Abstract
Nucleophosmin (NPM1)–mutated acute myeloid leukemia (AML), which is recognized as a provisional entity in the World Health Organization 2008 classification of myeloid neoplasms, accounts for 30% of AML. We analyzed 1227 diagnostic and follow-up samples in 252 NPM1-mutated AML patients with 17 different NPM1 mutation–specific real-time quantitative polymerase chain reaction (RQ-PCR) assays. Paired diagnostic/relapse samples of 84 patients revealed stable NPM1 mutations in all cases, suggesting that they are pathogenetically early events and thus applicable for minimal residual disease detection. A total of 47 relapses were predictable because of an NPM1 mutation level (%NPM1/ABL1) increase of at least 1 log or in 15 cases because of NPM1 mutation levels not decreasing less than 3 log ranges. A high prognostic value of NPM1 levels was shown for 4 different intervals after therapy was initiated. Furthermore, thresholds of 0.1 and 0.01%NPM1/ABL1 during/after treatment discriminated between prognostic subgroups. Univariate analyses, including age, white blood cell count, blast count, CD34 positivity, FLT3 mutations status, FAB type, karyotype, NPM1 mutation type, and pretreatment NPM1 mutational level, showed that, besides NPM1 mutation level, only age and FLT3-LM mutation status were prognostically significant for EFS. Multivariate analysis, including age, FLT3-LM status, and NPM1 mutation level at different time points, demonstrated that NPM1 level was the most relevant prognostic factor during first-line treatment. Similar results were obtained in patients undergoing second-line chemotherapy or allogeneic stem cell transplantation.
Introduction
Detection of minimal residual disease (MRD) is becoming increasingly important for risk stratification and early detection of relapse in patients with acute myeloid leukemia (AML), therefore providing a window for therapeutic intervention.1-4 Common targets for polymerase chain reaction (PCR)–based MRD detection are mainly fusion transcripts, eg, RUNX1-RUNX1T1 (previously AML1-ETO), CBFB-MYH11, PML-RARA, and MLL–gene fusions.1-8 Because fusion genes are present in only approximately 25% of AML, further markers like MLL-PTD, FLT3-LM,7,9 and ectopic expression of WT1 and EVI1 have recently been established as additional sensitive and valuable targets.10-12
Our potential to further expand the number of patients with AML that may benefit from analysis of MRD is closely dependent upon the availability of new molecular markers that allow us to identify new leukemia entities in the context of the poorly characterized group of AML with normal karyotype (NK; 40%-50% of cases in multicenter studies). Nucleophosmin (NPM1) mutations causing aberrant cytoplasmic dislocation of nucleophosmin were first described by Falini et al13 in approximately one-third of adult patients with AML. These cases usually carry an NK, show distinctive biological and clinical features, and are characterized by a relatively good prognosis (in the absence of a concomitant FLT3-LM).13-19 Because of these unique characteristics, AML with mutated NPM1 has been recently included in the 2008 World Health Organization (WHO) classification of myeloid neoplasms as a provisional entity.20 Approximately 85% of AML patients with mutated NPM1 show an NK, accounting for approximately 60% of adult AML with normal cytogenetics.18 Moreover, the authors of some studies21,22 indicate that NPM1 mutations and cytoplasmic dislocation of nucleophosmin are very stable at relapse, making the NPM1 mutation a potentially useful marker for MRD assessment in approximately one-third of patients with AML.
The applicability of this marker for PCR-based MRD detection recently has been demonstrated.23 This study showed that DNA- as well as RNA-based assays can be performed and that the MRD values obtained correlated with the clinical course in all 13 analyzed patients. The comparison of the latter DNA/cDNA data and our own unpublished results (S.S., unpublished data, 2005) showed that cDNA-based assays are more sensitive than DNA-based assays. Thus, we based our study on data generated by cDNA assays. We developed an independent and highly sensitive real-time quantitative polymerase chain reaction (RQ-PCR) assay with a common reverse primer and common hybridization probes and mutation-specific forward primers that are specific for 17 variant NPM1 mutations.
The aim of this study was to further evaluate the prognostic impact of NPM1-based MRD detection in comparison with other parameters such as age, FAB (French-American-British) subtype, cytogenetics, FLT3 mutations, and NPM1 mutation type. For this purpose 1227 samples of 252 NPM1-mutated AML cases were analyzed for NPM1 mutation expression at different time points before, during, and after treatment. We could show that expression of the NPM1 mutations can be used as a powerful MRD marker in patients with NPM1-mutated AML undergoing both first- and second-line chemotherapy as well as allogeneic stem cell transplantation (alloSCT).
Methods
Patients
Patient samples were referred to the MLL Munich Leukemia Laboratory for diagnostic or follow-up assessment. AML with mutated NPM1 was defined according to WHO classification.20 The patients received different treatment schedules and were in part included into controlled trials of German study groups (n = 86; 34.3%). The cohort was entirely unselected, but all patients received intensive treatment in trials or according to study protocols. All patients for whom an MRD analysis was requested were included regardless of age or treatment schedule or any other parameter. Before therapy all patients gave their informed consent for participation in the current evaluation after having been advised about the purpose and investigational nature of the study as well as of potential risks. The study design adhered to the Declaration of Helsinki, patient-informed consent was obtained, and the study was approved by the ethics committees of the participating institutions before its initiation. The characteristics of the 252 AML with mutated NPM1 at the time of initial diagnosis are summarized in Table 1.
Characteristics of NPM1-mutated patients with AML at diagnosis
| Total cohort, n = 252 . | . | . |
|---|---|---|
| Sex | Female, n = 132 | Male, n = 120 |
| Age, y | Median: 58.9 | Range: 20.1-79.3 |
| WBC, ×109/L | Median: 22.4 | Range: 0.5-300.0 |
| Platelets, ×109/L | Median: 72.5 | Range: 9.0-766.0 |
| De novo | n = 238 | 94.4% |
| After MDS (s-AML) | n = 8 | 3.2% |
| After treatment of a previous malignancy (t-AML) | n = 6 | 2.3% |
| FAB subtype | ||
| AML M0 | n = 5 | 2.0% |
| AML M1 | n = 55 | 21.9% |
| AML M2 | n = 63 | 25.1% |
| AML M4 | n = 70 | 27.9% |
| AML M5 | n = 32 | 12.7% |
| AML M6 | n = 7 | 2.8% |
| AML M7 | n = 1 | 0.4% |
| Not specified | n = 19 | 7.2% |
| Cytogenetics available | n = 247 | 98.0% |
| Normal karyotype | n = 205 | 83.0% |
| Trisomy 4 | n = 5 | 2.0% |
| Trisomy 8 | n = 6 | 2.4% |
| Trisomy 21 | n = 2 | 0.8% |
| Multiple trisomies | n = 6 | 2.4% |
| -7/7q- | n = 2 | 0.8% |
| -Y | n = 4 | 1.6% |
| Del(3p) | n = 1 | 0.4% |
| Del(9q) | n = 4 | 1.6% |
| Del(20q) | n = 2 | 0.8% |
| Nonrecurrent translocations | n = 10 | 4.1% |
| FLT3-LM (available: n = 251) | Mutated: 95 (37.8%) | Wild-type: 156 (62.2%) |
| FLT3-TKD (available: n = 246) | Mutated: 17 (6.9%) | Wild-type: 229 (93.1%) |
| Total cohort, n = 252 . | . | . |
|---|---|---|
| Sex | Female, n = 132 | Male, n = 120 |
| Age, y | Median: 58.9 | Range: 20.1-79.3 |
| WBC, ×109/L | Median: 22.4 | Range: 0.5-300.0 |
| Platelets, ×109/L | Median: 72.5 | Range: 9.0-766.0 |
| De novo | n = 238 | 94.4% |
| After MDS (s-AML) | n = 8 | 3.2% |
| After treatment of a previous malignancy (t-AML) | n = 6 | 2.3% |
| FAB subtype | ||
| AML M0 | n = 5 | 2.0% |
| AML M1 | n = 55 | 21.9% |
| AML M2 | n = 63 | 25.1% |
| AML M4 | n = 70 | 27.9% |
| AML M5 | n = 32 | 12.7% |
| AML M6 | n = 7 | 2.8% |
| AML M7 | n = 1 | 0.4% |
| Not specified | n = 19 | 7.2% |
| Cytogenetics available | n = 247 | 98.0% |
| Normal karyotype | n = 205 | 83.0% |
| Trisomy 4 | n = 5 | 2.0% |
| Trisomy 8 | n = 6 | 2.4% |
| Trisomy 21 | n = 2 | 0.8% |
| Multiple trisomies | n = 6 | 2.4% |
| -7/7q- | n = 2 | 0.8% |
| -Y | n = 4 | 1.6% |
| Del(3p) | n = 1 | 0.4% |
| Del(9q) | n = 4 | 1.6% |
| Del(20q) | n = 2 | 0.8% |
| Nonrecurrent translocations | n = 10 | 4.1% |
| FLT3-LM (available: n = 251) | Mutated: 95 (37.8%) | Wild-type: 156 (62.2%) |
| FLT3-TKD (available: n = 246) | Mutated: 17 (6.9%) | Wild-type: 229 (93.1%) |
AML indicates acute myeloid leukemia; MDS, myelodysplastic syndrome; and WBC, white blood cell count.
Cytomorphology, cytogenetics, and immunophenotyping
Cytomorphologic assessment was based on May-Grünwald-Giemsa stains, myeloperoxidase reaction, and nonspecific esterase by the use of alpha-naphtyl-acetate as described previously and were diagnosed according to the criteria defined in the FAB and the WHO classification.20,24-28 Cytogenetic studies were performed after short-term culture. Karyotypes, analyzed after G-banding, were described according to the International System for Human Cytogenetic nomenclature.29 Immunophenotyping was performed as described previously.30
Examination time points
We analyzed 1227 diagnostic and follow-up samples in 252 NPM1-mutated AML patients by the use of RQ-PCR. A pretreatment sample for NPM1 quantification was available in 229 cases. A total of 998 samples were from different time points during therapy and follow-up. Of these, 91 samples were investigated at relapses that occurred between days 61 and 1546 after diagnosis (median, 328 days). A paired analysis of samples from primary diagnosis and from time point of relapse was performed in 84 patients (no sample from primary diagnosis was available in 7 patients). A total of 2 to 16 follow-up samples (median, 4; mean, 4.9) were analyzed per patient. Median follow-up time of sampling was 282 days (mean, 390 days; range, 17-2529 days). Median available clinical follow-up was 298 days (mean, 423 days; range 20-2529 days). If multiple evaluations were performed within a specific time interval for 1 patient, all data were included in the respective calculations.
Molecular analysis
Isolation of mononuclear cells, mRNA extraction, and random primed cDNA synthesis was performed as previously reported.4 Initial screening for NPM1 mutations and further molecular characterization of the mutation were performed as described previously.14 Mutation type A could be identified by the melting of mutation-specific probe at 43°C. Rare types were identified by melting temperatures other than the wild type (53°C) and were further characterized by sequencing.
Analyses for FLT3-length mutation (FLT3-LM/ITD) and FLT3-TKD mutations also were described previously.31,32 Semiquantitative assessment of FLT3-LM was performed following the method as described by Thiede et al.33 In 200 cases bone marrow (BM) was used at diagnosis, whereas in 28 cases only peripheral blood was available. For MRD during follow-up, only data of BM samples were taken into account.
RQ-PCR
RQ-PCR was performed by the use of the LightCycler 1.5 or 2.0 System (Roche Diagnostics) with the application of hybridization probes as the detection format. The reaction was performed in a final volume of 20 μL by the use of 2 μL mastermix (LightCycler Fast Start DNA Master Hybridization Probes Plus; Roche Diagnostics), 4 mmol/L MgCL2, 0.25 μmol/L of each 3′ and 5′ fluorescent hybridization probe, 0.5 μmol/L of each 3′ and 5′ primer (Metabion), and 2 μL cDNA. Amplification was performed after initial incubation at 95°C for 10 minutes in a 3-step cycle procedure (denaturation 95°C, 1 second, ramp rate 20°C/s, annealing temperature 61°C, 10 seconds, ramp rate 20°C/s, and extension 72°C, 26 seconds, ramp rate 2°C/s) for 45 cycles.
Amplification was performed with 17 different mutation-specific forward primers (Table 2) and a common reverse primer Q-NPM1-R 5′-ACACGGTAGGGAAAGT-3′. Detection probes were as follows: Q-NPM-A, TTCCGTCTTATTTCATTTCTGTAACAGTTG-F and Q-NPM-S, Red640-ATCTGGCTGTCCTTTTTATAATGCAG-P.
Mutation-specific forward primers for quantitative detection of the mutated NPM1 allele
| Name . | Sequence . | Sensitivity . | Specificity . |
|---|---|---|---|
| Q-NPM1-F_Mut_A | TCAAGATCTCTGTCTG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_B | TCAAGATCTCTGCATG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_D | TCAAGATCTCTGCCTG | 1:10 000 | No |
| Q-NPM1-F_Mut_H | TCAAGATCTCTGAGGA | 1:10 000 | No |
| Q-NPM1-F_Mut_I | TCAAGATCTCTGCTTG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_L | TCAAGATCTCTGTTTG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_K | TCAAGATCTCTGCCAG | 1:10 000 | No |
| Q-NPM1-F_Mut_N | TCAAGATCTCTGTCGG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_R | TCAAGATCTCTGTATG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_S | TGGCAGTGTTTTTCTC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_T | TGGCAGTGCATGGCTC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_X | TCAAGATCTCTGTTCC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_Y | TCAAGATCTCTGCCGA | 1:100 000 | Yes |
| Q-NPM1-F_Mut_Z | TCAAGATCTCTGTAGG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_ZD | GATCTCTGGCAGAGGC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_ZF | GATCTCTGGCAGCTTCTCCA | 1:100 000 | Yes |
| Q-NPM1-F_Mut_ZG | CTCTGGCAGCGGATGGC | 1:10 000 | No |
| Name . | Sequence . | Sensitivity . | Specificity . |
|---|---|---|---|
| Q-NPM1-F_Mut_A | TCAAGATCTCTGTCTG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_B | TCAAGATCTCTGCATG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_D | TCAAGATCTCTGCCTG | 1:10 000 | No |
| Q-NPM1-F_Mut_H | TCAAGATCTCTGAGGA | 1:10 000 | No |
| Q-NPM1-F_Mut_I | TCAAGATCTCTGCTTG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_L | TCAAGATCTCTGTTTG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_K | TCAAGATCTCTGCCAG | 1:10 000 | No |
| Q-NPM1-F_Mut_N | TCAAGATCTCTGTCGG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_R | TCAAGATCTCTGTATG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_S | TGGCAGTGTTTTTCTC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_T | TGGCAGTGCATGGCTC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_X | TCAAGATCTCTGTTCC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_Y | TCAAGATCTCTGCCGA | 1:100 000 | Yes |
| Q-NPM1-F_Mut_Z | TCAAGATCTCTGTAGG | 1:100 000 | Yes |
| Q-NPM1-F_Mut_ZD | GATCTCTGGCAGAGGC | 1:100 000 | Yes |
| Q-NPM1-F_Mut_ZF | GATCTCTGGCAGCTTCTCCA | 1:100 000 | Yes |
| Q-NPM1-F_Mut_ZG | CTCTGGCAGCGGATGGC | 1:10 000 | No |
Names are according to laboratory-specific names of individual mutation. Sequences in bold are the sequences of the mutations.
The expression of the mutated NPM1 was normalized against the expression of the control gene ABL1 to adjust for variations in mRNA quality and efficiencies of cDNA synthesis. The NPM1 mutation levels are given as: %NPM1/ABL1. Quantification of ABL1 was performed as described previously.34
To analyze the efficiencies and sensitivities of the 17 different assays, we performed serial dilution experiments of NPM1-mutated diagnostic samples in NPM1-unmutated cDNA of individual patients. Efficiencies were calculated from the slope of the standard curves and ranged from 1.897 to 2.01, which is similar to the range of the efficiency of the ABL1 standard (1.99). Therefore, ABL1 and NPM1 estimations of a specific sample were performed in the same run with use of an ABL1 plasmid standard curve.
Statistical analysis
Survival curves were calculated for overall survival (OS) and event-free survival (EFS) according to Kaplan-Meier and compared by the use of the 2-sided log-rank test. OS was the time from diagnosis to death. EFS was the time from diagnosis to treatment failure, relapse, or death. Relapse was defined according to Cheson et al.35 Cox regression analysis was performed for OS and EFS with different analyzed parameters as covariates. NPM1 mutation levels were analyzed as a continuous variable in Cox analysis and as a dichotomous variable in Kaplan-Meier analysis. To investigate NPM1 mutation levels over the course of therapy, patients were grouped into 4 different time intervals (interval 1, early MRD assessments between days 18 and 60 after begin of therapy; interval 2, days 60-121; interval 3, days 121-365, interval 4, all assessments >1 year after start of treatment). The selection of cutoff levels for the use of NPM1 mutation levels as a dichotomous variable is described in “Results.”
Dichotomous variables were compared between different groups by the use of the χ2 test and continuous variables by the Student t test. Spearman rank correlation was used to analyze correlations between continuous parameters. Multivariate analysis for OS was not performed because of the limited impact of NPM1 mutation levels on OS. Multivariate analyses were performed for EFS only. They were performed for all 4 time intervals separately. In each time interval the dataset was analyzed univariately for parameters significantly related to EFS. Only these parameters were included as covariates into the respective multivariate analyses, whereas other parameters were not included. For all analyses, results were significant at a level of P less than .05 at both sides. SPSS (Version 14.0.1) software was used for statistical analysis.
Results
Characterization of NPM1 mutations at diagnosis
Seventeen different NPM1 mutations were identified in the cohort that was sent in for follow-up analysis (Table 3). Type A was the most frequent mutation (79.7%), followed by types B and D (6% each). Fourteen different rare mutations accounted for the remaining 8.3%. The exact numbers of analyzed patients and samples per mutation type also are indicated in Table 3.
Characterization of mutation types* used for follow-up studies
| Mutation ID . | Insertion . | Individual cases, n . | % . | Samples total, n . |
|---|---|---|---|---|
| A | 956 ins tctg | 201 | 79.7 | 972 |
| B | 956 ins catg | 15 | 6.0 | 77 |
| D | 956 ins cctg | 15 | 6.0 | 92 |
| HMLL | 960 ins agga | 1 | 0.4 | 2 |
| IMLL | 956 ins cttg | 5 | 2.0 | 21 |
| KMLL | 956 ins ccag | 1 | 0.4 | 3 |
| LMLL | 956 ins tttg | 1 | 0.4 | 2 |
| NMLL | 956 ins tcgg | 1 | 0.4 | 6 |
| RMLL | 956 ins tatg | 4 | 1.6 | 21 |
| SMLL | 962 ins tttttctc del 963-967 | 1 | 0.4 | 5 |
| TMLL | 962 ins catggctc del 963-967 | 1 | 0.4 | 4 |
| XMLL | 956 ins ttcc | 1 | 0.4 | 4 |
| YMLL | 956 ins ccga | 1 | 0.4 | 2 |
| ZMLL | 956 ins tagg | 1 | 0.4 | 4 |
| ZDMLL | 960 ins aggc | 1 | 0.4 | 2 |
| ZFMLL | 962 ins cttctcca del 963-967 | 1 | 0.4 | 5 |
| ZGMLL | 960 ins cggatggc del 961-963 | 1 | 0.4 | 5 |
| Total | 251 | 1224 |
| Mutation ID . | Insertion . | Individual cases, n . | % . | Samples total, n . |
|---|---|---|---|---|
| A | 956 ins tctg | 201 | 79.7 | 972 |
| B | 956 ins catg | 15 | 6.0 | 77 |
| D | 956 ins cctg | 15 | 6.0 | 92 |
| HMLL | 960 ins agga | 1 | 0.4 | 2 |
| IMLL | 956 ins cttg | 5 | 2.0 | 21 |
| KMLL | 956 ins ccag | 1 | 0.4 | 3 |
| LMLL | 956 ins tttg | 1 | 0.4 | 2 |
| NMLL | 956 ins tcgg | 1 | 0.4 | 6 |
| RMLL | 956 ins tatg | 4 | 1.6 | 21 |
| SMLL | 962 ins tttttctc del 963-967 | 1 | 0.4 | 5 |
| TMLL | 962 ins catggctc del 963-967 | 1 | 0.4 | 4 |
| XMLL | 956 ins ttcc | 1 | 0.4 | 4 |
| YMLL | 956 ins ccga | 1 | 0.4 | 2 |
| ZMLL | 956 ins tagg | 1 | 0.4 | 4 |
| ZDMLL | 960 ins aggc | 1 | 0.4 | 2 |
| ZFMLL | 962 ins cttctcca del 963-967 | 1 | 0.4 | 5 |
| ZGMLL | 960 ins cggatggc del 961-963 | 1 | 0.4 | 5 |
| Total | 251 | 1224 |
As assessed by melting curve analysis and sequencing.
Cytogenetic and further molecular characterization of patients
Cytogenetics was evaluable at diagnosis in 247 cases. A total of 205 cases (83%) had an NK, and 42 cases (17%) had aberrant karyotypes as described in Table 1. The most frequently observed cytogenetic alterations are single or multiple trisomies or nonrecurrent translocations.
An FLT3-LM was detected in 95 (37.7%) of 252 cases, and an FLT3-TKD mutation was detected in 17 (6.9%) of 246 cases. None of the cases had additional partial tandem duplications in the MLL gene (analyzed: n = 236) or a CEBPA mutation (analyzed: n = 144).
Sensitivity and specificity of the RQ-PCR assays
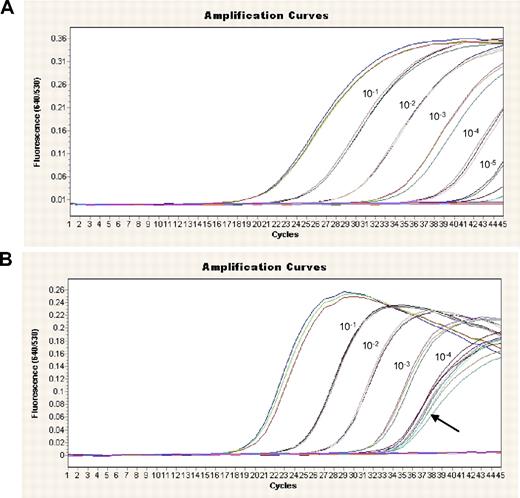
Sensitivity and specificity of the assays were estimated by serial dilutions of a diagnostic sample in NPM1-unmutated cDNA. The sensitivities for the individual assays ranged from 1:10 000 to 1:100 000, depending on the initial NPM1 mutation levels, the probe quality of the individual sample, and the specificity of the assay (Table 2). Indicated are the highest-possible sensitivities as obtained by good quality and high expressing samples at diagnosis. Most assays were specific (example in Figure 1A). In 3 assays (mutations D, H, K, and ZG) a coamplification of the wild type could not be avoided, resulting in a lower sensitivity of the assay (Table 2; Figure 1B).
Limited dilution series for the assessment of sensitivity and specificity of NPM1 mutations. cDNA from time point of diagnosis was diluted in an NPM1-unmutated cDNA. (A) An example of a patient with NPM1 mutation of type A is shown. This mutant shows high specificity and sensitivity. (B) In type D, a coamplification of wild-type NPM1 was observed (indicated by  ), leading to reduced sensitivity.
), leading to reduced sensitivity.
Limited dilution series for the assessment of sensitivity and specificity of NPM1 mutations. cDNA from time point of diagnosis was diluted in an NPM1-unmutated cDNA. (A) An example of a patient with NPM1 mutation of type A is shown. This mutant shows high specificity and sensitivity. (B) In type D, a coamplification of wild-type NPM1 was observed (indicated by  ), leading to reduced sensitivity.
), leading to reduced sensitivity.
Quantitative expression of NPM1 mutation at diagnosis
First, the quantitative expression of NPM1 mutations at diagnosis was analyzed. The %NPM1 mutation/ABL1 level at diagnosis showed a broad range between 10.7 and 8842.9 with a median of 982.5. OS or EFS as calculated by Cox regression analysis was not influenced by the expression levels at diagnosis (data not shown).
Molecular and cytogenetic studies in paired samples at diagnosis and relapse
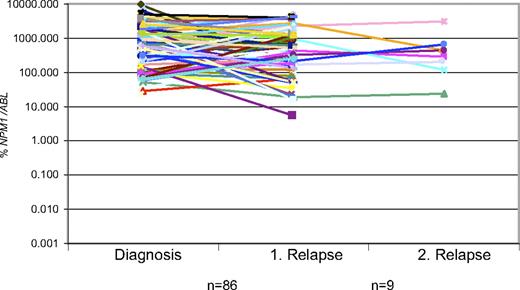
A total of 84 patients were analyzed both at diagnosis and relapse. For 9 of these patients a sample from a second relapse also was available. All patients showed the same NPM1 mutation at diagnosis and at first and second relapse. In addition, in all relapses the %NPM1/ABL1 level as assessed by RQ-PCR was in the same high 2 log ranges as measured at first diagnosis (Figure 2).
Comparison of NPM1 expression in paired samples at diagnosis and at first (n = 86) and second relapse (n = 9). These data show that NPM1 is expressed at all time points in the same high 2 log range for individual patients.
Comparison of NPM1 expression in paired samples at diagnosis and at first (n = 86) and second relapse (n = 9). These data show that NPM1 is expressed at all time points in the same high 2 log range for individual patients.
However, with regard to other genetic markers, we found that there was a tendency to clonal evolution at relapse. Karyotypes were available at both time points in 61 of 84 cases. Most cases still had a NK at relapse (n = 43). Two cases had the same aberrant karyotype at diagnosis and at relapse. Six of the NK AML and 2 of the aberrant cases gained additional aberrations at relapse. We found in only 4 patients a regression from aberrant karyotype at diagnosis to NK at relapse. In addition 4 patients had different aberrant karyotypes at diagnosis and at relapse.
Regarding the FLT3-LM status (n = 80, available for both time points), a stable unmutated status was found in 32 patients. A total of 31 patients retained the same FLT3-LM at relapse. However, in 18 of these patients a progression to a greater FLT3-LM/wild-type ratio was observed at relapse. In only 3 patients a regression from FLT3-LM to wild-type at relapse was observed. Thus, in 3 (8.8%) of 34 FLT3-LM mutated patients at diagnosis was the mutation lost at relapse. Data on NPM1 mutation level, FLT3-LM, and karyotype at diagnosis and relapse time points are summarized in Table 4.
Comparison of genetic parameters between paired samples of diagnosis, first, and second relapse
| . | Diagnosis n = 84 . | 1. Relapse n = 84 . | 2. Relapse n = 9 . |
|---|---|---|---|
| NPM1 level | Median: 546.4 | Median: 344.4 | Median: 359.5 |
| Range: 20.5-8842.9 | Range: 5.2-4532.9 | Range: 23.3-3040.6 | |
| FLT3-LM | + (n = 31) | + (n = 5) | |
| (n = 80) | + (n = 34) | ||
| − (n = 3) | |||
| − (n = 32) | − (n = 3) | ||
| − (n = 46) | |||
| + (n = 14) | |||
| Karyotype | NK (n = 43) | NK (n = 2) | |
| (n = 61) | NK (n = 49) | ||
| AK (n = 6) | AK (n = 2) | ||
| AK (n = 2) | AK (n = 2) | ||
| AK (n = 2) | AK + ACA (n = 2) | ||
| AK (n = 4) | Diff. AK (n = 4) | Diff. AK (n = 1) | |
| AK (n = 4) | NK (n = 4) |
| . | Diagnosis n = 84 . | 1. Relapse n = 84 . | 2. Relapse n = 9 . |
|---|---|---|---|
| NPM1 level | Median: 546.4 | Median: 344.4 | Median: 359.5 |
| Range: 20.5-8842.9 | Range: 5.2-4532.9 | Range: 23.3-3040.6 | |
| FLT3-LM | + (n = 31) | + (n = 5) | |
| (n = 80) | + (n = 34) | ||
| − (n = 3) | |||
| − (n = 32) | − (n = 3) | ||
| − (n = 46) | |||
| + (n = 14) | |||
| Karyotype | NK (n = 43) | NK (n = 2) | |
| (n = 61) | NK (n = 49) | ||
| AK (n = 6) | AK (n = 2) | ||
| AK (n = 2) | AK (n = 2) | ||
| AK (n = 2) | AK + ACA (n = 2) | ||
| AK (n = 4) | Diff. AK (n = 4) | Diff. AK (n = 1) | |
| AK (n = 4) | NK (n = 4) |
ACA indicates additional chromosomal aberrations; AK, aberrant karyotype; Diff, different; and NK, normal karyotype.
Molecular prediction of relapse
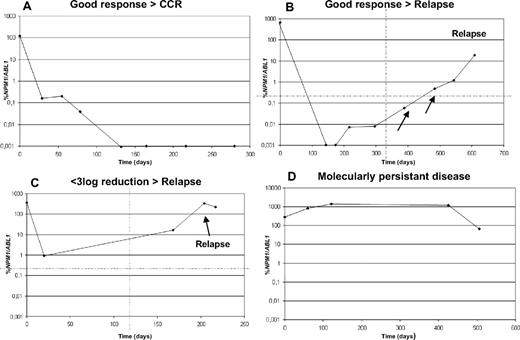
The total cohort included 93 relapses. Early detection of relapse based on increasing MRD levels (at least 1 log at any level) was possible in 47 patients at 15-221 days (median, 62 days) before overt clinical relapse. Significantly slower relapse kinetics were observed in cases with sole NPM1 mutation (n = 29) compared with the FLT3-LM/NPM1 double-mutated cases (n = 15) during first-line treatment (days from first >1 log increase to clinical relapse: 95 vs 55 days; P = .011). In an additional 15 patients, a relapse was predictable on the basis of no or low molecular response as defined by less than a 3-log reduction between ratio at diagnosis and any follow-up investigation. Remarkably, 9 (60%) of these 15 cases with highly persisting NPM1 mutation levels had an FLT3-LM. A total of 31 relapses occurring in the total cohort were not predictable based on MRD values because of missing samples at relevant follow-up time points (median time since last sampling 342 days; range, 143-807 days) in these cases. Thus, on the basis of these data it can be suggested that sample analysis of every 2 months (median of 62 days in the predictable relapses) would allow one to early predict 50% of all relapses, whereas on the basis of the 25 percentile, sampling intervals of 42 days would allow one to early predict 75% of all, and sampling intervals of 120 days (75th percentile) would allow one to early detect 25% of all relapses. Representative examples of early detection of relapse are shown in Figure 3. In addition, in a further 7 patients an increase in the last follow-up was observed, but a relapse was not yet diagnosed, and in an additional 13 cases follow-up was too short for us to draw any conclusions.
Four different response patterns defined according to the MRD values as assessed by RQ-PCR. Examples of individual patients are shown in panel A for good response, in panel B for good response and later relapse, which was detectable early as the result of increasing levels of the NPM1 mutation, in panel C for patients with less than 3-log reduction and relapse, and in panel D for patients with persistent disease. X-axis indicates time in days; y-axis indicates mutation level given as %NPM1/ABL1.
Four different response patterns defined according to the MRD values as assessed by RQ-PCR. Examples of individual patients are shown in panel A for good response, in panel B for good response and later relapse, which was detectable early as the result of increasing levels of the NPM1 mutation, in panel C for patients with less than 3-log reduction and relapse, and in panel D for patients with persistent disease. X-axis indicates time in days; y-axis indicates mutation level given as %NPM1/ABL1.
Response kinetics
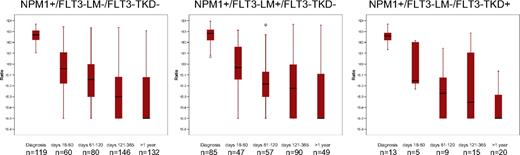
Patients with NPM1 sole-mutated AMLs have frequently been reported to have a better prognosis than patients with NPM1+/FLT3-LM+ (NPM1-mutated/FLT3 length-mutated) double-mutated AML.13-17,19,36 However, patients with NPM1+/FLT3-TKD double-mutated AML have been reported to do prognostically even better than the patients with NPM1 sole-mutated AML.14,32 To compare response kinetics of these different groups NPM1 expression levels at different time points were compared (Figure 4). The NPM1+/FLT3-LM− and NPM1+/FLT3-LM+ both had a median of 3-log reduction between diagnosis and at interval days 18 to 60. The NPM1+/FLT3-TKD+ (FLT3 tyrosine kinase mutated) group achieved a reduction of even 4 log during the same interval. However, this difference was not statistically significant because of small numbers. Also for all other time points, no significant differences in response kinetics reflecting different outcomes could be shown. As shown in Figure 4, at interval days 121-365 the NPM1 mutation levels in the NPM1+/FLT3-LM+ group were one log greater than in the NPM1+/FLT3-LM− group and nearly 2 logs greater than in the NPM1+/FLT3-TKD+ group, which may reflect the greater relapse rate in the NPM1+/FLT3-LM+ group; however, these values do not reach statistical significance and need further investigation (Figure 4).
Kinetics of therapy response compared in NPM1+/FLT3-LM−/FLT3-TKD−, NPM1+/FLT3-LM+/FLT3-TKD−, and NPM1+/FLT3-LM−/FLT3-TKD+ AML patients. Boxes represent the 50th percentile, red lines the 25th percentiles, and black bars the median of each group. These data show very similar kinetics in all 3 subgroups. Only the higher levels of the NPM1+/FLT3-LM+ between days 121 and 365 reflect the greater relapse rate in this group compared with the other 2 groups.
Kinetics of therapy response compared in NPM1+/FLT3-LM−/FLT3-TKD−, NPM1+/FLT3-LM+/FLT3-TKD−, and NPM1+/FLT3-LM−/FLT3-TKD+ AML patients. Boxes represent the 50th percentile, red lines the 25th percentiles, and black bars the median of each group. These data show very similar kinetics in all 3 subgroups. Only the higher levels of the NPM1+/FLT3-LM+ between days 121 and 365 reflect the greater relapse rate in this group compared with the other 2 groups.
Response patterns
On the basis of the MRD kinetics in 229 cases that were monitored during first-line treatment, 4 different response patterns were observed: (1) patients with a good response and at least a 5-log reduction of the NPM1 mutation level that remained in complete ongoing remission during the observation period (n = 137; example in Figure 3A); (2) patients with a good response (decrease of 5-7 log) but later relapse that were early detectable based on increasing MRD levels (n = 35; example in Figure 3B); (3) patients achieving complete remission (CR) but with a less than 3-log reduction and early relapse (n = 9; example in Figure 3C); (4) patients with “molecularly persistent disease” with no or less than 2-log reduction (n = 15; example in Figure 3D). Of the latter group, BM smears were evaluated in 12 cases: 4 had a morphologic CR, 5 had persistent disease, and 3 had very hypocellular BM after treatment, not allowing any remission evaluation.
Prognostic value of MRD
For evaluation of the prognostic value of NPM1 mutation–based MRD detection, our total cohort of 1227 samples was subdivided into 3 subcohorts. The first cohort was evaluated during and after intensive first-line therapy (678 samples of 229 patients in first CR). The second cohort was evaluated during second-line chemotherapy after relapse or persistence of leukemia (117 samples of 51 patients). The third cohort was analyzed after alloSCT (154 samples of 53 patients). Of these, 25 patients were transplanted in first and 28 in subsequent CR. Overall the median %NPM1/ABL1 MRD values were 0.007 after first-line therapy, 23.23 after second-line chemotherapy, and 0.001 after allogeneic transplantation.
For further evaluation, all follow-up samples were grouped into different time intervals: interval 1, early MRD assessments between day 18 and 60 after begin of therapy; interval 2, days 60-121; interval 3, 121-365; interval 4, all assessments longer than 1 year after the start of treatment.
Cohort 1.
The analysis for OS in the patients obtaining first-line treatment did not show significant values for time intervals 1 and 2 and was not evaluable for time intervals 3 and 4 as the result of single events. However, for EFS the NPM1 mutation level was found to be a strong prognostic factor for all different time intervals (Table 5).
Impact of NPM1 mutation levels on EFS in AML
| Interval . | Time, days . | Samples, n . | EFS, P . | RR for 100-fold increase of NPM1 level . | 95% CI . |
|---|---|---|---|---|---|
| First-line | |||||
| 1 | 18-60 | 108 | .014 | 1.07 | 1.013-1.121 |
| 2 | 61-120 | 140 | .001 | 1.08 | 1.032-1.128 |
| 3 | 121-365 | 234 | < .001 | 1.10 | 1.087-1.164 |
| 4 | > 365 | 196 | < .001 | 1.20 | 1.108-1.214 |
| Second-line | |||||
| 1 | 18-60 | 30 | n.s. | ||
| 2 | 61-120 | 36 | n.s. | ||
| 3 | 121-365 | 34 | .006 | 1.307 | 1.090-1.524 |
| 4 | > 361 | 11 | .024 | 1.266 | 1.035-1.498 |
| After alloSCT | |||||
| 1 | Up to 60 | 51 | n.s. | ||
| 2 | 61-120 | 38 | .012 | 1.061 | 1.013-1.108 |
| 3 | 121-365 | 42 | .025 | 1.060 | 1.075-1.113 |
| 4 | > 361 | 7 | n.s. |
| Interval . | Time, days . | Samples, n . | EFS, P . | RR for 100-fold increase of NPM1 level . | 95% CI . |
|---|---|---|---|---|---|
| First-line | |||||
| 1 | 18-60 | 108 | .014 | 1.07 | 1.013-1.121 |
| 2 | 61-120 | 140 | .001 | 1.08 | 1.032-1.128 |
| 3 | 121-365 | 234 | < .001 | 1.10 | 1.087-1.164 |
| 4 | > 365 | 196 | < .001 | 1.20 | 1.108-1.214 |
| Second-line | |||||
| 1 | 18-60 | 30 | n.s. | ||
| 2 | 61-120 | 36 | n.s. | ||
| 3 | 121-365 | 34 | .006 | 1.307 | 1.090-1.524 |
| 4 | > 361 | 11 | .024 | 1.266 | 1.035-1.498 |
| After alloSCT | |||||
| 1 | Up to 60 | 51 | n.s. | ||
| 2 | 61-120 | 38 | .012 | 1.061 | 1.013-1.108 |
| 3 | 121-365 | 42 | .025 | 1.060 | 1.075-1.113 |
| 4 | > 361 | 7 | n.s. |
Impact of NPM1 mutation levels on EFS in AML (Cox regression analysis) during and after first-line therapy (678 samples from 229 patients), after second-line chemotherapy (117 samples from 51 patients), and after alloSCT (154 samples from 53 patients).
alloSCT indicates allogeneic stem cell transplantation; CI, confidence interval; EFS, event-free survival; n.s., not significant; and RR, relative risk.
Cohort 2.
The same analysis was repeated for samples taken during second-line chemotherapy (n = 111). A significant effect of NPM1 mutation level on EFS was detected only for samples analyzed between day 121 and 365 and for those after 1 year of therapy. Results are summarized in Table 5.
Cohort 3.
In the third cohort with samples after allogeneic BM transplantation (n = 138), an effect of NPM1 mutation level on survival also could be shown for samples analyzed between days 61 and 120 after and between days 121 and 365 after alloSCT. Results are summarized in Table 5.
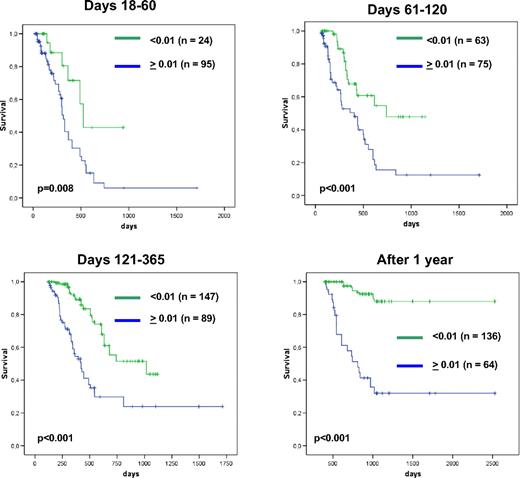
In a further analysis, we addressed the question whether there may be any kind of threshold that may differentiate different prognostic subgroups. The rationale for choosing the respective thresholds was based on the median MRD level found in different cohort. Thus, in patients undergoing first-line therapy, the median NPM1 mutation level amounted to 0.06% NPM1/ABL1 mutation, whereas the respective value for patients undergoing second-line therapy or stem cell transplantation was 0.08% NPM1/ABL1 mutation. Therefore, the 2 thresholds 0.1% NPM1/ABL1 mutation and 0.01% NPM1/ABL1 mutation were evaluated. In the first cohort the cutpoint 0.01% NPM1/ABL1 mutation resulted in larger differences compared with 0.1% NPM1/ABL1 mutation, whereas the opposite was true in the other cohorts.
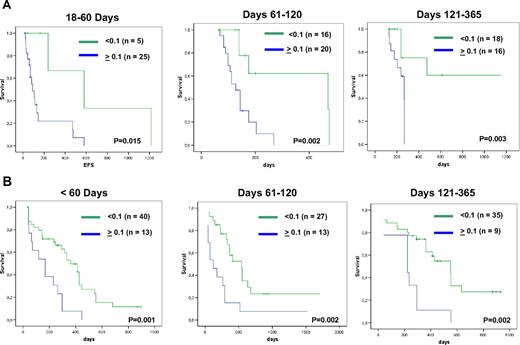
In the first cohort prognostically different groups could be defined with both thresholds. Results for thresholds 0.01% are presented in detail in Figure 5. By using a threshold 0.1%, we obtained similar results (day 18-60: P = .12; days 61-120: P < .001, days 121-365: P < .001; > 1 year: P < .001). Both thresholds are highly relevant for all time points. The threshold 0.1% could successfully also be applied to samples after second-line chemotherapy (Figure 6A) and after alloSCT (Figure 6B).
EFS in patients during first-line treatment. Kaplan-Meier plots show prognostically different groups as defined by threshold 0.01%NPM1/ABL expression at different intervals after the start of therapy.
EFS in patients during first-line treatment. Kaplan-Meier plots show prognostically different groups as defined by threshold 0.01%NPM1/ABL expression at different intervals after the start of therapy.
EFS in patients. EFS is shown during (A) second-line chemotherapy; and (B) after allogeneic BM transplantation. Kaplan-Meier plots show prognostically different groups as defined by a threshold of 0.1%NPM1/ABL expression at different intervals after start of therapy.
EFS in patients. EFS is shown during (A) second-line chemotherapy; and (B) after allogeneic BM transplantation. Kaplan-Meier plots show prognostically different groups as defined by a threshold of 0.1%NPM1/ABL expression at different intervals after start of therapy.
Influence of pretreatment parameters on prognosis
NPM1 has been described to be a favorable factor if not accompanied by an FLT3-LM. In addition, other parameters such as age, white blood cell count, percentage of blasts in the BM, and chromosome aberrations have been shown to influence prognosis in AML. Therefore, a Cox regression analysis that included all these parameters was performed in the presented cohort (Table 1) for OS and EFS. In addition, FAB subtype, NPM1 mutation type, and initial NPM1 mutation level at diagnosis were taken into account. Respective data were available for 226 patients undergoing first-line treatment. All results are summarized in Table 6. A significant unfavorable impact was observed only for increasing age (EFS) and FLT3-LM (OS and EFS).
Influence of different pretherapeutic parameters on OS and EFS in patients undergoing first-line treatment (data available for 227 patients)
| . | Number evaluable . | OS . | RR (OS) . | EFS . | RR (EFS) . |
|---|---|---|---|---|---|
| Age | 225 | 0.103 | 0.013 | 1.20* | |
| WBC | 177 | 0.793 | 0.323 | ||
| Blast count in BM | 146 | 0.232 | 0.228 | ||
| CD34+ | 120 | 0.217 | 0.297 | ||
| FLT3-LM | 227 | 0.010 | 4.12 | 0.013 | 1.69 |
| FLT3-TKD | 223 | 0.540 | 0.733 | ||
| FAB subtype | 213 | 0.366 | 0.346 | ||
| Chromosomal aberrations | 224 | 0.341 | 0.626 | ||
| NPM1 mutation type | 227 | 0.842 | 0.283 | ||
| Initial NPM1 level | 227 | 0.915 | 0.222 |
| . | Number evaluable . | OS . | RR (OS) . | EFS . | RR (EFS) . |
|---|---|---|---|---|---|
| Age | 225 | 0.103 | 0.013 | 1.20* | |
| WBC | 177 | 0.793 | 0.323 | ||
| Blast count in BM | 146 | 0.232 | 0.228 | ||
| CD34+ | 120 | 0.217 | 0.297 | ||
| FLT3-LM | 227 | 0.010 | 4.12 | 0.013 | 1.69 |
| FLT3-TKD | 223 | 0.540 | 0.733 | ||
| FAB subtype | 213 | 0.366 | 0.346 | ||
| Chromosomal aberrations | 224 | 0.341 | 0.626 | ||
| NPM1 mutation type | 227 | 0.842 | 0.283 | ||
| Initial NPM1 level | 227 | 0.915 | 0.222 |
Significant parameters are presented in bold type.
BM indicates bone marrow, EFS, event-free survival; FAB, French-American-British; OS, overall survival; RR, relative risk; and WBC, white blood cell count.
For 10-year intervals.
Multivariate analysis
For a multivariate analysis of EFS, age at diagnosis and pretreatment FLT3 mutation status were analyzed in combination with NPM1 mutation level at different time points. The NPM1 level was found to be an independent prognostic parameter at the time intervals 2, 3, and 4 (all after day 60; Table 7).
Multivariate analysis
| . | Days 18-60 . | Days 61-120 . | Days 121-365 . | More than 1 year . | ||||
|---|---|---|---|---|---|---|---|---|
| P . | RR . | P . | RR . | P . | RR . | P . | RR . | |
| First-line therapy | ||||||||
| Age* | .037 | 1.20 | .001 | 1.39 | <.001 | 1.40 | .513 | |
| FLT3-LM | .039 | 1.82 | .245 | 1.37 | .099 | 1.35 | .583 | |
| NPM1 level† | .097 | 0.021 | .021 | 1.10 | < .001 | 1.12 | < .001 | 1.15 |
| After second-line therapy | ||||||||
| FLT3* | na | na | na | na | .022 | 14.989 | ns | ns |
| NPM1 level* | na | na | na | na | .012 | 0.398 | .024 | 1.266 |
| After alloSCT | ||||||||
| Age* | na | na | .819 | na | .515 | ns | na | na |
| NPM1 ratio† | na | na | .065 | 1.065 | .024 | 1.062 | na | na |
| . | Days 18-60 . | Days 61-120 . | Days 121-365 . | More than 1 year . | ||||
|---|---|---|---|---|---|---|---|---|
| P . | RR . | P . | RR . | P . | RR . | P . | RR . | |
| First-line therapy | ||||||||
| Age* | .037 | 1.20 | .001 | 1.39 | <.001 | 1.40 | .513 | |
| FLT3-LM | .039 | 1.82 | .245 | 1.37 | .099 | 1.35 | .583 | |
| NPM1 level† | .097 | 0.021 | .021 | 1.10 | < .001 | 1.12 | < .001 | 1.15 |
| After second-line therapy | ||||||||
| FLT3* | na | na | na | na | .022 | 14.989 | ns | ns |
| NPM1 level* | na | na | na | na | .012 | 0.398 | .024 | 1.266 |
| After alloSCT | ||||||||
| Age* | na | na | .819 | na | .515 | ns | na | na |
| NPM1 ratio† | na | na | .065 | 1.065 | .024 | 1.062 | na | na |
Significant parameters are presented in bold type.
alloSCT indicates allogeneic stem cell transplantation; na: not applicable; ns, not significant; and RR, relative risk.
RR for 10-year period.
RR for 100-fold increase.
A similarly high impact for the NPM1 mutation level was found in the second cohort (after second-line chemotherapy samples taken between days 121 and 365 and after 1 year). In this cohort the FLT3-LM status also was found to have an adverse influence (P = .003). In a multivariate analysis, both parameters, FLT3 mutation status and NPM1 mutation level, were independent prognostic factors in the opposite direction for the days 121-365 (P = .022; RR, 14.989; 95% confidence interval [CI], 2.653-84.680 and P = 0.012; RR, 1.399; 95% CI, 1.088-1.711). However, after 1 year, the NPM1 mutation level was the only independent prognostic parameter (P = .024; RR, 1.266; 95% CI, 1.345-1.490; Table 7).
For samples after alloSCT the NPM1 mutation level in the time intervals 2 (days 61-120) and 3 (days 121-365) could be shown to be prognostically relevant. No other parameter with the exception of age was relevant for EFS in this cohort (P = .041; RR for 10-year interval, 1.588; 95% CI, 1.023-2.184). Taking NPM1 mutation level and age into account in a multivariate analysis, only the NPM1 mutation level emerged as an independent prognostic parameter (Table 7).
Additional information with the use of FLT3-LM as a further follow-up marker
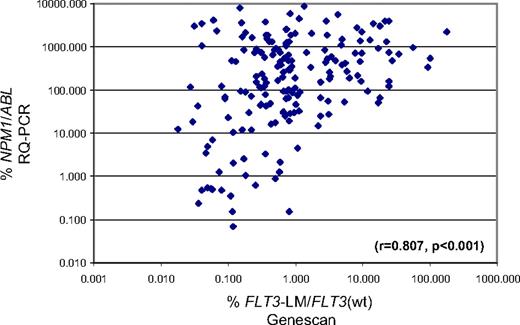
To evaluate a possible additional impact of the FLT3-LM during follow-up, this mutation was evaluated semiquantitatively by the use of FLT3-LM/FLT3wt ratio as assessed by genescan analysis and then compared with the %NPM1/ABL1 levels (Figure 7). In total, 324 parallel assessments were performed during follow-up of 87 patients who were positive for both markers at diagnosis. NPM1 was detectable in 264 and undetectable in 60 of these samples. In contrast, FLT3-LM was detectable in 179 and undetectable in 145 samples. Thus, although the correlation was high (r = 0.807; P < .001) the NPM1 mutation was the more sensitive marker because it was detectable in 85 cases in which FLT3-LM was already or still undetectable. On the contrary, there was no sample that was FLT3-LM+/NPM1−.
Parallel assessments of NPM1 and FLT3-LM. The %NPM1/ABL as assessed by RQ-PCR (Y-axis) is plotted against the FLT3-LM expression as assessed by gene scan analysis. Also, a high correlation was found the NPM1 marker was found positive in 85 samples that were already found to be negative for FLT3-LM.
Parallel assessments of NPM1 and FLT3-LM. The %NPM1/ABL as assessed by RQ-PCR (Y-axis) is plotted against the FLT3-LM expression as assessed by gene scan analysis. Also, a high correlation was found the NPM1 marker was found positive in 85 samples that were already found to be negative for FLT3-LM.
Discussion
The International Working group has included molecular remission as one of the criteria for treatment response35 in patients with AML, and RQ-PCR–based MRD detection has been found to be of high prognostic value in AML with fusion transcripts.4,37-39 However, with the exception of acute promyelocytic leukemia,40 the evaluation of MRD has not found broad entrance into clinical studies or patient management in AML. One of the major obstacles frequently reported has been the lack of markers for NK AML. However, because the detection of NPM1 gene mutations that can be found in approximately one-third of patients with AML, the assessment of MRD by RQ-PCR may become an important tool for individualized treatment, even including transplantation strategies.36
The applicability of RQ-PCR–based MRD detection for NPM1 recently has been shown22,23,41 ; these studies focused on the detection of the most common mutation type A that can be detected in approximately 80% of all NPM1-mutated AML. One of these groups23 also reported on the applicability of 3 rare variants. The use of a DNA-based assay for 4 other rare variants also has been shown.22 Here, we report for the first time on 17 different mutation-specific assays by using mutation-specific forward primers and one common reverse primer and common hybridization probes. Our results clearly demonstrate that nearly all rare NPM1 mutations can be monitored with a high sensitivity of up to 10−5.
All RQ-PCR analyses in this study were performed by the use of highly sensitive cDNA-based assays. At diagnosis, varying expression levels of up to 3 log ranges were detected. This finding is in accordance with data for PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11, also showing a broad range of expression levels at diagnosis. For these entities, it was shown that high initial expression of fusion genes are correlated with unfavorable outcome.4 In contrast to these findings, an effect of initial expression levels of the NPM1 mutations on any survival parameter could not be shown in the present study.
We then analyzed NPM1 expression during follow-up with respect to its influence on OS and could not show that %NPM1/ABL1 level had a significant influence for time intervals up to days 120 after start of treatment. Conclusions for OS were not possible for later time points because only very few deaths appeared after day 120 in the analyzed cohort. It has to be mentioned that the median OS for our patients was 518 days, which is very long but comparable with other NPM1-mutated cohorts.42 This overall favorable prognosis makes it difficult to find an additional prognostic factor and may account for the missing impact of MRD on OS in this prognostically good subgroup of AML. This may in part be the result of highly effective rescue therapies in NPM1 mutated AML after relapse. Furthermore, a bias may appear because patients were sent specifically for MRD analysis. Thus, there might be a selection to cases at risk that have been sent in for certain reasons. However, the distribution of age, FAB subtype, FLT3 mutations, and other parameters such as median survival shows that the cohort is highly comparable with other cohorts presented in different NPM1 studies.13-16,42 Subsequently, we concentrated our analysis on EFS. We could show that MRD for NPM1 mutations as assessed at 4 different time intervals after start of therapy is an independent and highly predictive parameter for EFS for patients receiving standard first-line therapy of AML.
In addition, it was possible to define thresholds of less than 0.1% and less than 0.01% NPM1/ABL1 that discriminate 2 groups of patients with significantly different outcome. The threshold value of 0.1% corresponds to that described previously for a time point after 6 weeks of treatment with a DNA-based assay.22 In the present study this value was not only confirmed in a much larger cohort but was also shown for 4 different time points and for different kind of therapies.
Even more, we were able to find similar information to the results of the first-line therapy for patients treated with second-line chemotherapy after resistance or relapse or allogeneic transplantation. The respective expression ratio of the NPM1 mutation was found to be highly relevant for EFS. Also for these samples the 0.1% threshold could be confirmed.
The median expression ratio in 51 patients after relapse chemotherapy was 23.23 compared with only 0.007 after first-line treatment. This finding clearly demonstrates that second-line chemotherapy, not including transplantation strategies, may not be very effective for relapsed NPM1-mutated patients. The finding of these very low levels after first-line chemotherapy supports the data of Schlenk et al,36 who have suggested that transplantation may not provide an advantage in first-line treatment of NPM1+/FLT3-ITD− patients but should be tried after relapse.
In addition, we focused on patient characteristics at diagnosis and relapse. Karyotype analysis of paired diagnostic and relapse samples revealed different patterns with either stable karyotype or clonal evolution with gain of additional chromosomal aberrations or even a total change of karyotype. Four patients each even showed a regression of chromosomal aberrations or total change of aberrant karyotype leading to different cytogenetic findings at diagnosis and at relapse. Interestingly, all these patients had the same NPM1 mutation at all time points, indicating that NPM1 is an early genetic event that appears to precede chromosomal aberrations, as we recently reported,43 and also FLT3 mutations as it has been suggested previously.21 In contrast to our data, the authors of 2 reports41,44 had suggested instability of NPM1 based on paired diagnostic and relapse samples. However, they found these shifts in 2 of only 17 and 2 of only 27 cases, respectively, that relapsed without NPM1 mutation. We never found such shift in 86 paired cases. One explanation may be that approximately 10% of relapses from the genetic point of view may be addressed as treatment-related AML.45 As an example, one case of AML with NK described by Papadaki et al41 that relapsed with a t(1;7) and also lost the FLT3-ITD may fulfill these criteria. Relapses showing genetic markers independent to the original clone also have been observed in patients with AML who have recurrent cytogenetic abnormalities. For instance we have described a patient with t(8;21) followed by AML1-ETO–specific real-time PCR that “relapsed” with a t(3;21)/AML1-EVI1–positive AML.46 Taken this into account one has to be aware that no molecular marker is 100% sensitive to detect a “relapse” if a t-AML appears. In our experience this is a very rare event and does not abrogate NPM1 as a very reliable MRD marker in AML.
Our data even show that NPM1 mutation is more stable and thus pathogenetically antecedent to the FLT3-ITD, which is also in accordance with previous observations.47,48 Thus, instability of FLT3-LM seems to be more relevant in the course of AML. However, in our cohort most patients had a progression from negative to positive status or positive cases to an even greater FLT3-LM/wild-type ratio at relapse. In only 3 cases a regression from FLT3-LM positivity to negativity was detected in our cohort. Thus, the possibility of overlooking an upcoming relapse as the result of the loss of the FLT3-LM is low (∼ 8.5%, based on 3 of 35 samples in this analysis) compared with the possible usefulness of FLT3-LM to better judge on an upcoming possible relapse based on increasing NPM1 mutation ratio. This information has to be taken into account, especially with respect to those NPM1-mutated AML that have increasing NPM1 mutation levels without showing clinically an overt relapse during a long period of time. Because FLT3-LM is a highly relevant progression marker, a parallel detection of FLT3-LM by the use of gene scan in combination with an increasing NPM1 mutation level is very meaningful to predict a relapse.
In conclusion, our data show that NPM1-based MRD assessment is useful to measure therapy response, persistence, or reappearance of the leukemic clone in AML. We also demonstrated that, by use of specific thresholds, it is possible to further subcategorize NPM1-mutated cases into prognostic groups based on treatment-related factors (MRD). This finding makes therapy monitoring by NPM1-based MRD detection the most relevant prognostic parameter in AML with mutated NPM1. These results should now be validated in randomized clinical studies. Such an approach will pave the way to improve monitoring and individualization of therapy in NPM1-mutated AML, even allowing the decision between conventional therapy and transplantation strategies based on the respective MRD load.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors from the Munich Leukemia Laboratory (MLL) thank all physicians and especially participants of the Acute Myeloid Leukemia Cooperative Group for sending bone marrow or blood samples to our laboratory for reference diagnosis and also for submitting clinical data.
Authorship
Contribution: S.S. served as principal investigator, designed the study, performed molecular genetic analysis, analyzed data, and wrote the manuscript; W.K. performed immunophenotyping, analyzed data, and compiled statistics; C.T. performed molecular genetic analysis; T.W. collected and documented clinical data; F.D. performed molecular genetic analysis; B.F. designed the study and wrote the manuscript; C.H. performed cytogenetic analysis; and T.H. performed cytomorphology and wrote the manuscript.
Conflict-of-interest disclosure: S.S., W.K., C.H., and T.H. own and are employed by the Munich Leukemia Laboratory (MLL) GmbH. C.T., T.W., and F.D. are employed by MLL GmbH. B.F. has applied for a patent on the clinical use of NPM1 mutants.
Correspondence: PD Dr rer. nat. Susanne Schnittger, PhD, MLL Munich Leukemia Laboratory, Max-Lebsche-Platz 31, 81377 Munich, Germany; e-mail: susanne.schnittger@mll-online.com.