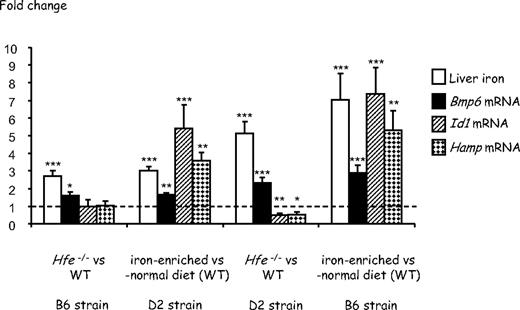
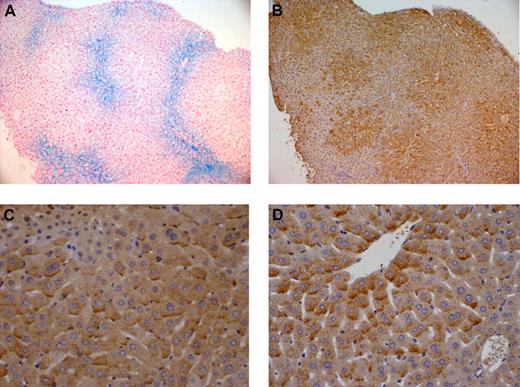
As previously observed,11,21 whereas 7-week-old Hfe-deficient mice of the DBA/2 (D2) background have a higher liver iron burden than Hfe-deficient mice of the C57BL/6 (B6) strain, wild-type mice of the B6 background fed an iron-enriched diet for 3 weeks are reproducibly more heavily iron-loaded than wild-type D2 mice fed the same iron-rich diet (Figure 1). This may reflect differences in the genetic susceptibility to iron loading in the presence or absence of functional Hfe. Real-time quantitative PCR shows that expression of Bmp6 is significantly up-regulated not only in the liver of wild-type mice with secondary iron overload but also in the liver of Hfe-deficient mice compared with that of wild-type controls (Figure 1). Noticeably, mice with the highest hepatic iron burden (B6 mice with secondary iron overload and Hfe-deficient D2 mice) have the highest induction of Bmp6 relative to control animals. We thus examined liver expression and cellular localization of Bmp6 by immunohistochemistry, using an antibody raised against a peptide mapping within the internal region of BMP6. Enhanced Bmp6 staining was observed in Hfe-deficient mice and in wild-type mice with secondary iron overload. Interestingly, the distribution of Bmp6 in the liver is zonal and, unlike iron deposits that are periportal (Figure 2A), Bmp6 staining is centrilobular (Figure 2B). This centrilobular layout of Bmp6 is observed in both Hfe-deficient mice and wild-type mice with secondary iron overload. BMP6 expression was previously shown to be confined to nonparenchymal liver cells, namely hepatic stellate cells and Kupffer cells.25 However, in iron-loaded livers, Bmp6 is also found in the hepatocytes, noticeably at the basolateral membrane domain as previously reported for hemojuvelin and TFR226 (Figure 2C-D). This staining was not observed in Bmp6-deficient mice or with control goat IgG.
Effect of Hfe deficiency or secondary iron overload on hepatic iron concentrations and Bmp6, Id1, and Hamp gene expression in 7-week-old B6 and D2 mice. Fold change in nonheme tissue iron content and expression ratio (and SE) of Bmp6, Id1, and Hamp transcripts normalized to the reference gene mRNA (Hprt) in Hfe-deficient mice relative to wild-type controls and in wild-type mice fed an iron-rich diet for 3 weeks relative to wild-type mice fed a standard rodent diet (5-10 mice per group). Statistical significance was determined using randomization tests. *P < .05; **P < .01; ***P < .001. Data are provided for 2 genetic backgrounds, C57BL/6 (B6) and DBA/2 (D2). At 7 weeks of age, wild-type mice of the 2 backgrounds have similar levels of Bmp6, Id1, and Hamp transcripts (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). However, Hfe-deficient mice of the D2 background have significantly more Bmp6 mRNA than Hfe-deficient mice of the B6 background (P = .001). Wild-type B6 mice fed the iron-rich diet for 3 weeks also have significantly more Bmp6 mRNA than wild-type D2 mice fed the same diet (P = .001). Absolute values (instead of fold changes) corresponding to the same data are provided in supplemental Figure 2.
Effect of Hfe deficiency or secondary iron overload on hepatic iron concentrations and Bmp6, Id1, and Hamp gene expression in 7-week-old B6 and D2 mice. Fold change in nonheme tissue iron content and expression ratio (and SE) of Bmp6, Id1, and Hamp transcripts normalized to the reference gene mRNA (Hprt) in Hfe-deficient mice relative to wild-type controls and in wild-type mice fed an iron-rich diet for 3 weeks relative to wild-type mice fed a standard rodent diet (5-10 mice per group). Statistical significance was determined using randomization tests. *P < .05; **P < .01; ***P < .001. Data are provided for 2 genetic backgrounds, C57BL/6 (B6) and DBA/2 (D2). At 7 weeks of age, wild-type mice of the 2 backgrounds have similar levels of Bmp6, Id1, and Hamp transcripts (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). However, Hfe-deficient mice of the D2 background have significantly more Bmp6 mRNA than Hfe-deficient mice of the B6 background (P = .001). Wild-type B6 mice fed the iron-rich diet for 3 weeks also have significantly more Bmp6 mRNA than wild-type D2 mice fed the same diet (P = .001). Absolute values (instead of fold changes) corresponding to the same data are provided in supplemental Figure 2.
Cellular localization of BMP6 in hepatic iron overload. BMP6 expression was detected by immunohistochemistry in (B-C) wild-type B6 mice with secondary iron overload and (D) Hfe-deficient D2 mice. These mice have similar degrees of iron loading. As seen in serial liver sections, whereas iron deposits visualized by Perls staining (A) are predominantly periportal, BMP6 staining is mostly centrilobular (B). Mutant animals and mice with secondary iron overload have intense staining at the basolateral membrane domain of hepatocytes (C-D). Original magnification, ×100 (A-B) or ×400 (C-D). Images were captured using a Leica DMR microscope equipped with an HC PL Fluotar 10×/1 (A-B) or 40×/1 (C-D) numeric aperture objective lens and a Leica DFC 300 Fx camera. They were processed using Leica IM50 image acquisition software.
Cellular localization of BMP6 in hepatic iron overload. BMP6 expression was detected by immunohistochemistry in (B-C) wild-type B6 mice with secondary iron overload and (D) Hfe-deficient D2 mice. These mice have similar degrees of iron loading. As seen in serial liver sections, whereas iron deposits visualized by Perls staining (A) are predominantly periportal, BMP6 staining is mostly centrilobular (B). Mutant animals and mice with secondary iron overload have intense staining at the basolateral membrane domain of hepatocytes (C-D). Original magnification, ×100 (A-B) or ×400 (C-D). Images were captured using a Leica DMR microscope equipped with an HC PL Fluotar 10×/1 (A-B) or 40×/1 (C-D) numeric aperture objective lens and a Leica DFC 300 Fx camera. They were processed using Leica IM50 image acquisition software.
Smad1/5/8 phosphorylation is not increased in Hfe-deficient mice
Because Bmp6 transmits signal through phosphorylation of Smads,27 we tested whether phosphorylation of Smad1/5/8 was increased in liver extracts of Hfe−/− mice. Total protein lysates from 3 groups of animals were obtained for the 2 strains B6 and D2: (1) wild-type controls fed a standard rodent diet; (2) Hfe−/− mice fed the same standard rodent diet; and (3) wild-type controls fed an iron-enriched diet to induce secondary iron overload. The amount of the phosphorylated forms of Smad1/5/8 in each group was determined by Western blot analysis. As shown in Figure 3, although the iron-enriched diet induced Smad1/5/8 phosphorylation in both strains, no significant increase in Smad1/5/8 phosphorylation was observed in 7-week-old B6 or D2 Hfe−/− mice compared with wild-type controls. Therefore, Hfe−/− mice do not appropriately respond to the increase in Bmp6. We also measured the levels of Id1 mRNA in the liver of the different mice. Id1 is a direct target gene for BMPs, and phosphorylated Smads 1 and 5 have been shown to regulate its transcription through direct binding to specific elements on its promoter.11 Its up-regulation therefore is an indicator of activation of the Bmp signaling cascade. As seen in Figure 1, whereas Id1 mRNA expression is very significantly up-regulated in the livers of mice with secondary iron overload, no such up-regulation is seen in the livers of Hfe-deficient mice, despite the increase in Bmp6 liver expression.
Smad1/5/8 phosphorylation is increased by secondary iron overload but unchanged by Hfe deficiency. (A) Liver lysates from wild-type controls fed a standard rodent diet (WT), Hfe-deficient mice (Hfe−/−), and mice with secondary iron overload (SIO) were analyzed by Western blot with antibodies to phosphorylated Smad1/5/8 and to β-actin as loading control. Membranes were scanned on the Odyssey Infrared Imaging System. One representative experiment is shown for each strain. (B) Band sizing was performed using the Odyssey 3.0 software (LI-COR Biosciences) and quantification of phosphorylated Smads was calculated by normalizing the specific probe band to β-actin. Mean ratio (p-Smad/β-actin) of 3 Hfe-deficient mouse samples (or 3 mice with secondary iron overload) ± SE are represented on this figure, relative to the mean ratio of 3 wild-type mice fed a standard rodent diet. Student t tests were used to compare mean ratios between Hfe-deficient mice and wild-type controls (P = .55 for B6 mice; P = .58 for D2 mice) or between mice with secondary iron overload and wild-type mice (**P = .01 for B6 mice; *P = .02 for D2 mice). (C) Liver lysates from the same mice were analyzed by Western blot with antibodies to Smad5 and to β-actin as loading control. (D) Quantification using the Odyssey 3.0 software was performed as in panel B. Student t tests were used to compare mean Smad5/β-actin ratios. The levels of Smad5 were not significantly different between Hfe-deficient mice and wild-type controls (P = .59 for B6 mice; P = .59 for D2 mice), or between mice with secondary iron overload and wild-type controls (P = .15 for B6 mice; P = .31 for D2 mice).
Smad1/5/8 phosphorylation is increased by secondary iron overload but unchanged by Hfe deficiency. (A) Liver lysates from wild-type controls fed a standard rodent diet (WT), Hfe-deficient mice (Hfe−/−), and mice with secondary iron overload (SIO) were analyzed by Western blot with antibodies to phosphorylated Smad1/5/8 and to β-actin as loading control. Membranes were scanned on the Odyssey Infrared Imaging System. One representative experiment is shown for each strain. (B) Band sizing was performed using the Odyssey 3.0 software (LI-COR Biosciences) and quantification of phosphorylated Smads was calculated by normalizing the specific probe band to β-actin. Mean ratio (p-Smad/β-actin) of 3 Hfe-deficient mouse samples (or 3 mice with secondary iron overload) ± SE are represented on this figure, relative to the mean ratio of 3 wild-type mice fed a standard rodent diet. Student t tests were used to compare mean ratios between Hfe-deficient mice and wild-type controls (P = .55 for B6 mice; P = .58 for D2 mice) or between mice with secondary iron overload and wild-type mice (**P = .01 for B6 mice; *P = .02 for D2 mice). (C) Liver lysates from the same mice were analyzed by Western blot with antibodies to Smad5 and to β-actin as loading control. (D) Quantification using the Odyssey 3.0 software was performed as in panel B. Student t tests were used to compare mean Smad5/β-actin ratios. The levels of Smad5 were not significantly different between Hfe-deficient mice and wild-type controls (P = .59 for B6 mice; P = .59 for D2 mice), or between mice with secondary iron overload and wild-type controls (P = .15 for B6 mice; P = .31 for D2 mice).
Up-regulation of Bmp6 is preceded by a marked down-regulation of hepcidin expression
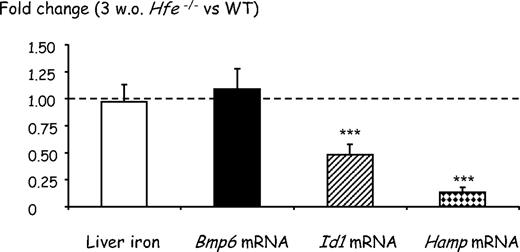
Because phosphorylation of Smad proteins 1/5/8 was not significantly different between 7-week-old Hfe-deficient mice and wild-type controls, we expected that hepcidin transcription would also be similar in the 2 groups of animals. Indeed, as shown in Figure 1, we found that Hamp mRNA levels in Hfe−/− mice of the B6 genetic background were equivalent to those in wild-type mice, and only slightly reduced in Hfe−/− mice of the D2 background. The excessive iron burden observed in 7-week-old Hfe-deficient mice is difficult to reconcile with quasi-normal levels of hepcidin. This led us to hypothesize that iron overload in 7-week-old Hfe-deficient mice results from reduced hepcidin production earlier in life. To test this hypothesis, we quantified liver iron as well as Bmp6 and Hamp mRNA levels in 3-week-old Hfe-deficient mice and wild-type controls. Weaning from a low-iron diet (milk) to the relatively high-iron diet provided by chow is associated with a rapid increase in transferrin saturation and in hepcidin expression within 1 week (data not shown). We suspected that this increase would be influenced by Hfe and therefore used Hfe-deficient mice and litter-matched controls to ensure that they were carefully matched for age. As seen in Figure 4, at 3 weeks of age, Hfe-deficient mice have liver iron content and Bmp6 gene expression similar to wild-type animals. However, their Hamp gene expression is approximately 8-fold lower than in control mice. This indicates that down-regulation of hepcidin expression is the first biologic manifestation of Hfe deficiency and precedes liver iron accumulation and increase in Bmp6 expression. Interestingly, although we were unable to detect a statistically significant decrease in Smad1/5/8 phosphorylation by Western blot analysis (supplemental Figure 3), the levels of Id1 mRNA, an indicator of activation of the BMP signaling cascade, are reduced by approximately 50% in these young Hfe-deficient mice compared with wild-type controls, further suggesting that Bmp6 signaling is impaired by lack of functional Hfe. In wild-type mice fed an iron-enriched diet (data shown here and in Kautz et al11 ) or an iron-deficient diet,11 modulation of Smad1/5/8 phosphorylation is always less pronounced than modulation of Id1 mRNA, which is itself often less pronounced than modulation of Hamp mRNA. Therefore, we cannot exclude an amplification of the response to Bmp6 between Smad1/5/8 phosphorylation and the transcription of the specific targets. Given that there is only a 2-fold decrease in Id1 mRNA expression in 3-week-old Hfe-deficient mice compared with wild-type mice (as seen in Figure 4), it is possible that modulation of Smad1/5/8 phosphorylation in these mice is too low to be visualized by Western blot analyses.
Effect of Hfe deficiency on hepatic iron concentrations and Bmp6, Id1, and Hamp gene expression in 3-week-old mice. Fold change in nonheme tissue iron content and expression ratio (and SE) of Bmp6, Id1, and Hamp transcripts normalized to the reference gene mRNA (Hprt) in 3-week-old Hfe-deficient mice relative to wild-type controls (8 mice per group). Statistical significance was determined using randomization tests. ***P < .001. At 3 weeks of age, wild-type mice have levels of Bmp6 and Id1 mRNAs similar to 7-week-old mice. Although they have slightly less Hamp gene expression than 7-week-old mice, the difference is not statistically significant (supplemental Figure 4).
Effect of Hfe deficiency on hepatic iron concentrations and Bmp6, Id1, and Hamp gene expression in 3-week-old mice. Fold change in nonheme tissue iron content and expression ratio (and SE) of Bmp6, Id1, and Hamp transcripts normalized to the reference gene mRNA (Hprt) in 3-week-old Hfe-deficient mice relative to wild-type controls (8 mice per group). Statistical significance was determined using randomization tests. ***P < .001. At 3 weeks of age, wild-type mice have levels of Bmp6 and Id1 mRNAs similar to 7-week-old mice. Although they have slightly less Hamp gene expression than 7-week-old mice, the difference is not statistically significant (supplemental Figure 4).
Discussion
Although the site of HFE regulatory function is the hepatocyte,17 the exact mechanisms by which HFE regulates iron homeostasis remain elusive. Our data suggest that lack of functional Hfe early in life severely impairs the Bmp/Smad signaling cascade, resulting in the down-regulation of hepcidin observed in 3-week-old mice in this and previously reported studies.28,29 As a consequence, there is no feedback mechanism to limit iron efflux from intestinal enterocytes. Between 3 and 7 weeks of age, Hfe-deficient mice progressively accumulate iron and, interestingly, retain their ability to increase Bmp6 in response to body iron excess, as do mice with secondary iron overload or mice with genetic iron overload due to inactivation of the Smad4 or the Hamp gene.11 However, due to the lack of functional Hfe, the response to increased Bmp6 expression is blunted compared with that of mice with secondary iron overload and, as shown in the present study, reaches only levels observed in wild-type controls fed a standard rodent diet. Given their iron burden, Smad1/5/8 phosphorylation and Id1 and hepcidin expression are all inappropriately low in 7-week-old Hfe-deficient mice. The age-related changes in Bmp6 and Hamp expression observed here in Hfe-deficient mice explain why, several weeks after birth, intestinal iron absorption decreases and hepatic iron concentrations reach a plateau.30 Of note, although Hfe-deficient D2 mice have higher Bmp6 gene expression than Hfe-deficient B6 mice (P = .001), they have slightly less hepcidin mRNA. Genetically determined differences22 in the maturation, secretion, or inhibition of Bmp6 between strains may affect the efficacy of signal transduction and explain these variations.
In hemochromatosis patients, iron absorption also declines as the iron load increases.31-33 Furthermore, hepcidin concentrations in the sera of iron-loaded patients with HH resulting from mutations in the HFE gene are similar to controls,34-36 suggesting a disease time course similar to that observed in mice although more spread out over time. Interestingly, hepcidin concentrations are lower than controls in patients who have been iron depleted by phlebotomy treatment.34-36 As liver biopsies are no longer required for the diagnosis of HH, the relationship between liver iron content and Bmp6 expression is difficult to assess in humans. However, it might be expected that BMP6 levels are high in untreated patients and that therapeutic venesections, by removing excess iron stores, restore these levels to those seen in controls, thus reducing the efficacy of signal transduction. The consequent decrease in hepcidin expression could then explain the reaccumulation of iron in the absence of maintenance phlebotomies. As already suggested in the literature,35 clinical guidelines for the treatment of C282Y homozygotes should probably be revised and the currently recommended serum ferritin thresholds for therapeutic venesections corrected upward.
This is the first demonstration that lack of HFE impairs propagation of the signaling cascade induced by the BMP6 ligand and suggests that HFE and the BMP type I and II serine/threonine kinase receptors are associated at the hepatocyte cell membrane and that this association is required to ensure proper signal transduction. Hemojuvelin, TFR2,26 and, as demonstrated here by immunohistochemistry, BMP6 all localize to the hepatocyte basolateral membrane domain, suggesting a functional interaction of these molecules in the context of iron metabolism regulation. HFE, TFR2, and other proteins such as BMP6, its receptors, and hemojuvelin would then form in this functional membrane domain an iron signaling complex that induces hepcidin transcription via Smad proteins. Interestingly, there are previous reports of physical associations of major histocompatibility complex class I molecules to tetrameric membrane receptors such as the insulin receptor in mouse liver membranes.37,38
In summary, our data demonstrate that the role of HFE is not solely limited to iron sensing by a mechanism involving a competition between HFE and diferric transferrin for TFR1 binding.19 Indeed, we showed that HFE is necessary for correct signal transduction from BMP6, suggesting that, when dissociated from TFR1, HFE participates in the BMPRI/II molecular complex. In the presence of HFE, basal levels of BMP6 are probably sufficient for physiologic modulation of hepcidin outside of massive iron overload. Indeed, wild-type D2 mice fed an iron-enriched diet for a short period have increased transferrin saturation and elevated hepcidin expression, but no increase in hepatic iron or in Bmp6 mRNA expression (data not shown). Furthermore, at weaning from milk to the relatively high-iron diet provided by chow, wild-type mice have a rapid increase in transferrin saturation and in hepcidin expression, but again no increase in hepatic iron or in Bmp6 mRNA expression (data not shown). Therefore, the ability to increase Bmp6 expression seems restricted to animals with liver iron accumulation, whether due to Hfe deficiency or to an iron-enriched diet for several weeks. A greater amount of the Bmp6 ligand then allows a more efficient propagation of the signaling cascade that clearly improves the status of Hfe-deficient animals and hopefully that of hemochromatosis patients. This may explain why a plateau in iron loading is reached over time and why hepcidin decreases after iron depletion in human patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Julie Seumois and Maryline Calise (Service de Zootechnie, IFR30) for their help in the mouse breeding, and Sophie Allart (Cellular Imaging platform, IFR30) and Florence Capilla (Experimental Histopathology platform, IFR30) for skilled advice.
This work was supported in part by grants from the European Commission (LSHM-CT-2006-037296: EUROIRON1), the Association pour la Recherche sur le Cancer (ARC), the Ligue contre le Cancer, and the Fondation pour la Recherche Médicale (L.K.).
Authorship
Contribution: L.K., D.M., C.B.-F., and V.D. performed research, analyzed data, and reviewed the paper; T.A.S. provided expert advice on immunohistochemistry; and H.C. and M.-P.R. conceived the project, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marie-Paule Roth, Inserm U563, CHU Purpan, BP 3028, F-31024 Toulouse Cedex 3, France; e-mail: marie-paule.roth@inserm.fr.
References
Author notes
*H.C. and M.-P.R. contributed equally to this work.