Abstract
FANCM is a component of the Fanconi anemia (FA) core complex and one FA patient (EUFA867) with biallelic mutations in FANCM has been described. Strikingly, we found that EUFA867 also carries biallelic mutations in FANCA. After correcting the FANCA defect in EUFA867 lymphoblasts, a “clean” FA-M cell line was generated. These cells were hypersensitive to mitomycin C, but unlike cells defective in other core complex members, FANCM−/− cells were proficient in monoubiquitinating FANCD2 and were sensitive to the topoisomerase inhibitor camptothecin, a feature shared only with the FA subtype D1 and N. In addition, FANCM−/− cells were sensitive to UV light. FANCM and a C-terminal deletion mutant rescued the cross-linker sensitivity of FANCM−/− cells, whereas a FANCM ATPase mutant did not. Because both mutants restored the formation of FANCD2 foci, we conclude that FANCM functions in an FA core complex–dependent and –independent manner.
Introduction
Fanconi anemia (FA) is a recessive genetic instability syndrome that has uncovered a cellular pathway involved in the protection against replication-blocking lesions. Inactivation of this pathway, as seen in FA patients, results in hypersensitivity to DNA cross-linking agents and cancer susceptibility.1 Defects in 13 different genes have been found in FA patients,2 and the proteins encoded by these genes cooperate in a pathway that can be subdivided in an upstream and downstream part based upon the monoubiquitination of FANCD2 and FANCI.1 The upstream part of the pathway consists of a nuclear core complex formed by the FA proteins FANCA, -B, -C, -E, -F, -G, -L, and -M and 2 FA-associated proteins FAAP100 and FAAP24. This complex monoubiquitinates FANCD2 through the E3-ubiquitin ligase FANCL in conjunction with the ubiquitin-conjugating enzyme.3,4 The FA core complex, UBE2T, and FANCD2 are independently recruited to the stalled replication fork.5 For FANCD2, this relies on the ATR-mediated phosphorylation of its binding partner FANCI,6 whereas the recruitment of the FA core complex seems to depend on FANCM.7 Like FANCD2, FANCI is also monoubiquitinated by the FA core complex and these modified proteins colocalize with Rad51 and BRCA1 in nuclear foci.8,9 The link between FA and BRCA proteins was further strengthened by the discovery of FA patients with mutations in BRCA2,10 and in the BRCA1- and BRCA2-interacting proteins BRIP111,12 and PALB2.13,14 FA patients with a defect in any of these genes have normal FANCD2 monoubiquitination and therefore these proteins are considered as downstream players in the FA pathway.
Despite the identification of the various components of the FA core complex, its role in the maintenance of genome stability remains unclear, because of the absence of functional domains in most of the core complex members. A notable exception is FANCM, an ortholog of the archaeal DNA repair protein HEF, which contains 2 conserved domains: a DEAH helicase domain in the N-terminus and an endonuclease domain in the C-terminus.15,16 The helicase domain is shared with yeast orthologs MPH1 (Saccharomyces cerevisiae) and FML1 (Schizosaccharomyces pombe), which play a regulatory role in homologous recombination repair by replication fork reversal and D-loop disruption.17-19 HEF and MPH1 possess helicase activity,20,21 whereas for FANCM only translocase activity was observed.15 Purified FANCM also promotes branch migration of Holliday junctions and replication forks22 and remodels DNA replication structures,23 establishing a possible link between the FA pathway and DNA repair.
FANCM, initially known as FAAP250, was identified as a FANCA-interacting protein in an attempt to purify the FA core complex.24 FAAP250 was named FANCM after an unclassified FA patient (EUFA867) with biallelic mutations in the FAAP250-encoding gene was found.15 Lymphoblasts from this patient lacked FANCM protein expression and FANCD2 monoubiquitination. In addition, FANCA and FANCG levels were reduced and the nuclear localization of FANCA and FANCL was disturbed. EUFA867 had been excluded from other known complementation groups by linkage analysis, cell fusion, and cDNA transfection and was therefore assigned to a new complementation group FA-M. Curiously, experiments designed to complement cross-linker sensitivity in EUFA867 lymphoblasts with FANCM cDNA, have reproducibly failed.
We now report that EUFA867, in addition to biallelic FANCM mutations, also has biallelic mutations in FANCA. Correction of the FANCA defect revealed the features specific for FA-M cells, that is, hypersensitivity to DNA cross-linking agents and to the topoisomerase I inhibitor camptothecin, which could be rescued by ectopic expression of FANCM. At the molecular level, FANCM-deficient lymphoblasts show residual FANCD2 monoubiquitination, indicating that FANCM is required both for efficient FANCD2 monoubiquitination and in DNA repairs, later in the FA pathway.
Methods
Cell lines and culture conditions
Epstein-Barr virus–transformed lymphoblasts were cultured in RPMI 1640 medium or Iscove modified Dulbecco medium supplemented with 10% fetal calf serum. EUFA867 lymphoblasts stably expressing FANCA were generated by electroporation of an episomal pMEP4 construct encoding FANCA with a C-terminal flag-tag and selection on hygromycin, by transduction with retroviral vector pMMP-FANCA and selection on puromycin, or by transduction with bicistronic retrovirus SF91-FANCA and sorting transduced EGFP positive by fluorescence-activated cell sorting (FACS). The mitomycin C (MMC)–, camptothecin-, and UV-induced growth inhibition assays as well as the cell fusions were performed as previously described.25
Retroviral infections
The pMIEG3 bicistronic retroviral vector, bicistronic retroviral vectors expressing FA proteins, and pMMP-FANCA have been described.26,27 FANCM was cloned into BamHI and XhoI sites of pMIEG3 to generate pMIEG3-FANCM. Similarly K117R-FANCM and delC-FANCM were subcloned into pMIEG3. Retroviral vectors SF91-FANCM (without IRES-EGFP) and SF91-FANCA (with IRES-EGFP) were also constructed.
To prepare transient amphotropic virus stocks, 293T cells (1.5 × 106) were plated in 10-cm dishes. The next day, cells were transfected with the retroviral expression vectors and the appropriate helper plasmid (gag-pol and RD114) using Lipofectamine 2000. The medium was changed 5 hours after transfection and retrovirus-containing medium was harvested in 12-hour increments at 24 hours after transfection.
Lymphoblasts were transduced in 6-well plates coated with fibronectin fragment CH-296 (8 Ag/cm2; Takara Shuzo). Approximately 5 × 105 cells per well were exposed twice to the virus supernatant for 6 hours.26 Subsequently, the transduced cells were removed from the wells using cell dissociation buffer (Invitrogen) and transferred to new plates containing fresh medium. Two days after transduction, the transduced cells were sorted based on the EGFP expression using a FACSVantage cell sorter (BD Biosciences).
Antibodies
FANCD2 immunofluorescence
Cells were fixed with 2% paraformaldehyde in PBS for 20 minutes at room temperature (RT) and then deposited on 12-mm diameter glass coverslips coated with poly-d-lysine by centrifugation in a Thermo Shandon Cytospin apparatus (163g, 2 minutes). After washing with PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 3 minutes, and washed again with PBS. Then anti-FANCD2 antibody E35 (1:200) in antibody buffer (PBS/3% BSA/0.05% Tween 20/0.04% sodium azide) was added for 1 hour at 37°C with. Cells were washed with PBS and incubated for 30 minutes at 37°C with rhodamine B–conjugated donkey anti–rabbit IgG antibody (Jackson ImmunoResearch Laboratories; 1:200). Cells were washed with PBS, mounted by Vectashield with 4′,6-diamino-2-phenylindole (DAPI) to stain DNA, and analyzed with a Zeiss Axiovert 200M microscope.
RAD51 immunofluorescence
Lymphoblasts were transferred to 9-cm culture dishes at a density of 0.5 × 106 cells/mL and either mock-treated or treated with MMC (2.4 μg/mL for 1 hour) or x-ray irradiation (12 Gy). Cells (106) were seeded on poly-d-lysine–coated glass slides 7 or 24 hours after recovery from the treatment and left to attach for 15 minutes before fixation with 2% formaldehyde in PBS. Then cells were permeabilized with PBS/0.1% Triton X-100. Subsequently, the slides were blocked for 30 minutes in PBS/0.5% BSA/0.15% glycine and incubated with anti-Rad51 antiserum FBE2 for 90 minutes. The slides were washed in PBS/0.1% Triton X-100 and incubated with Alexa 488–conjugated goat anti–rabbit (Molecular Probes) for 1 hour at 37°C. After 3 washes with PBS/0.1% Triton X-100, the cells were counterstained with DAPI in Vectashield mounting medium and analyzed.
Preparation of cellular fractions
Cellular fractionation was as described.7 The cells were trypsinized and washed with ice cold PBS. Pellets were resuspended in cold buffer A (10 mM PIPES [pH 7.0], 100 mM NaCl, 300 mM sucrose, 1 mM EGTA, 0.5% Triton X-100, 50 mM sodium fluoride, 0.5 mM sodium orthovanadate, protease inhibitor cocktail, 1 mM phenylmethanesulphonylfluoride) at 5 times the volume of the cell pellet and incubated for 2 minutes at RT to permeabilize the cells. The suspension was centrifuged at 200g for 3 minutes, and the supernatant S100 (detergent-soluble cytonucleoplasmic proteins) was collected. Pellets were washed with cold buffer A and extracted with buffer B (buffer A with 400 mM NaCl) for 5 minutes to obtain supernatant S400 (chromatin fraction).
Cell-cycle analysis
To induce cell-cycle arrest, approximately 106 cells were plated in culture medium containing 0.5 μg/mL of the cross-linking agent melphalan (Sigma-Aldrich) for 48 hours. Cells were fixed for 20 minutes at RT in PBS/1% paraformaldehyde. Subsequently, cells were washed with PBS and resuspended for 10 minutes at RT in PBS/0.1% Triton X-100. After washing with PBS, cells were stained for 1 hour at 4°C with a solution of PBS containing 2 mg/mL RNase A (QIAGEN) and 50 mg/mL propidium iodide (Sigma-Aldrich). Flow cytometry was performed on a FACS Calibur flow cytometer (BD Biosciences) and analysis of the percentage of cells in each phase of the cell cycle was performed using the MODFIT-LT software (Verity Software House).
Drug sensitivity assay
Lymphoblasts (104) were cultured continuously for 5 days in 100 μL medium containing different concentrations of drug. Cell viability was measured using the chromogenic Cell Titer 96 Proliferation Assay (Promega Corporation) recording the optical density at 490 nm.
Results
FANCA mutations in patient EUFA867
The reference FA-M patient EUFA867 carries 2 pathogenic mutations in FANCM. As expected for cells with a defect in core complex proteins, lymphoblasts from this patient, which do not express FANCM protein, lack monoubiquitinated FANCD2.15 Surprisingly, ectopic FANCM expression was unable to restore FANCD2 monoubiquitination (Figure 1A lane 1 and supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article), suggesting an additional defect in this cell line. Expression of FANCA, but not of other FA proteins, in FANCM-expressing EUFA867 cells completely rescued FANCD2 monoubiquitination (Figure 1A lane 2), pointing to a FANCA defect in these lymphoblasts. This idea was strengthened by cell fusion experiments between EUFA867 lymphoblasts and FANCA-deficient HSC72 lymphoblasts, resulting in tetraploid cell hydrids that remained MMC sensitive (supplemental Figure 1B), whereas fusion hybrids with the FANCC-deficient cell line HSC536 were MMC resistant (supplemental Figure 1C). Indeed, sequencing revealed biallelic FANCA mutations in the EUFA867 cell line: a nonsense mutation (2557C>T, R853X), inherited from the father (Figure 1B and supplemental Figure 2), and a splice-site mutation in intron 7 (IVS7 + 5 G>A), inherited from the mother (Figure 1C and supplemental Figure 2). These mutations were confirmed in blood DNA of the patient and the parents, as well as in DNA from lymphoblastoid cell lines of the parents. The splice-site mutation, which has been observed previously,29,30 leads to a 30-bp insertion in the FANCA mRNA (Figure 1C). The insertion does not affect the FANCA open reading frame and adds 10 amino acids between amino acids 236 and 237 of the FANCA protein. However, the mutant FANCA protein was unable to restore FANCD2 monoubiquitination in FANCA-deficient lymphoblasts (Figure 1D), demonstrating that the splice-site mutation is pathogenic. The insertion is near the amino acids L210 and L274, which when mutated result in loss of function by abolishing the nuclear accumulation of FANCA.28 Because subcellular fractionation experiments in EUFA867 cells failed to detect FANCA in the nuclear fraction,15 the insertion probably affects the nuclear localization of FANCA and the interaction with other FA core complex members.
EUFA867 has biallelic FANCA mutations. (A) Introduction of FANCA in EUFA867 lymphoblasts stably expressing FANCM restores FANCD2 monoubiquitination. EUFA867 lymphoblasts were transduced with SF91-FANCM to obtain EUFA867 + FANCM. Subsequently these cells were transduced with bicistronic retroviruses encoding the indicated FA proteins. Cells were treated with 2 mM HU for 16 hours and immunoblotted for FANCD2 and FANCM. (B) EUFA867 has a nonsense mutation 2557C>T (R853X) in FANCA. (C) Mutation IVS7 + 5G>A affects the normal splice donor in FANCA intron 7 and results in a 30-bp insertion of intron 7 sequence in the cDNAs of EUFA867 and her mother. The sequence shown is from an isolated cDNA clone of EUFA867. (D) FANCA cDNA derived from the IVS7 + 5G>A mutation does not correct FANCD2 monoubiquitination. FANCA-deficient HSC72 lymphoblasts were transduced with bicistronic retrovirus encoding either wild-type FANCA or the FANCA mutant derived from the mutation IVS7 + 5G>A. Cells were treated with 1 mM HU to stimulate FANCD2 monoubiquitination.
EUFA867 has biallelic FANCA mutations. (A) Introduction of FANCA in EUFA867 lymphoblasts stably expressing FANCM restores FANCD2 monoubiquitination. EUFA867 lymphoblasts were transduced with SF91-FANCM to obtain EUFA867 + FANCM. Subsequently these cells were transduced with bicistronic retroviruses encoding the indicated FA proteins. Cells were treated with 2 mM HU for 16 hours and immunoblotted for FANCD2 and FANCM. (B) EUFA867 has a nonsense mutation 2557C>T (R853X) in FANCA. (C) Mutation IVS7 + 5G>A affects the normal splice donor in FANCA intron 7 and results in a 30-bp insertion of intron 7 sequence in the cDNAs of EUFA867 and her mother. The sequence shown is from an isolated cDNA clone of EUFA867. (D) FANCA cDNA derived from the IVS7 + 5G>A mutation does not correct FANCD2 monoubiquitination. FANCA-deficient HSC72 lymphoblasts were transduced with bicistronic retrovirus encoding either wild-type FANCA or the FANCA mutant derived from the mutation IVS7 + 5G>A. Cells were treated with 1 mM HU to stimulate FANCD2 monoubiquitination.
FANCM deficiency leads to cross-linker sensitivity and reduced FANCD2 monoubiquitination
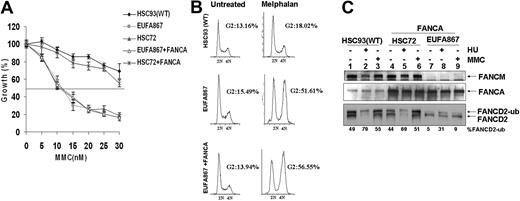
These new genetic data led us to re-examine the role of FANCM in the FA pathway. We first corrected the FANCA defect in EUFA867 lymphoblasts to create a cell line that is deficient only in FANCM. FANCA expression by either retroviral transduction or cDNA transfection did not restore MMC hypersensitivity in EUFA867 cells (Figure 2A and supplemental Figure 3A). In addition, the G2 arrest observed in EUFA867 cells after melphalan treatment was still present in the FANCA-expressing cells (Figure 2B), suggesting that FANCM deficiency is associated with cross-linker hypersensitivity. FANCA expression partially rescued the FANCD2 monoubiquitination defect seen in EUFA867 cells (Figure 2C and supplemental Figure 3B), which is very similar to the situation observed in FANCM-deficient mouse embryonic fibroblasts (S.T.B., J.P.d.W., H.J., and Hein te Riele, manuscript submitted, March 20, 2009). Strikingly MMC-induced monoubiquitination of FANCD2 is more severely impaired in FANCA-expressing EUFA867 cells than HU-induced monoubiquitination. (Figure 2C lanes 8-9 and Figure 3B lanes 1-2). These data indicate that FANCM is not essential for FANCD2 monoubiquitination, but may increase the efficiency of this process.
Stable expression of FANCA in EUFA867 lymphoblasts partially corrects the phenotype of this cell line. (A) EUFA867 lymphoblasts stably expressing wild-type FANCA are still hypersensitive to growth inhibition by mitomycin C. FANCA-deficient HSC72 and EUFA867 lymphoblasts were transduced with SF91-FANCA. Viable cells were measured with the Cell Titer 96 Proliferation Assay. The data represent the percentage growth compared with untreated cells and show 1 representative result of 3 independent experiments with standard deviations. (B) EUFA867 lymphoblasts stably expressing wild-type FANCA show melphalan-induced G2 arrest. (C) EUFA867 lymphoblasts stably expressing wild-type FANCA have reduced FANCD2 monoubiquitination. Cells were treated with either 2 mM HU or 240 nM MMC for 16 hours or left untreated. Total lysates were immunoblotted for FANCM, FANCD2, and FANCA.
Stable expression of FANCA in EUFA867 lymphoblasts partially corrects the phenotype of this cell line. (A) EUFA867 lymphoblasts stably expressing wild-type FANCA are still hypersensitive to growth inhibition by mitomycin C. FANCA-deficient HSC72 and EUFA867 lymphoblasts were transduced with SF91-FANCA. Viable cells were measured with the Cell Titer 96 Proliferation Assay. The data represent the percentage growth compared with untreated cells and show 1 representative result of 3 independent experiments with standard deviations. (B) EUFA867 lymphoblasts stably expressing wild-type FANCA show melphalan-induced G2 arrest. (C) EUFA867 lymphoblasts stably expressing wild-type FANCA have reduced FANCD2 monoubiquitination. Cells were treated with either 2 mM HU or 240 nM MMC for 16 hours or left untreated. Total lysates were immunoblotted for FANCM, FANCD2, and FANCA.
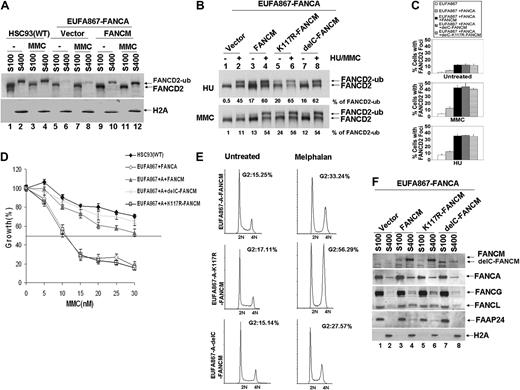
Ectopic expression of both FANCA and FANCM can correct the FA phenotype of EUFA867 lymphoblasts. (A) Ectopic expression of FANCA and FANCM in EUFA867 restores the chromatin localization of monoubiquitinated FANCD2, which coincides with an enhanced FANCD2 monoubiquitination. EUFA867 cells stably expressing FANCA (EUFA867-FANCA) were generated with pMMP-FANCA. Subsequently, these cells were transduced with either MIEG3 bicistronic retroviral vector or MIEG3 encoding wild-type FANCM. EGFP-positive cells were treated with 240 nM MMC for 16 hours or left untreated and subcellular fractions were made: S100 cytoplasmic and nucleoplasmic proteins, S400 chromatin-bound proteins. (B) The ATPase activity and the C-terminus of FANCM are not required for efficient FANCD2 monoubiquitination. EUFA867 lymphoblasts stably expressing FANCA were transduced with either MIEG3 retroviral vector or MIEG3 encoding wild-type FANCM, an ATPase-dead FANCM mutant (K117R-FANCM) or a C-terminal FANCM deletion mutant (delC-FANCM). EGFP-positive cells were treated with either 2 mM HU (top panel) or 240 mM MMC (bottom panel) for 16 hours. Total lysates were immunoblotted for FANCD2. (C) FANCM, but not its ATPase activity or its C-terminus, is required for the assembly of FANCD2 foci. Cells were either left untreated or exposed to 450 nM MMC or 2 mM HU for 24 hours, and the percentage of cells with 5 or more FANCD2 foci was determined in at least 150 cells. The result shows the average of 3 independent experiments with standard deviations. (D) The ATPase activity, but not the C-terminus of FANCM, is required for MMC resistance. Viable cells were measured with the Cell Titer 96 Proliferation Assay. The data represent the percentage growth compared with untreated cells and show 1 representative result of 3 independent experiments with standard deviations. (E) The FANCM ATPase mutant does not rescue the melphalan-induced G2 arrest in EUFA867 lymphoblasts. (F) FANCM is required for chromatin targeting of the FA core complex proteins. Subcellular fractions of EUFA867 lymphoblast and stably transduced derivatives were immunoblotted for FANCM, FANCA, FANCG, FANCL, and FAAP24. H2A was used as a loading control for the chromatin fraction.
Ectopic expression of both FANCA and FANCM can correct the FA phenotype of EUFA867 lymphoblasts. (A) Ectopic expression of FANCA and FANCM in EUFA867 restores the chromatin localization of monoubiquitinated FANCD2, which coincides with an enhanced FANCD2 monoubiquitination. EUFA867 cells stably expressing FANCA (EUFA867-FANCA) were generated with pMMP-FANCA. Subsequently, these cells were transduced with either MIEG3 bicistronic retroviral vector or MIEG3 encoding wild-type FANCM. EGFP-positive cells were treated with 240 nM MMC for 16 hours or left untreated and subcellular fractions were made: S100 cytoplasmic and nucleoplasmic proteins, S400 chromatin-bound proteins. (B) The ATPase activity and the C-terminus of FANCM are not required for efficient FANCD2 monoubiquitination. EUFA867 lymphoblasts stably expressing FANCA were transduced with either MIEG3 retroviral vector or MIEG3 encoding wild-type FANCM, an ATPase-dead FANCM mutant (K117R-FANCM) or a C-terminal FANCM deletion mutant (delC-FANCM). EGFP-positive cells were treated with either 2 mM HU (top panel) or 240 mM MMC (bottom panel) for 16 hours. Total lysates were immunoblotted for FANCD2. (C) FANCM, but not its ATPase activity or its C-terminus, is required for the assembly of FANCD2 foci. Cells were either left untreated or exposed to 450 nM MMC or 2 mM HU for 24 hours, and the percentage of cells with 5 or more FANCD2 foci was determined in at least 150 cells. The result shows the average of 3 independent experiments with standard deviations. (D) The ATPase activity, but not the C-terminus of FANCM, is required for MMC resistance. Viable cells were measured with the Cell Titer 96 Proliferation Assay. The data represent the percentage growth compared with untreated cells and show 1 representative result of 3 independent experiments with standard deviations. (E) The FANCM ATPase mutant does not rescue the melphalan-induced G2 arrest in EUFA867 lymphoblasts. (F) FANCM is required for chromatin targeting of the FA core complex proteins. Subcellular fractions of EUFA867 lymphoblast and stably transduced derivatives were immunoblotted for FANCM, FANCA, FANCG, FANCL, and FAAP24. H2A was used as a loading control for the chromatin fraction.
To further investigate the function of FANCM, chromatin fractions of FANCA-transfected EUFA867 cells were analyzed and compared with EUFA867 cells in which both FANCA and FANCM defects were corrected. In the absence of FANCM, less monoubiquitinated FANCD2 was found in the chromatin fraction, which could be restored by FANCM expression (Figure 3A). This increased chromatin binding coincided with an enhanced FANCD2 monoubiquitination (Figure 3B), the appearance of FANCD2 foci (Figure 3C), a restoration of the MMC hypersensitivity (Figure 3D), and a reduction of melphalan-induced G2 arrest (Figure 3E). In addition, FANCG, FANCL, and FAAP24 levels, which were decreased in the chromatin fraction of FANCA-corrected EUFA867 cells, were normalized by FANCM expression (Figure 3F). As has been suggested from siRNA experiments in HeLa cells,7 our data show that FANCM assists the recruitment of the FA core complex to the chromatin, which then may increase FANCD2 in chromatin-bound foci through monoubiquitination.
The ATPase activity of FANCM is essential for cross-linker tolerance
FANCM contains 2 interesting motifs: an N-terminal DEAH helicase domain and a C-terminal ERCC4-like nuclease domain. The involvement of these domains in cross-linker resistance was investigated by expressing either a FANCM ATPase point mutant (K117R) or a C-terminal FANCM deletion mutant in FANCA-corrected EUFA867 cells. Like wild-type FANCM, both mutants restored the monoubiquitination of FANCD2 and its ability to form nuclear foci (Figure 3B-C). However, only the C-terminal deletion mutant was able to restore MMC resistance (Figure 3D) and melphalan-induced G2 arrest (Figure 3E). These data confirm previous siRNA experiments in HeLa cells, showing that the K117R mutant supports normal FANCD2 and FANCI monoubiquitination, but is unable to induce cross-linker resistance.31 Interestingly, this ATPase point mutant assisted in the recruitment of FANCG, FANCL, and FAAP24 to the chromatin (Figure 3F), indicating that the ATPase activity of FANCM is essential for cross-linker resistance by supporting a step that is independent of the FA core complex.
The C-terminal deletion mutant seems to have a reduced chromatin association and only partially supports the chromatin binding of FANCG and FANCL (Figure 3F), but apparently this is sufficient to complement cross-linker sensitivity. In line with the role of the FANCM C-terminus in binding FAAP24,32 FAAP24 is absent in the chromatin fraction of cells transfected with the C-terminal deletion mutant (Figure 3F). This could imply that, in contrast to HeLa cells, in which knockdown of FAAP24 by siRNA resulted in a cross-linker–sensitive phenotype,32 lymphoblasts may not depend on FAAP24 for cross-linker resistance. The same could hold for the chicken B-cell line DT40, because ablation of the FANCM C-terminus in these cells resulted in a moderate level of cross-linker sensitivity.16
FANCM deficiency is associated with camptothecin and UV sensitivity
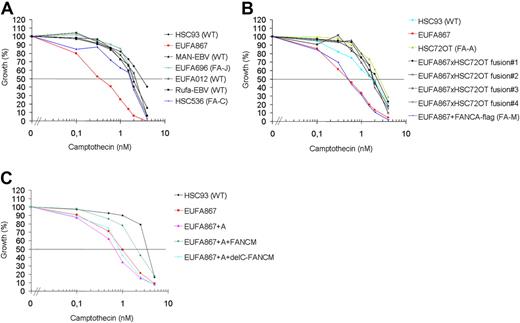
Recently it has been shown that purified FANCM can promote branch migration of Holliday junctions and replication forks.22 Therefore, it was postulated that FANCM, apart from its function in the FA pathway, might have additional functions in the remodeling of replication intermediates. We speculated that if FANCM indeed has additional functions, cells deficient for FANCM might be sensitive to a broader range of genotoxic agents. Therefore, we tested various DNA-damaging agents in growth inhibition assays on EUFA867 lymphoblasts and found that this cell line was hypersensitive to the topoisomerase I inhibitor camptothecin (Figure 4A). This hypersensitivity was specific for FANCM deficiency, because FANCA overexpression in EUFA867 cells did not restore camptothecin resistance, whereas introduction of FANCM by fusion of EUFA867 lymphoblasts with FANCA- or FANCC-deficient lymphoblasts corrected this phenotype (Figure 4B and supplemental Figure 4C). As expected from these cell fusion data, wild-type FANCM restored the camptothecin resistance, but surprisingly the C-terminal deletion mutant did not correct this defect (Figure 4C). This indicates that the C-terminus of FANCM is involved in the recovery of replication forks stalled by topoisomerase I-DNA cleavage intermediates fixed on the DNA by topoisomerase I inhibitors.
Camptothecin sensitivity in human lymphoblasts. (A) EUFA867 lymphoblasts are sensitive to the topoisomerase I inhibitor camptothecin. Lymphoblasts were continuously exposed to different doses of camptothecin and cell growth was compared with untreated cells by cell counting. Wild-type, and FANCC- and FANCJ-deficient lymphoblasts were included as camptothecin-resistant controls. (B) Camptothecin sensitivity of EUFA867 lymphoblasts is due to a defect in FANCM. The FANCA defect in EUFA867 was corrected by stable transfection of flag-tagged FANCA (EUFA867+FANCA-flag); the FANCM defect was corrected by cell fusion with FANCA-deficient lymphoblasts HSC72 (EUFA867xHSC72OT fusion). Four independent cell fusions are depicted. (C) The C-terminus of FANCM is involved in camptothecin resistance. EUFA867 cells were stably transduced with FANCA and wild-type FANCM (FANCM) or a C-terminal deletion mutant of FANCM (delC-FANCM).
Camptothecin sensitivity in human lymphoblasts. (A) EUFA867 lymphoblasts are sensitive to the topoisomerase I inhibitor camptothecin. Lymphoblasts were continuously exposed to different doses of camptothecin and cell growth was compared with untreated cells by cell counting. Wild-type, and FANCC- and FANCJ-deficient lymphoblasts were included as camptothecin-resistant controls. (B) Camptothecin sensitivity of EUFA867 lymphoblasts is due to a defect in FANCM. The FANCA defect in EUFA867 was corrected by stable transfection of flag-tagged FANCA (EUFA867+FANCA-flag); the FANCM defect was corrected by cell fusion with FANCA-deficient lymphoblasts HSC72 (EUFA867xHSC72OT fusion). Four independent cell fusions are depicted. (C) The C-terminus of FANCM is involved in camptothecin resistance. EUFA867 cells were stably transduced with FANCA and wild-type FANCM (FANCM) or a C-terminal deletion mutant of FANCM (delC-FANCM).
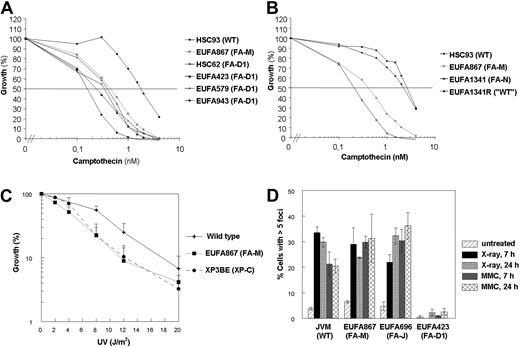
The camptothecin sensitivity is specific for EUFA867 lymphoblasts and was not found in lymphoblasts with a defect in other FA core complex members or in FANCD2, FANCI, or FANCJ (Figure 4A, supplemental Figure 4D,G). In contrast, we observed camptothecin sensitivity in 4 independent FANCD1/BRCA2-deficient lymphoblasts (Figure 5A) and in FANCN/PALB2-deficient lymphoblasts (Figure 5B). These results suggest that FANCJ and BRCA2/PALB2 function in 2 different branches of the FA pathway and that FANCM might have a role in the BRCA2/PALB2 branch, which is thought to be connected to homologous recombination. Like BRCA2-deficient cells,33 EUFA867 cells were found to be UV sensitive (Figure 5C), which is probably not due to the FANCA defect in this cell line because FANCA-deficient cells were shown to be UV resistant.33 However, the normal formation of DNA damage–induced Rad51 foci in EUFA867 cells (Figure 5D) indicates that, unlike BRCA2 and PALB2, FANCM does not play an essential role in homologous recombination repair.
Camptothecin and UV sensitivity and Rad51 focus formation in human lymphoblasts. (A) FA-D1 lymphoblasts with a defect in FANCD1/BRCA2 are as sensitive to growth inhibition by camptothecin as EUFA867 lymphoblasts. (B) FA-N lymphoblasts with a defect in FANCN/PALB2 are as sensitive to growth inhibition by camptothecin as EUFA867 lymphoblasts. EUFA1341R is a MMC-resistant reverted derivative of EUFA1341 lymphoblasts. (C) EUFA867 lymphoblasts are sensitive to UV. Lymphoblasts were exposed to different doses of UVC light and cell growth was compared with untreated cells by cell counting. Lymphoblasts of xeroderma pigmentosum patient XP3BE were used as a positive control. Results show mean values of at least 5 experiments with SEM. (D) Normal Rad51 focus formation in EUFA867 lymphoblasts. Kinetics of Rad51 foci formation in response to x-ray irradiation (12 Gy) or MMC treatment (2.4 μg/mL for 1 hour) analyzed 7 and 24 hours after treatment. A cell with more than 5 distinct foci in the nucleus was considered positive. Results show the mean values of at least 2 experiments with standard error of the mean.
Camptothecin and UV sensitivity and Rad51 focus formation in human lymphoblasts. (A) FA-D1 lymphoblasts with a defect in FANCD1/BRCA2 are as sensitive to growth inhibition by camptothecin as EUFA867 lymphoblasts. (B) FA-N lymphoblasts with a defect in FANCN/PALB2 are as sensitive to growth inhibition by camptothecin as EUFA867 lymphoblasts. EUFA1341R is a MMC-resistant reverted derivative of EUFA1341 lymphoblasts. (C) EUFA867 lymphoblasts are sensitive to UV. Lymphoblasts were exposed to different doses of UVC light and cell growth was compared with untreated cells by cell counting. Lymphoblasts of xeroderma pigmentosum patient XP3BE were used as a positive control. Results show mean values of at least 5 experiments with SEM. (D) Normal Rad51 focus formation in EUFA867 lymphoblasts. Kinetics of Rad51 foci formation in response to x-ray irradiation (12 Gy) or MMC treatment (2.4 μg/mL for 1 hour) analyzed 7 and 24 hours after treatment. A cell with more than 5 distinct foci in the nucleus was considered positive. Results show the mean values of at least 2 experiments with standard error of the mean.
Discussion
FANCM was classified as an FA gene based upon the discovery of pathogenic mutations in FA patient EUFA867, but ectopic FANCM expression in lymphoblasts from the patient has been unable to rescue its cellular phenotype.15 Here we show that this was probably due to an additional FANCA defect in this patient. Stable FANCA expression in EUFA867 cells revealed the consequences of FANCM deficiency, for example, hypersensitivity to DNA cross-linking agents and to the topoisomerase I inhibitor camptothecin as well as impaired FANCD2 monoubiquitination. In addition, correction of the FANCA defect now allowed functional complementation studies with FANCM and FANCM mutants, which showed that the C-terminus of FANCM is essential for camptothecin resistance, but dispensable for cross-linker tolerance.
Our data indicate that patient EUFA867 is a very exceptional case, with 2 pathogenic FANCA mutations besides the biallelic FANCM mutations. Remarkably, the sibling of EUFA867, who was the first to be diagnosed with FA after the development of typical FA features, carried the same biallelic mutations in FANCA, but carried only one of the FANCM mutations, which in retrospect classifies this patient as an FA-A patient (supplemental Figure 2). EUFA867 was not suspected to have FA, because she failed to display any conspicuous clinical symptoms typical for FA. The basis of her diagnosis was the result of a chromosomal breakage assay performed because of the diagnosis of her brother. Although in general clinical features of FA patients are highly variable, the atypical appearance of EUFA867 may suggest that patients with a FANCM defect present with a different phenotype. As FANCM appears to be essential for the core complex functions.7 the FANCM deficiency in EUFA867 may have overruled the FANCA defect and changed the clinical outcome in this patient. A similar situation has been observed in DT40 cells, in which disruption of FANCM in a FANCC-deficient background attenuates cisplatin toxicity.16 This hypothesis is strengthened by the atypical phenotype of FANCM-deficient mice (S.T.B., J.P.d.W., H.J., and Hein te Riele, manuscript submitted, March 20, 2009) and may explain why the number of FA patients diagnosed with a FANCM defect is still limited to patient EUFA867. Our data warrant FANCM mutation screening in related syndromes of which the disease gene has not been identified.
Although the absence of FANCM may lead to a phenotype different from defects in other FA core complex members, FANCM-deficient cells showed cross-linker sensitivity and lacked FANCD2 foci, indicating that FANCM plays an essential role in the FA pathway. The residual monoubiquitination observed in EUFA867 lymphoblasts stably expressing FANCA suggests that this role does not involve the formation of the FA core complex. Chromatin fractionation studies in the FANCA-corrected EUFA867 cells indicated that FANCM is rather responsible for the recruitment of the FA core complex to the chromatin, which may than allow efficient FANCD2 monoubiquitination and focus formation. This function does not require ATP hydrolysis, because the K117R FANCM mutant was able to support this step and also the C-terminus of FANCM appeared to be dispensable for this process. The FANCM mutants do point to a role for FANCM in DNA repair at the later part of the FA pathway after FANCD2 monoubiquitination. The K117R mutant did not restore MMC resistance, indicating that an ATP-dependent step is needed after FANCD2 is monoubiquitinated, which probably involves the ATPase-dependent remodeling of the stalled replication fork.23 The C-terminus of FANCM was not critical for cross-linker resistance, but seemed to be essential for the repair of camptothecin-induced DNA damage, possibly through its interaction with FAAP24.
The camptothecin sensitivity of FANCM-deficient lymphoblasts was shared with lymphoblasts defective in BRCA2 or PALB2, thus suggesting a role for FANCM in the DNA repair branch of the FA pathway connected to homologous recombination repair. However, unlike lymphoblasts with a mutation in BRCA2 or PALB2, EUFA867 lymphoblasts showed normal Rad51 foci, indicating that FANCM is not essential for homologous recombination repair. FANCM may have a more regulatory function in the homologous recombination process as suggested by the role of the yeast FANCM orthologs MPH1 (S cerevisiae) and FML1 (S pombe), in replication fork reversal and D-loop disruption.17-19 The increased sister chromatid exchanges in FANCM-deficient DT40 cells16 and FANCM-deficient mouse embryonic fibroblasts (S.T.B., J.P.d.W., H.J., and Hein te Riele, manuscript submitted, March 20, 2009) hint at a role for FANCM in the suppression of crossover recombination.
In summary, we have shown that EUFA867 lymphoblasts are defective in 2 distinct FA core complex components, FANCA and FANCM, which explains why FANCM expression has been ineffective in complementing this cell line. This discovery also clarifies the apparent discrepancies with regard to FA core complex stability and FANCD2 monoubiquitination between EUFA867 lymphoblasts and HeLa cells depleted of FANCM by siRNA7,31 as well as FANCM DT40 knockouts.16 Correction of the FANCA defect in EUFA867 cells partially restored FANCD2 monoubiquitination, but these cells are still cross-linker sensitive. EUFA867 lymphoblasts were also hypersensitive to camptothecin and UV light, a phenotype not previously known to be associated with FANCM deficiency. FANCD2 monoubiquitination and focus formation as well as cross-linker and camptothecin sensitivity were restored by ectopic FANCM expression. However, a C-terminal FANCM deletion mutant normalized only FANCD2 monoubiquitination and cross-linker sensitivity, but not the camptothecin sensitivity. The ATPase activity of FANCM is required for cross-linker resistance but not for FANCD2 monoubiquitination and focus formation. Our data thus suggest specific roles for different parts of the FANCM protein in the DNA damage response, both for efficient FANCD2 monoubiquitination and DNA repair steps later in the FA pathway.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Weidong Wang, Alan D'Andrea, Steve West, and Fiona Benson for reagents. We thank Drs Chris Mathew and Inderjeet Dokal for the clinical data of the FA-M patients.
This work was supported by National Institutes of Health research grants R01 HL084082 (A.R.M.) and R01 HL081499 (D.A.W.), the Fanconi Anemia Research Fund (A.R.M.), the American Society of Hematology Junior Faculty Award (A.R.M.), the Dutch Cancer Society (H.J.), and the Netherlands Organisation for Health Research and Development (J.P.d.W.).
National Institutes of Health
Authorship
Contribution: T.R.S. designed and performed research; S.T.B., S.A., and B.C.G. performed research and edited the paper; M.J., E.G., A.M.A., C.-h.D., M.A.R., Q.F., K.W., J.S., and P.R.A. performed research; D.A.W. designed research and controlled data; H.J. controlled data and edited the paper; and J.P.d.W. and A.R.M. designed research, controlled data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Johan P. de Winter, Department of Clinical Genetics, VU University Medical Center, Van der Boechorststraat 7, 1081 BT Amsterdam, The Netherlands; e-mail: j.dewinter@vumc.nl; or Amom Ruhikanta Meetei, Division of Experimental Hematology, Cincinnati Children's Research Foundation, University of Cincinnati College of Medicine, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: ruhikanta.meetei@cchmc.org.