Xanthohumol (XN), a prenylated chalcone isolated from hop plant, exhibits anti-inflammatory, antiproliferative, and antiangiogenic properties through an undefined mechanism. Whether examined by intracellular esterase activity, phosphatidylserine externalization, DNA strand breaks, or caspase activation, we found that XN potentiated tumor necrosis factor–induced apoptosis in leukemia and myeloma cells. This enhancement of apoptosis correlated with down-regulation of nuclear factor-κB (NF-κB) survivin, bcl-xL, XIAP, cIAP1, cIAP2, cylin D1, and c-myc. XN down-regulated both constitutive and inducible NF-κB activation, inhibition of phosphorylation and degradation of IκBα, suppression of p65 nuclear translocation, and NF-κB–dependent reporter gene transcription. XN directly inhibited tumor necrosis factor-induced IκBα kinase (IKK) activation and a reducing agent abolished this inhibition, indicating the role of cysteine residue. XN had no effect on the IKK activity when cysteine residue 179 of IKK was mutated to alanine. XN also directly inhibited binding of p65 to DNA, a reducing agent reversed this effect, and mutation of cysteine residue 38 to serine of p65 abolished this effect. Thus, our results show that modification of cysteine residues of IKK and p65 by XN leads to inhibition of the NF-κB activation pathway, suppression of antiapoptotic gene products, and potentiation of apoptosis in leukemia cells.
Introduction
Although traditional therapies using natural sources have been used for thousands of years, neither the active components nor their molecular targets have been very well defined. Identification of the active chemical entities and molecular targets of these natural products is an active area of research. Up to 70% of all drugs currently used for the treatment of cancer were derived from natural sources.1 In particular, studies have shown that xanthohumol (XN; 2′,4′,6′,4-tetrahydroxy-3′-prenylchalcone), a prenylated chalcone isolated from the hop plant (Humulus lupulus L.),2 inhibits the growth of different types of human cancer cells (including breast, colon, ovarian, and prostate), leukemia cells, and adipocytes,3,,,,,,,–11 and prevents the development of carcinogen-induced preneoplastic lesions in mouse mammary gland organ culture.5 Researchers also showed that this chalcone inhibits tumor-cell invasion,12 angiogenesis,13 and bone resorption.7 How XN mediates these effects is not fully understood. XN has been shown to inhibit nuclear factor-κB (NF-κB) activation,13,14 suppress the activity of diacylglycerol acyltransferase, which is involved in triglyceride synthesis,15,16 down-regulate topoisomerase I17 and aromatase,18 and inhibit nitric oxide19 and prostaglandin E2 production.5 In addition, others have described both caspase-dependent3 and -independent6 activation of apoptosis by XN. Furthermore, this agent inhibits phase 1 cytochrome P450 enzyme, which is involved in metabolic activation of carcinogens20 and induces phase 2 enzyme NAD(P)H:quinone reductase.21 XN was found to activate the farnesoid X receptor (FXR),22 inhibits triglyceride and apolipoprotein B secretion,23 and exhibits antidiabetic activity through the inhibition of lipid and glucose metabolism.22
Because the ability of XN to control cellular proliferation, cell survival, invasion, angiogenesis, and inflammation is closely associated with expression of gene products regulated by NF-κB, we postulated that XN must mediate most of these effects by regulating the NF-κB signaling cascade. Thus, in this study, we investigated in detail the effects of XN on different steps leading to NF-κB activation, NF-κB regulation of gene products, and NF-κB–regulated cellular responses. The results showed, for the first time, that modification of the cysteine residues in IκBα kinase (IKK) and p65 by XN leads directly to suppression of NF-κB–regulated gene products and potentiation of apoptosis in human leukemia and myeloma cells. We also examined these effects of XN to determine whether they are mediated through activation of FXR.
Methods
Reagents
A 50-mM solution of XN (Axxora Life Sciences, San Diego, CA) was prepared initially in dimethyl sulfoxide, stored as small aliquots at −20°C, and then thawed and diluted in a cell-culture medium as required. Bacteria-derived human recombinant tumor necrosis factor (TNF), purified to homogeneity with a specific activity of 5 × 107 U/mg, was provided by Genentech (South San Francisco, CA). Penicillin, streptomycin, RPMI 1640, Iscove modified Dulbecco medium, and Dulbecco modified Eagle medium were obtained from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was supplied by Atlanta Biologicals (Norcross, GA). Antibodies against p65, p50, IκBα, cyclin D1, cyclooxygenase-2, matrix mellatoproteinase-9 (MMP-9), poly (ADP-ribose) polymerase (PARP), inhibitor of apoptosis protein-1 (IAP-1), IAP-2, Bcl-2, Bcl-xL, and intercellular adhesion molecule-1 and the Annexin V Staining Kit were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). For immunocytochemistry, an antibody against p65 was obtained from Abcam (Cambridge, MA). An anti–vascular endothelial growth factor (VEGF) antibody was purchased from NeoMarkers (Fremont, CA). Phosphospecific anti-IκBα (serine 32 and 36) and phosphospecific anti-p65 (Ser536) antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-FXR, anti–IKK-α, and anti–IKK-β antibodies were provided by Imgenex (San Diego, CA). Expression plasmids for transforming growth factor-β–activated kinase 1 (TAK1) were described previously.24
Cell lines
The cell lines KBM-5 (human chronic myeloid leukemia), U937 (human histocytic leukemic), HL-60 (human promyelocytic leukemia), Jurkat (T-cell leukemia), A293 (human embryonic kidney carcinoma), H1299 (human lung adenocarcinoma), U266 (human multiple myeloma), and MCF-7 (breast cancer) were obtained from the ATCC (Manassas, VA). K562 (human chronic myeloid leukemia) was a gift from Dr Hesham Amin (University of Texas M. D. Anderson Cancer Center, Houston, TX). KBM-5 cells were cultured in Iscove modified Dulbecco medium with 15% FBS; H1299, MCF-7, K562, U937, HL-60, Jurkat, and U266 cells were cultured in RPMI 1640; and A293 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% FBS. Culture media were supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin.
Plasmids
The pcDNA3.1 and pcDNA expression vectors for murine p65 and murine p65C38S were kindly provided by T. D. Gilmore (Boston University, Boston, MA).
Electrophoretic mobility shift assay
To assess NF-κB, AP-1 and SP-1 activation, nuclear extracts were prepared, and electrophoretic mobility shift assay (EMSA) was performed as described previously.25
Western blot analysis
Western blot analysis was performed as described previously.24
IKK assay
To determine the effect of XN on TNF-induced IKK activation, we analyzed IKK activation using an immunocomplex kinase assay with glutathione S-transferase–IκBα as the substrate as described previously.25
Immunocytochemical analysis for NF-κB p65 localization
The effect of XN on TNF-induced nuclear translocation of p65 was examined using an immunocytochemical method. Slides were analyzed under a fluorescence microscope (Labophot-2; Nikon, Tokyo, Japan), and images were captured using a Photometrics Coolsnap CF color camera (Nikon) as described previously.26
NF-κB–dependent reporter gene expression assay
An NF-κB–dependent reporter gene expression assay was performed as described previously.27 The effect of XN on TNF-dependent, TNF-receptor associated factor-2 (TRAF-2)–dependent, NF-κB–inducing kinase (NIK)–dependent, TAK1-dependent, and TNF receptor–associated death domain (TRADD)–dependent reporter gene expression was analyzed using a secretory alkaline phosphatase (SEAP) assay.
Live/dead assay
The live/dead assay (Invitrogen), which assesses plasma membrane integrity, was used to measure the intracellular esterase activity. This assay was performed as described previously.25
Annexin V assay
To identify phosphatidylserine externalization during the process of apoptosis, cells were stained with an annexin V antibody conjugated with the fluorescent dye fluorescein isothiocyanate (FITC). In brief, 5 × 105 cells were coincubated with 50 μM XN and 1 nM TNF for 6 hours, stained with annexin-FITC conjugate, and then analyzed using a flow cytometer (FACSCalibur; BD Biosciences, San Jose, CA).
Terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling
To measure the DNA-strand breaks during apoptosis, we used the terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) assay, which uses the In Situ Cell Death Detection Reagent (Roche Molecular Biochemicals, Indianapolis, IN). In brief, 5 × 105 cells were coincubated with 50 μM XN and 1 nM TNF for 12 hours and then incubated with a reaction mixture. Stained cells were analyzed using a flow cytometer (FACSCalibur; BD Biosciences).
Invasion assay
Invasion through the extracellular matrix is a crucial step in tumor-cell metastasis. To determine the effect of XN on TNF-induced invasion, we used the BD BioCoat tumor invasion system (BD Biosciences) and the assay protocol followed was similar to that described previously.25
Statistical analysis
Experiments were repeated minimum 3 times with consistent results. Data are expressed as the mean plus or minus SD. Analysis of statistical significance between groups was made using a 2-tailed unpaired Student t test (*P ≤ .01, **P ≤ .05, ***P ≤ .001).
Results
The aim of the present study was to determine whether XN modulates NF-κB–mediated cellular responses, NF-κB–regulated gene expression, and the NF-κB–signaling pathway. We conducted most of our experiments using KBM-5 with TNF as an inducer of biologic response. These cells express both types of TNF receptors. Moreover, the TNF-induced NF-κB activation cascade is well characterized.
XN enhances TNF-induced apoptosis in leukemia and myeloma-cells
To determine whether XN modulates TNF-induced apoptosis in KBM-5-cells, we performed the live/dead assay, which measures intracellular esterase activity and assesses plasma membrane integrity. We found that XN increased the TNF-induced apoptosis from 6% to 75% (Figure 1A). We also evaluated XN to determine whether it modulates TNF-induced apoptosis in other cell types. We observed that XN increased the TNF-induced apoptosis rate in U266 cells from 7% to 80% (Figure 1B). In contrast to tumor cells, normal human peripheral blood mononuclear cells when treated with XN (50 μM for 12 hours) had no effect on cell viability (data not shown).
XN potentiates the TNF-induced apoptosis in chronic myeloid leukemia cells. (A) XN potentiates the TNF-induced apoptosis in chronic myeloid leukemia cell line KBM-5 as determined by live/dead assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 24 hours. The cells were stained with a live/dead assay reagent for 30 minutes and then analyzed under a fluorescence microscope. (B) XN increases TNF-induced apoptosis in human multiple myeloma U-266 cells as determined by live/dead assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 24 hours. The cells were stained with a live/dead assay reagent for 30 minutes and then analyzed under a fluorescence microscope. (C) XN potentiates TNF-induced apoptosis in KBM-5 cells as determined by annexin V assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 6 hours. The cells were incubated with a fluorescein isothiocyanate-conjugated annexin V antibody and then analyzed using flow cytometry. (D) XN potentiates TNF-induced apoptosis in KBM-5 cells as determined using a TUNEL assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 12 hours. The cells were stained for TUNEL-positive cells and then analyzed using flow cytometry. (E) XN potentiates TNF-induced apoptosis in KBM-5 cells as determined by caspase-3 activation. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed using Western blotting with an anti-PARP antibody. (F) XN suppresses TNF-induced tumor-cell invasion. H1299 cells (2.5 × 104 cells/mL) were seeded in the top chamber of a Matrigel invasion chamber system overnight in the absence of serum and then treated with 50 μM XN. After incubation, the cells were treated with TNF in the presence of 1% serum and then assayed for invasion. The results are expressed as the fold activity of the untreated control.
XN potentiates the TNF-induced apoptosis in chronic myeloid leukemia cells. (A) XN potentiates the TNF-induced apoptosis in chronic myeloid leukemia cell line KBM-5 as determined by live/dead assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 24 hours. The cells were stained with a live/dead assay reagent for 30 minutes and then analyzed under a fluorescence microscope. (B) XN increases TNF-induced apoptosis in human multiple myeloma U-266 cells as determined by live/dead assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 24 hours. The cells were stained with a live/dead assay reagent for 30 minutes and then analyzed under a fluorescence microscope. (C) XN potentiates TNF-induced apoptosis in KBM-5 cells as determined by annexin V assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 6 hours. The cells were incubated with a fluorescein isothiocyanate-conjugated annexin V antibody and then analyzed using flow cytometry. (D) XN potentiates TNF-induced apoptosis in KBM-5 cells as determined using a TUNEL assay. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for 12 hours. The cells were stained for TUNEL-positive cells and then analyzed using flow cytometry. (E) XN potentiates TNF-induced apoptosis in KBM-5 cells as determined by caspase-3 activation. Cells (106/mL) were pretreated with 50 μM XN for 4 hours and then incubated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed using Western blotting with an anti-PARP antibody. (F) XN suppresses TNF-induced tumor-cell invasion. H1299 cells (2.5 × 104 cells/mL) were seeded in the top chamber of a Matrigel invasion chamber system overnight in the absence of serum and then treated with 50 μM XN. After incubation, the cells were treated with TNF in the presence of 1% serum and then assayed for invasion. The results are expressed as the fold activity of the untreated control.
We also examined XN to determine whether it potentiates TNF-induced apoptosis in KBM-5 cells when assessed according to phosphatidylserine externalization using the annexin V assay. The results shown in Figure 1C indicate that XN increased the TNF-induced apoptosis from 5% to 50%. Similarly, a DNA-strand break assay using TUNEL revealed that XN induced an increase in the apoptosis to the same extent (Figure 1D). In addition, we found that caspase-3–mediated PARP cleavage induced by TNF was accelerated substantially by the presence of XN (Figure 1E). These results indicated that XN significantly increased the apoptotic activity of TNF in myeloma and leukemia cells.
XN suppresses TNF-induced tumor-cell invasion
Whether XN can modulate TNF-induced tumor-cell invasion in vitro was determined using a Matrigel invasion assay. As shown in Figure 1F, XN inhibited the TNF-induced invasion of tumor cells.
XN inhibits the TNF-induced expression of cell-proliferative gene products
Studies have shown that NF-κB activation regulates the expression of genes, such as cyclin D1 and c-myc, which are involved in the proliferation of different types of tumor cells. Thus, we sought to determine whether XN affects the expression of cyclin D1 and c-myc induced by TNF- treatment in KBM-5 cells. We observed that TNF induced the expression of these gene products and the treatment with XN inhibited this expression (Figure 2A).
XN down-regulates the TNF-induced gene products linked to proliferation, apoptosis, and invasion of chronic myeloid leukemia cells. (A) XN suppresses the TNF-induced expression of proliferative proteins. KBM-5 cells were incubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed by Western blotting with the indicated antibodies. (B) XN inhibits the expression of TNF-induced antiapoptotic proteins. KBM-5 cells were incubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed by Western blotting with relevant antibodies. (C) XN inhibits the expression of TNF-induced metastatic proteins. KBM-5 cells were incubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed by Western blotting with relevant antibodies.
XN down-regulates the TNF-induced gene products linked to proliferation, apoptosis, and invasion of chronic myeloid leukemia cells. (A) XN suppresses the TNF-induced expression of proliferative proteins. KBM-5 cells were incubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed by Western blotting with the indicated antibodies. (B) XN inhibits the expression of TNF-induced antiapoptotic proteins. KBM-5 cells were incubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed by Western blotting with relevant antibodies. (C) XN inhibits the expression of TNF-induced metastatic proteins. KBM-5 cells were incubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and analyzed by Western blotting with relevant antibodies.
XN suppresses the TNF-induced expression of antiapoptotic genes
The activated form of NF-κB is a survival signal for tumor cells because the activation of NF-κB leads to expression of several antiapoptotic genes. We examined whether XN can modulate the expression of antiapoptotic genes, which are under the control of NF-κB, such as Bcl-xL, cIAP-1, cIAP-2, XIAP, survivin, and TRAF-2. The results showed that XN inhibited the expression of all of these gene products (Figure 2B).
XN suppresses the TNF-induced gene products involved in invasion and angiogenesis
Intercellular adhesion molecule-1 (ICAM-1) and MMP-9 have been implicated in the tumor-cell invasion and are regulated by NF-κB. Therefore, we evaluated the effect of XN on TNF-induced ICAM-1 and MMP-9 expression in KBM-5 cells. We found that XN inhibited the expression of both of these gene products (Figure 2C). Furthermore, VEGF is an angiogenic factor, and its expression is also regulated by NF-κB. We found that TNF-induced expression of VEGF was inhibited by XN (Figure 2C).
XN suppresses TNF-induced NF-κB activation
Both cellular response and gene products modulated by XN as described above are regulated by NF-κB activation. We investigated XN in detail to determine whether it modulates TNF-induced NF-κB activation using EMSA. We first treated cells with XN for different times and then exposed them to TNF for the activation of NF-κB. We found that XN by itself had no effect on NF-κB activation, but it suppressed NF-κB activation induced by TNF in a time-dependent manner, with optimum inhibition occurring at 4 hours (Figure 3A).
XN down-regulates the TNF-induced NF-κB activation induced by different agents and in different cell lines. (A) Time-dependent effect of XN on TNF-induced NF-κB activation. KBM-5 cells were preincubated with 50 μM XN for the indicated times and then treated with 0.1 nM TNF for 30 minutes. Nuclear extracts were prepared and assayed for NF-κB activation using EMSA. The fold activation of NF-κB and cell viability (CV) are shown at the bottom. (B) Dose-dependent effect of XN on TNF-induced NF-κB activation. KBM-5 cells were incubated with XN at the indicated concentrations for 4 hours and treated with 0.1 nM TNF for 30 minutes. Nuclear extracts were assayed for NF-κB activation using EMSA. The fold activation of NF-κB and cell viability (CV) are shown at the bottom. (C) XN inhibits NF-κB activation induced by CSC, H2O2, IL-1β, PMA, epidermal growth factor (EGF), LPS, okadaic acid (OA), and TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours and then treated with 0.1 nM TNF for 30 minutes, 500 nM okadaic acid for 4 hours, 250 μM/mL H2O2 for 2 hours, 25 ng/mL PMA for 2 hours, and 10 μg/mL LPS, 10 μg/mL CSC, and 100 nM IL-1β for 1 hour each. Nuclear extracts were analyzed for NF-κB activation using EMSA. The fold activation of NF-κB is shown at the bottom. (D) Effect of XN on constitutive NF-κB activation. Multiple myeloma U266 cells were incubated with XN at the indicated concentrations for 4 hours. Nuclear extracts were prepared and analyzed for NF-κB activation by EMSA. The fold activation of NF-κB is shown at the bottom. (E) Effect of XN on TNF induced NF-κB activation in other types of leukemia cells. K562, HL-60, U937, and Jurkat cells were incubated with XN at the indicated concentrations for 4 hours and treated with 0.1 nM TNF for 30 minutes. Nuclear extracts were assayed for NF-κB activation using EMSA.
XN down-regulates the TNF-induced NF-κB activation induced by different agents and in different cell lines. (A) Time-dependent effect of XN on TNF-induced NF-κB activation. KBM-5 cells were preincubated with 50 μM XN for the indicated times and then treated with 0.1 nM TNF for 30 minutes. Nuclear extracts were prepared and assayed for NF-κB activation using EMSA. The fold activation of NF-κB and cell viability (CV) are shown at the bottom. (B) Dose-dependent effect of XN on TNF-induced NF-κB activation. KBM-5 cells were incubated with XN at the indicated concentrations for 4 hours and treated with 0.1 nM TNF for 30 minutes. Nuclear extracts were assayed for NF-κB activation using EMSA. The fold activation of NF-κB and cell viability (CV) are shown at the bottom. (C) XN inhibits NF-κB activation induced by CSC, H2O2, IL-1β, PMA, epidermal growth factor (EGF), LPS, okadaic acid (OA), and TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours and then treated with 0.1 nM TNF for 30 minutes, 500 nM okadaic acid for 4 hours, 250 μM/mL H2O2 for 2 hours, 25 ng/mL PMA for 2 hours, and 10 μg/mL LPS, 10 μg/mL CSC, and 100 nM IL-1β for 1 hour each. Nuclear extracts were analyzed for NF-κB activation using EMSA. The fold activation of NF-κB is shown at the bottom. (D) Effect of XN on constitutive NF-κB activation. Multiple myeloma U266 cells were incubated with XN at the indicated concentrations for 4 hours. Nuclear extracts were prepared and analyzed for NF-κB activation by EMSA. The fold activation of NF-κB is shown at the bottom. (E) Effect of XN on TNF induced NF-κB activation in other types of leukemia cells. K562, HL-60, U937, and Jurkat cells were incubated with XN at the indicated concentrations for 4 hours and treated with 0.1 nM TNF for 30 minutes. Nuclear extracts were assayed for NF-κB activation using EMSA.
In addition, we sought to determine the minimum dose of XN required to inhibit TNF-induced NF-κB activation. We exposed KBM-5 cells to XN at different concentrations and then exposed them to TNF for the activation of NF-κB. XN suppressed the TNF-induced NF-κB activation in a dose-dependent manner, with maximum inhibition occurring at 50 μM (Figure 3B).
NF-κB is a complex protein in which various combinations of members of the Rel family constitute NF-κB heterodimers with the ability to bind to specific sequence elements in DNA. Thus, to show that the band visualized in TNF-treated cells using EMSA was indeed NF-κB, we incubated nuclear extracts prepared from KBM-5 cells treated with TNF alone with antibodies specific for p50 and p65 followed by EMSA. We found that bands had shifted to higher molecular masses, indicating that the NF-κB complex activated by TNF consists of p65 and p50 subunits (data not shown).
XN was found to have no effect on TNF-induced AP-1 or SP-1 activation (Figure S1A,B, available on the Blood website; see the Supplemental Materials link at the top of the online article), thus indicating that the effect of XN on TNF-induced NF-κB activation is specific.
XN inhibits NF-κB activation induced by carcinogens and other inflammatory stimuli
Numerous agents, including cigarette smoke condensate (CSC), tumor promoters (eg, okadaic acid [OA], phorbol myristate acetate [PMA]), inflammatory agents such as hydrogen peroxide, lipopolysaccharide (LPS), interleukin-1β (IL-1β), and growth factors (eg, epidermal growth factor), are known to activate NF-κB. Work performed in our laboratory and others has shown that the mechanisms by which these agents induce activation of NF-κB vary significantly.28,,–31 Thus, we investigated whether XN affects NF-κB activation induced by these agents. The results showed that all of these agents activated NF-κB in KBM-5 cells and that XN suppressed the activation induced by these agents (Figure 3C), suggesting that XN acts at a step in the NF-κB activation pathway that is common to all of these agents.
XN inhibits constitutive NF-κB expression
Several tumor-cell types are known to constitutively express NF-κB through a mechanism yet to be fully defined.32 U266 multiple myeloma cells in particular are known to have constitutively active NF-κB. To determine whether XN affects NF-κB expression in these cells, we exposed them to XN at different concentrations for 4 hours and then analyzed them using EMSA. As shown Figure 3D, XN completely suppressed constitutive NF-κB activation in U266 cells, indicating that XN can suppress both inducible and constitutive NF-κB activation.
XN inhibits NF-κB activation in different type of leukemia cells
Whether XN modulates TNF-induced NF-κB activation in other types of leukemic cells was also examined. We found that the effect of XN is not limited to KBM-5 cells only but is also observed in chronic myeloid leukemia (K562), promyelomonocytic leukemia (HL-60), histiocytic lymphoma (U-937), and T-cell leukemia (Jurkat) as well (Figure 3E).
XN inhibits TNF-dependent IκBα phosphorylation and degradation
IκBα is the inhibitory subunit present in the NF-κB complex. Translocation of NF-κB to the nucleus is accompanied by phosphorylation, ubiquitination, and degradation of IκBα. To determine whether inhibition of TNF-induced NF-κB activation is caused by suppression of IκBα degradation, we pretreated KBM-5 cells with XN and then exposed them to TNF at various time points. In addition, we examined nuclear extracts for NF-κB activation using EMSA and cytoplasmic extracts for IκBα degradation using Western blotting. TNF activated NF-κB in a time-dependent manner; however, we observed no activation of NF-κB in XN-pretreated cells (Figure 4A).
XN down-regulates the TNF-induced NF-κB activation through inhibition of IκBα kinase. (A) XN inhibits TNF-induced activation of NF-κB. KBM-5 cells were incubated with 50 μM XN for 4 hours, treated with 0.1 nM TNF for the indicated times, and then analyzed for NF-κB activation by EMSA. The fold activation of NF-κB is shown at the bottom. (B) Effect of XN on TNF-induced degradation of IκBα. KBM-5 cells were incubated with 50 μM XN for 4 hours and treated with 0.1 nM TNF for the indicated times. Cytoplasmic extracts were prepared and analyzed by Western blotting using antibody against IκBα. Equal protein loading was evaluated using β-actin. (C) Effect of XN on phosphorylation of IκBα induced by TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours, incubated with 50 μg/mL N-acetyl-leucyl-leucyl-norleucinal (ALLN) for 30 minutes, and then treated with 0.1 nM TNF for 10 minutes. Cytoplasmic extracts were fractionated and then subjected to Western blot analysis with a phosphospecific anti-IκBα antibody (P-IκBα). Ser32/36. (D) Effect of XN on activation of IKK by TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were immunoprecipitated with an antibody against IKK-α and analyzed using an immune complex kinase assay. To determine the effect of XN on the level of expression of IKK proteins, whole-cell extracts were fractionated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and examined by Western blot analysis with anti–IKK-α and anti–IKK-β antibodies. (E) Direct effect of XN on IKK activation induced by TNF. Whole-cell extracts were prepared from KBM-5 cells treated with 1 nM TNF and immunoprecipitated with an anti–IKK-α antibody. The immunocomplex kinase assay was performed in the absence or presence of XN at the indicated concentrations. (F) Reversal of XN-induced suppression of TNF-induced IKK activation by the reducing agent DTT. Assays were performed as indicated in Figure 4E, except that the IKK activity was determined after treatment with DTT (100 μM), XN (50 μM), or both in kinase assay buffer. (G) The kinase activity of mutated IKK is unaffected by XN. For this, A293 cells were transfected with wild-type FLAG-IKK-β (IKK-β WT) or mutated FLAG-IKK-β (IKK-β MT [C179A]). Whole-cell extracts were prepared, and XN at the indicated concentrations was added in vitro. Immunocomplexes were analyzed for IKK activity.
XN down-regulates the TNF-induced NF-κB activation through inhibition of IκBα kinase. (A) XN inhibits TNF-induced activation of NF-κB. KBM-5 cells were incubated with 50 μM XN for 4 hours, treated with 0.1 nM TNF for the indicated times, and then analyzed for NF-κB activation by EMSA. The fold activation of NF-κB is shown at the bottom. (B) Effect of XN on TNF-induced degradation of IκBα. KBM-5 cells were incubated with 50 μM XN for 4 hours and treated with 0.1 nM TNF for the indicated times. Cytoplasmic extracts were prepared and analyzed by Western blotting using antibody against IκBα. Equal protein loading was evaluated using β-actin. (C) Effect of XN on phosphorylation of IκBα induced by TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours, incubated with 50 μg/mL N-acetyl-leucyl-leucyl-norleucinal (ALLN) for 30 minutes, and then treated with 0.1 nM TNF for 10 minutes. Cytoplasmic extracts were fractionated and then subjected to Western blot analysis with a phosphospecific anti-IκBα antibody (P-IκBα). Ser32/36. (D) Effect of XN on activation of IKK by TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were immunoprecipitated with an antibody against IKK-α and analyzed using an immune complex kinase assay. To determine the effect of XN on the level of expression of IKK proteins, whole-cell extracts were fractionated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and examined by Western blot analysis with anti–IKK-α and anti–IKK-β antibodies. (E) Direct effect of XN on IKK activation induced by TNF. Whole-cell extracts were prepared from KBM-5 cells treated with 1 nM TNF and immunoprecipitated with an anti–IKK-α antibody. The immunocomplex kinase assay was performed in the absence or presence of XN at the indicated concentrations. (F) Reversal of XN-induced suppression of TNF-induced IKK activation by the reducing agent DTT. Assays were performed as indicated in Figure 4E, except that the IKK activity was determined after treatment with DTT (100 μM), XN (50 μM), or both in kinase assay buffer. (G) The kinase activity of mutated IKK is unaffected by XN. For this, A293 cells were transfected with wild-type FLAG-IKK-β (IKK-β WT) or mutated FLAG-IKK-β (IKK-β MT [C179A]). Whole-cell extracts were prepared, and XN at the indicated concentrations was added in vitro. Immunocomplexes were analyzed for IKK activity.
When examined for cytoplasmic IκBα, we found that TNF-induced IκBα degradation started at 10 minutes after TNF treatment and reached the maximum level at 30 minutes and that resynthesis occurred 60 to 120 minutes after TNF treatment (Figure 4B left panel). In contrast, we found no degradation of IκBα in XN-pretreated cells (Figure 4B right panel). These results indicated that XN can suppress TNF-induced IκBα degradation, which leads to suppression of NF-κB activation.
To determine whether inhibition of TNF-induced degradation of IκBα was caused by inhibition of phosphorylation of IκBα, we used the proteosome inhibitor N-acetyl-leucyl-leucyl-norleucinal to block this degradation. We performed Western blot analysis using an antibody that specifically recognized the serine 32–phosphorylated form of IκBα. The results of this analysis showed that TNF induced IκBα phosphorylation and that XN strongly inhibited this phosphorylation (Figure 4C).
XN inhibits TNF-induced IKK activation
TNF-induced phosphorylation of IκBα requires activation of IKK. We investigated whether XN inhibits TNF-induced activation of IKK in KBM-5 cells using immune complex assays. These assays showed that TNF activated IKK in a time-dependent manner and that XN suppressed TNF-induced activation of IKK. Neither TNF nor XN affected the expression of IKK-α or IKK-β protein (Figure 4D).
To elucidate whether XN suppresses IKK activity directly by binding to IKK or indirectly by suppressing its activation, we incubated the immune complexes with XN at various concentrations and then examined the kinase activity. The results showed that XN directly inhibited the activity of IKK in a dose-dependent manner (Figure 4E). This indicated that XN directly modulates TNF-induced activation of IKK.
The IKK-β subunit of the IKK complex is essential for activation of NF-κB in response to various proinflammatory stimuli. Reports have described the critical involvement of a cysteine residue in the activation of IKK. This cysteine residue can be easily modified by thiol-reactive or oxidizing agents, such as dithiothreitol (DTT). We sought to determine whether DTT could reverse XN-induced inhibition of IKK activation. We found that the addition of DTT to immune complex reversed XN-mediated inhibition of IKK activity induced by TNF (Figure 4F).
The cysteine residue at position 179 in the activation loop of IKK-β has been shown to be a target residue for thiol-modifying agents.33 To determine whether this cysteine residue is involved in XN-mediated inhibition, we transfected A293 cells with wild-type FLAG–IKK-β or FLAG–IKK-β with a C179A mutation. The results showed that XN inhibited the activity of the wild-type IKK-β but did not affect the activity of mutated FLAG–IKK-β (C179A) (Figure 4G). These findings collectively indicated that XN inhibits the IKK activity by directly reacting with cysteine residue 179 in the IKK-β subunit.
XN inhibits TNF-induced p65 subunit translocation
Because IκBα degradation is required for nuclear translocation of p65, we sought to determine whether XN also suppresses TNF-induced nuclear translocation of p65. Immunocytochemical analysis showed that XN suppressed the TNF-induced translocation of p65 to the nucleus in KBM-5 cells (Figure 5A). In both untreated cells and cells treated with XN, p65 was localized to the cytoplasm, whereas in cells treated with TNF alone, p65 was translocated to the nucleus. These results confirmed that XN inhibited translocation of p65.
XN down-regulates the TNF-induced NF-κB activation through modification of p65 subunit of NF-κB. (A) XN inhibits TNF-induced nuclear translocation of p65 assayed by immunocytochemical analysis. KBM-5 cells were first treated with 50 μM XN for 4 hours at 37°C and then exposed to 0.1 nM TNF for 15 minutes. After cytospinning, immunocytochemical analysis was performed as described. (B) XN inhibits TNF-induced phosphorylation of p65. KBM-5 cells were either left untreated or pretreated with 50 μM XN for 4 hours at 37°C and then treated with 0.1 nM TNF for the indicated times. Cell extracts were prepared and analyzed by Western blotting with phosphospecific p65 antibodies. Cell extracts blotted with an anti–β-actin antibody were used as loading controls. (C) XN directly inhibits p65 binding to DNA. Nuclear extracts (NE) were prepared from KBM-5 cells treated with 0.1 nM TNF for 30 minutes, incubated with XN at indicated concentrations for 30 minutes, and EMSA was performed. (D) Reversal of XN-induced suppression of DNA binding in whole cells by DTT. Nuclear extracts were prepared from untreated KBM-5 cells or cells treated with 0.1 nM TNF for 30 minutes, incubated with 50 μM XN for 30 minutes in the presence or absence of 100 μM DTT, and then assayed for NF-κB binding to DNA by EMSA. (E) DTT reverses XN-induced suppression of DNA binding of recombinant p65 and inhibits XN-mediated suppression of recombinant p65 in vitro in A293 cells. Nuclear extracts from A293 cells transfected with p65 plasmid were incubated with 50 μM XN with or without 100 μM DTT for 30 minutes and then assayed for NF-κB binding to DNA by EMSA. (F) XN has no effect on DNA binding of p65 mutated at Cys-38 position. A293 cells were transfected with wild-type p65 and mutated p65C38S in vitro nuclear extracts were prepared and EMSA was performed.
XN down-regulates the TNF-induced NF-κB activation through modification of p65 subunit of NF-κB. (A) XN inhibits TNF-induced nuclear translocation of p65 assayed by immunocytochemical analysis. KBM-5 cells were first treated with 50 μM XN for 4 hours at 37°C and then exposed to 0.1 nM TNF for 15 minutes. After cytospinning, immunocytochemical analysis was performed as described. (B) XN inhibits TNF-induced phosphorylation of p65. KBM-5 cells were either left untreated or pretreated with 50 μM XN for 4 hours at 37°C and then treated with 0.1 nM TNF for the indicated times. Cell extracts were prepared and analyzed by Western blotting with phosphospecific p65 antibodies. Cell extracts blotted with an anti–β-actin antibody were used as loading controls. (C) XN directly inhibits p65 binding to DNA. Nuclear extracts (NE) were prepared from KBM-5 cells treated with 0.1 nM TNF for 30 minutes, incubated with XN at indicated concentrations for 30 minutes, and EMSA was performed. (D) Reversal of XN-induced suppression of DNA binding in whole cells by DTT. Nuclear extracts were prepared from untreated KBM-5 cells or cells treated with 0.1 nM TNF for 30 minutes, incubated with 50 μM XN for 30 minutes in the presence or absence of 100 μM DTT, and then assayed for NF-κB binding to DNA by EMSA. (E) DTT reverses XN-induced suppression of DNA binding of recombinant p65 and inhibits XN-mediated suppression of recombinant p65 in vitro in A293 cells. Nuclear extracts from A293 cells transfected with p65 plasmid were incubated with 50 μM XN with or without 100 μM DTT for 30 minutes and then assayed for NF-κB binding to DNA by EMSA. (F) XN has no effect on DNA binding of p65 mutated at Cys-38 position. A293 cells were transfected with wild-type p65 and mutated p65C38S in vitro nuclear extracts were prepared and EMSA was performed.
XN inhibits TNF-induced p65 phosphorylation
Transcriptional activation of p65 requires phosphorylation at the serine 536 residue. Thus, we also investigated the effect of XN on TNF-induced phosphorylation of p65. We found that phosphorylation of p65 occurred in a time-dependent manner in TNF-treated KBM-5 cells but not in cells that have also been treated with XN (Figure 5B).
XN directly modulates the binding of NF-κB to the DNA
We also examined whether XN can directly interact with the p65 subunit of NF-κB and abolish its binding to DNA. To do so, we incubated nuclear extracts isolated from TNF-treated KBM-5 cells with XN at different concentrations and then examined its binding to DNA. We found that XN directly inhibited p65 binding to DNA in a dose-dependent manner (Figure 5C). XN was found to have no effect on DNA binding of AP-1 or SP-1 (Figure S1C,D), thus indicating that the effect of XN on p65 is specific.
We also investigated the possibility that XN modifies the binding properties of p65 subunits through interaction with critical cysteine residues. To determine this, TNF-treated nuclear extracts were exposed to XN in the presence and absence of DTT and then examined for DNA binding. The results showed that DTT completely reversed XN-induced suppression of p65 binding to DNA (Figure 5D).
Whether XN inhibits the activity of the recombinant p65 subunit and whether DTT reverses it were also investigated. For this, we transfected A293 cells with p65 plasmids, prepared the nuclear extracts, and then treated with XN in the presence and absence of DTT. The results showed that nuclear extracts of untransfected cells did not bind to DNA, nuclear extracts of transfected cells bound to DNA, treatment with XN abolished binding to DNA, and DTT reversed this inhibition (Figure 5E).
It has been reported previously that the cysteine residue present at the 38th position (cys-38) of p65 participates in DNA binding by forming a hydrogen bond with the phosphate-sugar backbone of DNA and that cys-38 in p65 is highly susceptible to various agents.34,35 Whether XN targets Cys38 in p65 was investigated. To do so, we transfected A293 cells with plasmids either wild-type or carrying a cysteine to alanine mutation at position 38. After 48 hours of transfection, we prepared nuclear extracts and treated them with XN and evaluated DNA binding using EMSA. The results indicated that XN inhibited the DNA binding of wild-type p65 but not that of mutated p65 (Figure 5F). Thus, these series of results indicated that XN could inhibit NF-κB activation by targeting Cys-38 in p65 subunit.
Because XN targets cys179 of IKK and cys38 of p65, whether apoptotic effects of XN differ in cells transfected with mutated versus wild-type plasmids was examined. Using transient transfection (18%-20% transfection efficiency), we found that A293 cells transfected with wild-type IKK showed more XN-induced apoptosis (62% ± 1.6% apoptosis) than those transfected with mutant IKK (48% ± 2.5%). Similar results were obtained when A293 cells were transfected with wild-type p65 (59% ± 1.8%) versus mutant (45% ± 2.4%). These results further confirm the role of Cys residues in IKK and p65 for the action of XN.
XN suppresses TNF-induced, NF-κB–dependent reporter gene expression
Although we determined, using EMSA, that XN inhibited TNF-induced NF-κB expression. DNA binding alone does not always correlate with NF-κB–dependent gene transcription, suggesting that additional regulatory steps are involved. Thus, we also decided to determine whether XN affects TNF-induced reporter gene transcription. We transiently transfected A293 cells with an NF-κB–regulated SEAP reporter construct, treated them with XN, and then exposed them to TNF. As shown in Figure 6A, TNF induced NF-κB reporter activity and XN inhibited the TNF-induced NF-κB reporter activity in a dose-dependent manner.
XN down-regulates the NF-κB reporter activity induced by TNF and TNF-signaling components. (A) XN inhibits TNF-induced NF-κB–dependent reporter gene (SEAP) expression. A293 cells were transiently transfected with an NF-κB–containing plasmid linked with the SEAP gene. Cells were treated with XN for 4 hours at the indicated concentrations followed by 1 nM TNF for 24 hours, cell supernatants were collected and assayed for SEAP activity. The results are expressed as the fold activity over the activity of the vector control. DN indicates dominant-negative. (B) XN inhibits NF-κB–dependent reporter gene expression induced by TNF, NIK, TAK1, TRADD, and TRAF-2. A293 cells were transiently transfected with the indicated plasmids along with an NF-κB–containing plasmid linked with the SEAP gene for 24 hours. After medium change, cells were treated with XN (50 μM) for 4 hours. Where indicated, cells were exposed to 1 nM TNF for 24 hours. Cell supernatants were assayed for SEAP activity. The results are expressed as the fold activity over the activity of the vector control.
XN down-regulates the NF-κB reporter activity induced by TNF and TNF-signaling components. (A) XN inhibits TNF-induced NF-κB–dependent reporter gene (SEAP) expression. A293 cells were transiently transfected with an NF-κB–containing plasmid linked with the SEAP gene. Cells were treated with XN for 4 hours at the indicated concentrations followed by 1 nM TNF for 24 hours, cell supernatants were collected and assayed for SEAP activity. The results are expressed as the fold activity over the activity of the vector control. DN indicates dominant-negative. (B) XN inhibits NF-κB–dependent reporter gene expression induced by TNF, NIK, TAK1, TRADD, and TRAF-2. A293 cells were transiently transfected with the indicated plasmids along with an NF-κB–containing plasmid linked with the SEAP gene for 24 hours. After medium change, cells were treated with XN (50 μM) for 4 hours. Where indicated, cells were exposed to 1 nM TNF for 24 hours. Cell supernatants were assayed for SEAP activity. The results are expressed as the fold activity over the activity of the vector control.
XN inhibits NF-κB activation stimulated by NIK, TAK1, TRADD, and TRAF-2
NF-κB activation by TNF and the other agents investigated in this study is mediated through NIK, TAK1, TRADD, and TRAF-2.24 We investigated whether NF-κB reporter activity induced by these intermediates is also inhibited by XN. The results presented in Figure 6B show that all of these plasmids induced NF-κB reporter activity and that XN substantially inhibited the reporter activity.
Inhibition of the NF-κB–signaling cascade by XN is independent of FXR activation
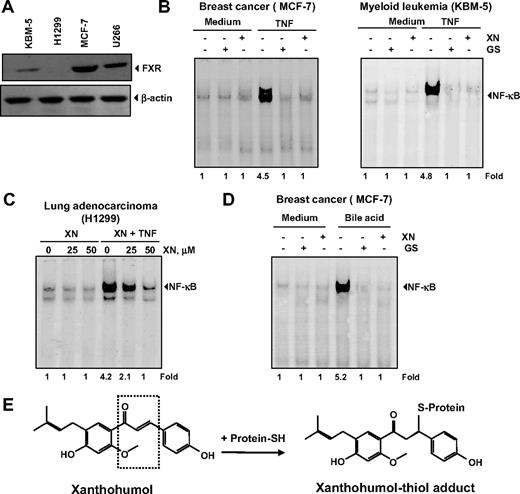
Previous studies in our laboratory showed that the FXR ligand guggulsterone could inhibit TNF-induced NF-κB activation.36 In addition, researchers have shown that XN is a natural ligand for FXR and can activate FXR and modulate genes involved in lipid and glucose metabolism.22 We sought to determine whether the effects of XN on the NF-κB pathway are mediated through interaction with FXR. First, we used Western blot analysis to examine the expression of FXR in different cell lines used in our studies and in those known to express FXR (such as MCF-7). Indeed, we found that U266, KBM-5, and MCF-7 cells expressed FXR to various degrees, with MCF-7 cells having maximum expression; H1299 cells did not express FXR (Figure 7A). We then examined the ability of XN to suppress TNF-induced NF-κB activation in all of these cell lines. We used guggulsterone as a positive control in this experiment. The results presented in Figure 7B show that, like guggulsterone, XN suppressed TNF-induced NF-κB activation in all the cell lines. XN suppressed TNF-induced NF-κB activation in H1299 cells, which lack FXR (Figure 7C), thus suggesting that the effects of XN on NF-κB activation are independent of FXR.
XN down-regulates the TNF-induced NF-κB activation through FXR-independent mechanism. (A) Expression of the FXR receptor in various cell types. Whole-cell extracts were prepared and analyzed by Western blotting with an anti-FXR antibody. (B) XN and guggulsterone (GS) inhibit TNF-induced NF-κB expression in both MCF-7 and KBM-5 cells. Cells were pretreated with either 50 μM XN for 4 hours or 50 μM GS for 4 hours and then incubated with 0.1 nM TNF. Nuclear extracts were prepared and EMSA was performed. (C) Effect of XN on TNF-induced NF-κB expression in FXR-negative H1299 cells. Cells were pretreated with 50 μM XN for 4 hours and then incubated with 0.1 nM TNF. Nuclear extracts were prepared and EMSA was performed. (D) XN and GS inhibit bile acid induced NF-κB expression in MCF-7 cells. Cells were pretreated with either 50 μM XN for 4 hours or 50 μM GS for 4 hours and then incubated with 250 μM bile acids for 3 hours. Nuclear extracts were prepared and EMSA was performed. (E) The chemical structure of XN, an FXR ligand. A schematic representation of XN′s interaction with thiol group is also shown.
XN down-regulates the TNF-induced NF-κB activation through FXR-independent mechanism. (A) Expression of the FXR receptor in various cell types. Whole-cell extracts were prepared and analyzed by Western blotting with an anti-FXR antibody. (B) XN and guggulsterone (GS) inhibit TNF-induced NF-κB expression in both MCF-7 and KBM-5 cells. Cells were pretreated with either 50 μM XN for 4 hours or 50 μM GS for 4 hours and then incubated with 0.1 nM TNF. Nuclear extracts were prepared and EMSA was performed. (C) Effect of XN on TNF-induced NF-κB expression in FXR-negative H1299 cells. Cells were pretreated with 50 μM XN for 4 hours and then incubated with 0.1 nM TNF. Nuclear extracts were prepared and EMSA was performed. (D) XN and GS inhibit bile acid induced NF-κB expression in MCF-7 cells. Cells were pretreated with either 50 μM XN for 4 hours or 50 μM GS for 4 hours and then incubated with 250 μM bile acids for 3 hours. Nuclear extracts were prepared and EMSA was performed. (E) The chemical structure of XN, an FXR ligand. A schematic representation of XN′s interaction with thiol group is also shown.
Bile acids are the natural ligands for FXR.37 We found that bile acids can activate NF-κB in MCF-7 cells and that XN can suppress NF-κB activation (Figure 7D). Like XN, we found that guggulsterone also suppressed bile acid–induced NF-κB activation. These results show that FXR can activate NF-κB. However, XN mediates suppression of NF-κB activation independent of FXR expression.
Discussion
NF-κB activation pathway has been linked with tumor-cell survival, proliferation, invasion, and angiogenesis.38 Because XN modulates these responses, whether it does so through the regulation of the NF-κB–signaling pathway was the focus of the present study. TNF is known to activate both the NF-κB and apoptosis pathways simultaneously. We found that suppression of the NF-κB pathway by XN potentiated TNF-induced apoptosis several-fold in both myeloid and leukemic cells, as examined by intracellular esterase activity, phosphatidylserine externalization, DNA strand breaks, or caspase activation. Similarly, apoptosis induced by chemotherapeutic agents was also enhanced by XN (data not shown). These results provide an opportunity to use XN in combination with existing drugs to induce apoptosis in tumor cells. Although the antiproliferative effects of this chalcone alone against tumor cells has been reported,3,,,,,,,–11 this is the first report on the effect of XN in combination with cytokines and chemotherapeutic agents.
We also investigated how XN potentiates apoptosis in detail. We found that it down-regulated the expression of survivin, Bcl-xL, XIAP, cIAP-1, cIAP-2, and TRAF-2, all of which are known to suppress apoptosis. We also found that XN inhibited the expression of cyclin D1 and c-myc, involved in cell proliferation. Suppression of proliferation of various tumor cells by XN could be the result of inhibition of antiapoptotic and cell proliferation gene products as described herein. Because this chalcone has been reported to exhibit anti-invasive and antiangiogenic activities,12,13 these effects can be linked with ability of XN to inhibit the expression of ICAM-1, MMP-9, and VEGF, respectively.
Because all the gene products suppressed by XN are regulated by NF-κB, we investigated the effect of this agent on the NF-κB pathway in detail. We found that XN suppressed TNF-induced NF-κB activation whether examined by DNA binding, or immunocytochemistry, or by NF-κB reporter assay. Our results in leukemia cells, in particular, are in agreement with that reported by Albini et al,13 who described suppression of TNF-induced NF-κB activation in human endothelial cells by immunocytochemistry. Besides TNF-inducible NF-κB activation, we found that XN abrogated constitutive NF-κB activation in human multiple myeloma U266 cells. The suppression of constitutive NF-κB activation by XN agrees with prostate cancer cells reported previously.10
We also found, for the first time, that XN inhibits NF-κB expression induced by inflammatory stimuli, such as TNF, IL-1β, LPS, and PMA, pro-oxidants, such as H2O2, carcinogens, such as okadaic acid, and tumor promoters, such as CSC. The inhibition of NF-κB expression induced by these agents suggests that XN must act at a step that is common to all of these agents. We found that XN inhibited TNF-induced IκBα phosphorylation and degradation, thus leading to suppression of nuclear translocation of p65. Albini et al13 showed that XN can inhibit TNF-induced phosphorylation of IκBα, but a mechanism for the effect was not identified. We found that XN inhibits TNF-induced IKK activation. Our evidence indicates that XN affects IKK through direct interaction. We also found that the presence of a reducing agent reversed the effect of XN on IKK, suggesting a role for cysteine residues. Furthermore, mutation of IKK-β from cysteine to alanine at position 179 abolished the effect of XN on inhibition of IKK activity. How XN modifies cysteine 179 of IKK is not fully understood. Most polyphenols mediate their cellular effects through 2 different mechanisms, redox recycling or reaction with sulfhydryl groups. Redox cycling results in the generation of the semiquinone radicals followed by formation of superoxide radical and H2O2. Because XN directly modified IKK not only in vivo but also in vitro (Figure 4F), it is doubtful that the effect of XN is mediated through generation of ROS. Moreover, this XN is known to be a scavenger of free radicals.19,39 It is also less probable that ROS is being produced by the nuclear extracts in vitro conditions used for the modification of IKK by XN. All these results suggest that XN is interacting with cysteine residue of IKK directly. XN contains one Michael acceptor and an electrophilic carbonyl group, which could interact directly with the sulfhydryl group of cysteine residue, thus leading to an adduct formation (Figure 7E). Similar mechanism has been reported for cyclopentenone prostaglandins,40 arsenite,41 butein,26 and nitric oxide.42
Besides its interaction with IKK, we found that XN also inhibited NF-κB through its interaction with the p65 subunit of NF-κB. Specifically, we observed that XN inhibited the binding of p65 to DNA both in vivo and in vitro. This inhibition of binding by XN again could be reversed by reducing agents, suggesting a role for cysteine residue. XN inhibited the DNA binding of the recombinant wild-type p65 but not that of p65 in which cysteine residue 38 had been mutated. This suggests that cysteine residue 38 is another major target of XN. A mechanism similar to IKK may apply to the modification of p65. This is consistent with reports on polyphenols, such as sesquiterpene lactones35 epoxyquinone A34 and plumbagin,43 shown to directly alkylate Cys38 of p65. This is a general mechanism for polyphenols, which possess α, β, or α, β, γ-unsaturated carbonyl structures, such as α methylene-γ-lactones or α, β-unsubstituted cyclopentenones. These functional groups are known to react with nucleophiles, especially with cysteine sulfhydryl groups, in a Michael-type addition.44,45
XN has been shown to be a natural ligand for FXR.37 However, we found that XN inhibits TNF-induced NF-κB activation in both FXR-positive and -negative cell lines, suggesting that FXR is not required for the NF-κB–inhibitory activity of XN. However, this chalcone also inhibited bile acid-induced NF-κB activation, which is linked with FXR.
Whether levels of XN used in our studies in vitro are achievable in vivo is not clear. Albini et al13 showed that XN at 25 μM dose could inhibit the growth of human endothelial cells in culture. Comparable dose (20 μM), when given orally to mice, inhibited the angiogenesis and tumor growth in mice.13 Monteiro et al also showed that treatment of breast cancer cells with 50 μM XN for 72 hours was required for significant inhibition of the growth of breast cancer in vitro.46 In vivo, they administered orally 100 μM XN to breast cancer bearing nude mice and showed significant inhibition of tumor weight. Because doses used in both of these studies in vitro were comparable with ours, it is possible that these doses are achievable in vivo.
XN is a component of regular beer, although in very low amounts (100 μg/L). Investigators have found that XN is quite safe when given to rats at doses as high as 1 g/kg body weight per day.47 The pharmacologic safety of XN, combined with the fact that it can suppress NF-κB and NF-κB–mediated cellular responses, has major implications. In addition, XN is able to inhibit osteoporosis, diabetes, and atherosclerosis,7,23 which may also be linked with suppression of the NF-κB pathway as reported here. More animal studies are required to fully explore the anticancer potential of this fascinating beer-derived molecule.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Donald R. Norwood for editing the manuscript.
This work was supported by grants from the Clayton Foundation and by a National Institutes of Health Core grant. B.B.A. is the Ransom Horne Jr Professor of Cancer Research.
National Institutes of Health
Authorship
Contribution: K.B.H., A.B.K., K.S.A., and P.A conducted all the experiments; S.K. and S.G. supervised the experiments; and B.B.A. supervised the experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bharat B. Aggarwal, Cytokine Research Laboratory, Department of Experimental Therapeutics, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Box 143, Houston, TX 77030; e-mail: aggarwal@mdanderson.org.




![Figure 4. XN down-regulates the TNF-induced NF-κB activation through inhibition of IκBα kinase. (A) XN inhibits TNF-induced activation of NF-κB. KBM-5 cells were incubated with 50 μM XN for 4 hours, treated with 0.1 nM TNF for the indicated times, and then analyzed for NF-κB activation by EMSA. The fold activation of NF-κB is shown at the bottom. (B) Effect of XN on TNF-induced degradation of IκBα. KBM-5 cells were incubated with 50 μM XN for 4 hours and treated with 0.1 nM TNF for the indicated times. Cytoplasmic extracts were prepared and analyzed by Western blotting using antibody against IκBα. Equal protein loading was evaluated using β-actin. (C) Effect of XN on phosphorylation of IκBα induced by TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours, incubated with 50 μg/mL N-acetyl-leucyl-leucyl-norleucinal (ALLN) for 30 minutes, and then treated with 0.1 nM TNF for 10 minutes. Cytoplasmic extracts were fractionated and then subjected to Western blot analysis with a phosphospecific anti-IκBα antibody (P-IκBα). Ser32/36. (D) Effect of XN on activation of IKK by TNF. KBM-5 cells were preincubated with 50 μM XN for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were immunoprecipitated with an antibody against IKK-α and analyzed using an immune complex kinase assay. To determine the effect of XN on the level of expression of IKK proteins, whole-cell extracts were fractionated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and examined by Western blot analysis with anti–IKK-α and anti–IKK-β antibodies. (E) Direct effect of XN on IKK activation induced by TNF. Whole-cell extracts were prepared from KBM-5 cells treated with 1 nM TNF and immunoprecipitated with an anti–IKK-α antibody. The immunocomplex kinase assay was performed in the absence or presence of XN at the indicated concentrations. (F) Reversal of XN-induced suppression of TNF-induced IKK activation by the reducing agent DTT. Assays were performed as indicated in Figure 4E, except that the IKK activity was determined after treatment with DTT (100 μM), XN (50 μM), or both in kinase assay buffer. (G) The kinase activity of mutated IKK is unaffected by XN. For this, A293 cells were transfected with wild-type FLAG-IKK-β (IKK-β WT) or mutated FLAG-IKK-β (IKK-β MT [C179A]). Whole-cell extracts were prepared, and XN at the indicated concentrations was added in vitro. Immunocomplexes were analyzed for IKK activity.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/9/10.1182_blood-2008-04-151944/6/m_zh80030929550004.jpeg?Expires=1767744903&Signature=kz0SzoupDG6yHOjMJ2A~em2KfsYSSVtbFLuA-ReUUM9oDesDGpSNKVEUwD4jgl5k6Eb5szfcWGH78Jgc7kp~3f6NCPR0lqcYVT7AkAwXof2~1061XWAZOytuBA26d8OjHeu0yt3~FsgHC30HcpOxslbrJ~rcmhbRMO5zjS8MzJrmfMUr3bjSiB-Na0Dnab4VtI~2s9WzpEW~lRnFGbk9akHxgiMpQMxu3CqkqoVUfFEtaW1JNVhce3rmJMqSE0Nt1s3jh1Y5oxrSUrxY53eYHccoYWaO0NEsCA0a8y4HWymxQLcNc-8htV6Pb2-Sx9Ww~STqrgj8qwiC57Kd5ZJBzg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal