Abstract
The plasma membrane glycoprotein receptor CD163 is a member of the scavenger receptor cystein-rich (SRCR) superfamily class B that is highly expressed on resident tissue macrophages in vivo. Previously, the molecule has been shown to act as a receptor for hemoglobin-haptoglobin complexes and to mediate cell-cell interactions between macrophages and developing erythroblasts in erythroblastic islands. Here, we provide evidence for a potential role for CD163 in host defense. In particular, we demonstrate that CD163 can function as a macrophage receptor for bacteria. CD163 was shown to bind both Gram-positive and -negative bacteria, and a previously identified cell-binding motif in the second scavenger domain of CD163 was sufficient to mediate this binding. Expression of CD163 in monocytic cells promoted bacteria-induced proinflammatory cytokine production. Finally, newly generated antagonistic antibodies against CD163 were able to potently inhibit cytokine production elicited by bacteria in freshly isolated human monocytes. These findings identify CD163 as a macrophage receptor for bacteria and suggest that, during bacterial infection, CD163 on resident tissue macrophages acts as an innate immune sensor and inducer of local inflammation.
Introduction
CD163 is a cell-surface glycoprotein receptor that is highly expressed on most subsets of resident tissue macrophages.1 The molecule is a member of the scavenger receptor cystein-rich (SRCR) family class B2,3 and has been identified as an endocytic receptor for hemoglobin-haptoglobin (Hb-Hp) complexes.4 As such it has been proposed to mediate the clearance of free Hb from the circulation, by eg splenic red pulp and liver macrophages, offering protection against hemoglobin-mediated oxidative tissue damage during conditions of excessive hemolysis.5 Furthermore, CD163 is expressed on resident bone-marrow macrophages and other macrophages involved in the formation of erythroblastic islands.6 Here, it functions as an erythroblast adhesion receptor and may play a regulatory role during erythropoiesis. Apart from these homeostatic functions, the cross-linking of CD163 with monoclonal antibodies directed against CD163 triggers the production of inflammatory mediators, such as nitric oxide, tumor necrosis factor (TNF)–α, interleukin (IL)–1β, IL-6, and IL-10, suggesting a potential role in immunity and/or host defense.7-9 Interestingly, several members of the SRCR class B family, including the secreted glycoproteins gp-340/SAG/DMBT110,11 and Spα,12 and the lymphocyte surface receptor CD6,13 were recently reported to mediate bacterial binding
In the present report we provide evidence that CD163 functions as a macrophage surface receptor for bacteria. CD163, expressed on cells or as an immobilized protein, supported the binding of bacteria, including both Gram-negative and Gram-positive species. A peptide motif within the second scavenger receptor domain of CD163, that was previously shown to mediate erythroblast binding,6 was sufficient to establish bacterial binding, suggesting that this domain harbours a binding site for both cellular and bacterial adhesion. Recognition of bacteria by CD163 potently enhanced inflammatory cytokine production in monocytic THP-1 cells, and cytokine production by freshly isolated human monocytes was strongly suppressed by novel agonistic mAb against CD163.
Methods
Antibodies and peptides
Bacteria
Streptococcus mutans (Ingbritt strain), Escherichia coli (K4), Staphylococcus aureus (clinical isolate), and E coli Dsred15 were cultured on blood agar plates under anaerobic conditions with 5% CO2 for 24 hours at 37°C. Bacteria were harvested and washed twice in Tris-buffered saline (TBS)–Ca2+: 50 mM Tris, pH 7.5, containing 150 mM sodium chloride and 2 mM calcium chloride). Bacteria were adjusted to their final concentration by measuring the optical density (OD) at 600 nm. Where indicated, bacteria were fluorescein isothiocyanate (FITC)–labeled as described.16 Bacteria from overnight cultures were suspended into TBS-Ca2+, and adjusted to an A600 of 1 (corresponding to approximately 109 bacteria/mL). One milliliter of bacteria was transferred to a microcentrifuge tube, pelleted, and resuspended in 1 mL FITC isomer I (Molecular Probes, Eugene, OR; 0.5-2 mg/mL) in TBS-Ca2+. Bacteria were incubated for 60 minutes at 37°C, washed at least 3 times in TBS-Ca2+ at 3000g for 7 minutes at 4°C, and then suspended in 660 μL TBS-Ca2+.
Cell culture and retroviral transductions
Monocytes were isolated by Ficoll density centrifugation to obtain peripheral blood mononuclear cells (PBMCs), followed by magnetic cell sorting using CD14 beads (Miltenyi Biotec) according to instructions provided by the manufacturer. Chinese hamster ovary (CHO) cells stably expressing the full-length human CD163 or the pCDNA3.1/zeo empty vector (EV) were cultured as described.6 Monocytes were isolated by counterflow elutriation. THP-1 cells expressing the full-length human CD163 were generated as follows. DNA encoding the full-length human CD163 was excised from a pCDNA3.1/zeo-based plasmid4 by means of KpnI and NotI and was cloned into pBacPAK-His1 and subsequently into the modified retroviral vector LZRS-IRES-GFP17 by means of XhoI and NotI. The resulting construct, pLZRS-CD163-IRES-GFP, was transfected with calcium phosphate into amphotropic Phoenix retrovirus producer cells17 for the generation of helper-free amphotropic retroviruses. Virus-containing supernatant was used to transduce THP-1 cells in a plastic culture dish pretreated with 30 μg/mL retronectin (Takara Biomedicals, Shiga, Japan). After 48 hours at 32°C, cells were transferred to fresh medium. Transduction was repeated 3 times until approximately 5% of the THP-1 cells expressed CD163-IRES-GFP as determined by flow cytometry. Cells were sorted on green fluorescent protein (GFP) expression by flow cytometric cell sorting (MoFlo cell sorter; Dako, Heverlee, Belgium) and enriched until THP-1 CD163-GFP cells were 95% or more GFP-positive. As a control, an empty vector (EV) LZRS-IRES-GFP construct was introduced into THP-1 cells and used throughout these experiments. Cells were cultured in RPMI medium (PAA, Pashing, Austria) containing 10% fetal calf serum (Bodinco, Alkmaar, The Netherlands) and antibiotics (penicillin/streptomycin; Cambrex, Verviers, Belgium) and CD163 expression was checked regularly by flow cytometry.
Flow cytometry
Surface expression of CD163 was determined as follows. Cells were stained with the indicated monoclonal antibodies (10 μg/mL) for 60 minutes at 4°C in phosphate-buffered saline (PBS)–0.1% bovine serum albumin (BSA; Boehringer-Mannheim, Mannheim, Germany), and washed. Subsequently, cells were stained with goat anti–mouse Ig labeled with Alexa Fluor 633 (Molecular Probes/Invitrogen, Breda, The Netherlands) for 45 minutes. After washing, cells were resuspended in PBS-0.1% BSA and fluorescence intensity was determined on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA).
Antibody generation and selection
BALB/c mice were immunized 3 to 4 times with 5 to 10 × 106 human CD163-expressing CHO cells according to an immunization protocol as described previously.18 Spleen cells were then fused with the mouse myeloma cell line Sp2/0 and the resulting hybridomas were plated at a density of approximately 1 cell/well under hypoxanthine-aminopterin-thymidine selection. Hybridoma supernatants were collected and screened in the first round for specific reactivity with CD163 by means of (1) cytospins of CD163-transfected CHO cells and (2) human spleen frozen sections by immunohistochemistry as described.18 In the second round of screening the antibodies were tested for blocking of bacterial binding to CD163-CHO cells by flow cytometry and fluorescent bead adhesion assay as described below. Two blocking antibodies, EDhu2 and EDhu3, and one nonblocking antibody EDhu4, all of the IgG2a subclass, were selected for further use. Selected hybridomas were subcloned by limiting dilution, produced in bulk, and antibodies were purified by protein-A-sepharose chromatography. Generally, antibody incubations were performed at saturating concentrations as determined by flow cytometry or functional assay. All animal experiments have been approved by the animal and human ethical committees of the VU University Medical Center Amsterdam.
Cell binding assays
Adherent CD163 CHO cells and WT CHO cells (2.5 × 105 cells) were incubated with 100 μL FITC-labeled bacteria (corresponding to 2.5 × 106 bacteria) in TBS-Ca2+ at 4°C for 30 minutes. Samples were washed 3 times and analyzed by flow cytometry. For the nonadherent THP-1 cells the following assay for analyzing the binding of bacteria was performed. THP-EV-GFP and THP-163-GFP cells (105 cells in 100 μL TBS-Ca2+) were incubated with DsRed-transfected E coli (at a ratio of 100 bacteria/cell) and incubated for 60 minutes at 4°C. Samples were washed 3 times (centrifuging them at 500g for 3 minutes) and analyzed by flow cytometry. Where applicable, saturating concentrations of antibodies against CD163 or isotype-matched controls were preincubated with the cells and washed twice before bacteria were added. Where indicated bacterial binding was estimated by calculating the geometric mean fluorescence intensity values corrected for measurements performed in the absence of bacteria.
Phagocytosis assay
Monocytes were pretreated with saturating concentrations (10 μg/mL) of the various antibodies directed against CD163 for 30 minutes on ice. Subsequently, 106 monocytes were incubated with 108 FITC-labeled bacteria at 37°C. Samples were taken at 0 minutes, 30 minutes, 1 hour, and 2 hours, and then diluted 20× in ice-cold PBS containing 0.5% paraformaldehyde. After the last time point, all samples were centrifuged and resuspended in 300 μL ice-cold PBS and analyzed by flow cytometry in a Becton Dickinson LSRII (Palo Alto, CA).
Solid-phase adhesion assay
Solid-phase binding of bacteria to synthetic peptides was performed as described previously.10,11 Briefly, 40 μg/mL synthetic peptide was diluted serially in coating buffer (100 mM sodium carbonate, pH 9.6) in microtiter plates (Fluotrac 600; Greiner, Recklinghausen, Germany). After overnight incubation at 4°C plates were washed and 100 μL bacteria suspension (5 × 108 bacteria/mL in TBS-Ca2+ containing 0.1% Tween 20) were added and incubated for 2 hours at 37°C. After washing with TBS-Ca2+ containing 0.1% Tween 20, adhesion of bacteria was quantified by labeling with the cell-permeable DNA-binding probe SYTO-13 (1:1000; Molecular Probes, Leiden, The Netherlands) and measured in a fluorescence microplate reader (Fluostar Galaxy; BMG Laboratories, Offenburg, Germany) at 488 nm excitation and 509 nm emission.
Bacterial aggregation assay
The bacterial aggregation assay was performed as described.10 A suspension of bacteria (150 μL; 5 × 108 bacteria/mL TBS-Ca2+) was mixed with 150 μL peptide solution (at final concentrations of 0-200 μg/mL in TBS-Ca2+) for 2 hours at 37°C. After agglutination 10 μL sediment was transferred to a microscopic slide. After heat fixation, bacteria were stained with a 20% crystal violet solution (Merck) and examined by light microscopy.
Fluorescent bead adhesion assay
CD163-Fc protein derived from stably transfected CD163-Fc CHO cells was generated and purified as described previously6 and coated onto carboxylate-modified TransFluoSpheres (488/645 nm, 1.0 μm; Molecular Probes, Eugene, OR). Briefly, streptavidin was covalently coupled onto TransFluoSpheres as described by the manufacturer. To enable coupling of CD163-Fc, streptavidin-coated beads were allowed to bind to Fab2 fragments of biotinylated goat anti–human IgG-Fc (6 μg/mL; Jackson ImmunoResearch Laboratories, West Grove, PA) in 0.5 mL TBS containing 2 mM Ca2+ for 2 hours at 37°C. The beads were washed once with TBS-Ca2+ and incubated with human CD163-Fc (1 μg) for 2 days at 4°C. The ligand-coated beads were washed, resuspended in 100 μL TBS-Ca2+, and stored at 4°C. As a control human ICAM-3-Fc19 –coated beads were used. For bead adhesion to bacteria, bacteria were resuspended in TBS-Ca2+ in a concentration of 109 bacteria/mL. Screening for blocking activity of bead adhesion to bacteria by monoclonal antibodies was performed by preincubation of beads with 1 mL hybridoma supernatant for 30 minutes at 37°C. The beads were then washed twice with TBS-Ca2+ by centrifuging at 14 000 rpm for 3 minutes. The ligand-coated beads were added to 50 μL bacteria-suspension at a ratio of 1 bead/20 bacteria and incubated for 60 minutes at 37°C. The percentage of bacteria bound to beads was measured by flow cytometry in a FACSCalibur (BD Biosciences).
Cytokine measurements
THP-CD163-GFP cells, THP-EV-GFP cells, and monocytes were cultured, in triplicate, in a concentration of 105 cells/mL in 96-well tissue-culture plates. Bacteria were added to the cells at a ratio of 100:1. The supernatants were harvested after 4 hours (TNF-α measurements) or 18 hours (IL-1β and IL-6 measurements) of culture and stored at −20°C until further use. Where indicated, saturating concentrations of antibodies against CD163 or isotype-matched controls were added to the wells before the bacteria. TNF-α, IL-1β, and IL-6 concentrations were determined by enzyme-linked immunosorbent assays (ELISAs; BioSource/Invitrogen).
Results
CD163 functions as a macrophage receptor for bacteria
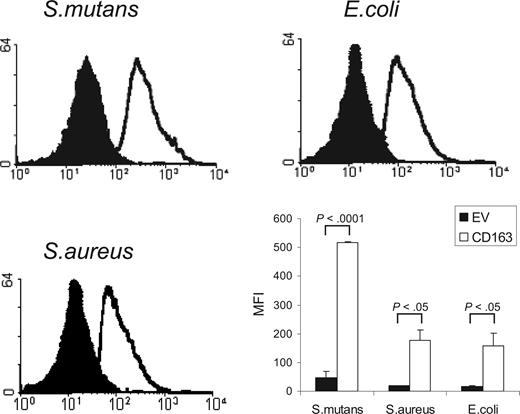
CD163 is a member of the SRCR group B family, characterized by the presence of 9 extracellular SRCR domains. Several other members of this family, including the secreted gp340/SAG/DMBT110,11 and Spα,12 and recently also the lymphocyte surface receptor CD6,13 have been shown to mediate bacterial recognition through their scavenger receptor domains. We investigated whether CD163 would also be able to mediate binding of bacteria. We used a flow cytometric assay to test the binding to CD163-expressing CHO cells of various FITC-labeled species of bacteria, including Gram-positive S mutans and S aureus, and Gram-negative E coli (Figure 1). CHO cells transfected with empty vector were used as a control. For all of these bacterial strains a clear binding to CD163-expressing cells was observed. Evaluation by confocal microscopy revealed that bacteria were bound to the surface of CD163-expressing cells, with individual cells binding approximately 3 to 15 bacteria/cell, but there was no evidence for phagocytosis (data not shown). These experiments indicated that cell surface CD163 supports the recognition of bacteria.
Binding of Gram-positive and Gram-negative bacteria to CD163-expressing CHO cells. FITC-labeled S mutans, S aureus, and E coli were incubated with CD163-expressing CHO cells or cells expressing empty vector (EV) and bacterial binding was assessed by flow cytometry as described in “Cell binding assays.” CD163-CHO cells (open histograms) show clearly increased bacterial binding as compared with CHO EV (filled histograms). The panel at bottom right shows the average mean fluorescence intensities (MFIs) plus or minus standard deviation of 3 independent experiments. Statistics: 2-tailed Student t test.
Binding of Gram-positive and Gram-negative bacteria to CD163-expressing CHO cells. FITC-labeled S mutans, S aureus, and E coli were incubated with CD163-expressing CHO cells or cells expressing empty vector (EV) and bacterial binding was assessed by flow cytometry as described in “Cell binding assays.” CD163-CHO cells (open histograms) show clearly increased bacterial binding as compared with CHO EV (filled histograms). The panel at bottom right shows the average mean fluorescence intensities (MFIs) plus or minus standard deviation of 3 independent experiments. Statistics: 2-tailed Student t test.
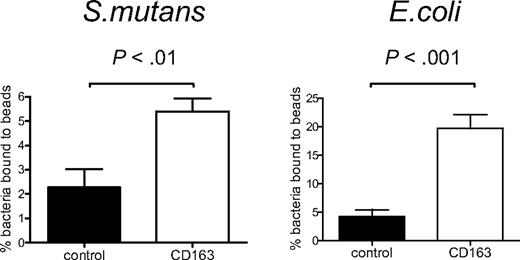
To provide evidence for a possible direct interaction between CD163 and bacteria, we tested binding of bacteria to a recombinant CD163 protein composed of the 9 extracellular SRCR domains of CD163 fused to the Fc-portion of human IgG1 (CD163-Fc).6 CD163-Fc was coated onto fluorescent beads, to create a high-density ligand, and these were then used to test bacterial binding by flow cytometry. As compared with the ICAM-3-coated beads used as the control, the CD163-coated beads mediated clearly enhanced binding to both S mutans and E coli (Figure 2A,B), indicating that CD163 can directly mediate the recognition of intact bacteria.
CD163 supports direct binding of Gram-positive and Gram-negative bacteria. CD163-Fc-coated beads (□) or control ICAM3-Fc–coated beads (■) were incubated with S mutans or with E coli bacteria and binding was assessed by flow cytometry as described in “Fluorescent bead adhesion assay.” Bars represent averages plus or minus SD of 2 (S mutans) or 3 (E coli) independent experiments, respectively. Statistics: 2-tailed Student t test.
CD163 supports direct binding of Gram-positive and Gram-negative bacteria. CD163-Fc-coated beads (□) or control ICAM3-Fc–coated beads (■) were incubated with S mutans or with E coli bacteria and binding was assessed by flow cytometry as described in “Fluorescent bead adhesion assay.” Bars represent averages plus or minus SD of 2 (S mutans) or 3 (E coli) independent experiments, respectively. Statistics: 2-tailed Student t test.
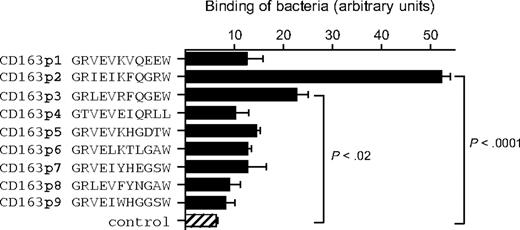
Previous work on gp340/SAG/DMBT1 has mapped a bacterial-binding site to a GRVEVxxxxxW motif within the scavenger domain(s),11 and a similar bacterial-binding region is found in the class A SRCR family member MARCO.20 We wanted to test whether the corresponding motifs within the CD163 scavenger domains also mediated bacterial binding. For each of the 9 scavenger domains of human CD163, 11-amino acid peptides representing the GRVEVxxxxxW motif were generated and tested for binding to bacteria. Among these, only the one from domain 2 (CD163p2) displayed high binding to S mutans (Figure 3). The peptide from domain 3 (CD163p3) also showed a significant but consistently lower binding. In line with these results, CD163p2 was the only among these peptides that mediated detectable bacterial aggregation (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), as was previously shown for the gp340/SAG/DMBT1 bacterial-binding motif.11 Collectively, these observations suggest that the CD163p2 motif from the second SRCR domain, and perhaps to a lesser extent also the CD163p3 motif from the third domain, acts as a bacterial-binding site within the CD163 extracellular region. Of interest, the same motif from domain 2 appears to mediate cellular adhesion of erythroblasts as well.6 In contrast, the available evidence points to an essential role for the third SRCR domain in Hp-Hb complex binding.14 The idea that CD163 uses at least 2 structurally different binding sites to interact with distinct ligands is also supported by the observation that some monoclonal antibodies, such as EDhu1 directed against the third scavenger domain of CD163, strongly inhibit Hp-Hb binding,14 but have no detectable effect on bacterial binding (see below). At present no structural information is available for CD163 that could provide direct insight into the molecular basis of bacterial recognition. However, we used the recently reported X-ray structure of the third domain of CD5,21 which constitutes the first SCRC class B domain for which structural data are available, to generate a model for the second CD163 SRCR domain (data not shown). This identified the CD163p2 peptide motif in a short loop extending from the surface of the scavenger domain, which is consistent with a role in bacterial and cellular recognition.
Binding of S mutans to 11-mer peptide motifs from the extracellular scavenger domains 1 through 9. Microtiter plates were coated with indicated peptides (40 μg/mL) and incubated with S mutans bacteria, and binding was quantified as described in “Solid-phase adhesion assay.” The combined means plus or minus SD from 3 independent experiments are shown. Statistics: 2-tailed Student t test.
Binding of S mutans to 11-mer peptide motifs from the extracellular scavenger domains 1 through 9. Microtiter plates were coated with indicated peptides (40 μg/mL) and incubated with S mutans bacteria, and binding was quantified as described in “Solid-phase adhesion assay.” The combined means plus or minus SD from 3 independent experiments are shown. Statistics: 2-tailed Student t test.
Recognition of bacteria by CD163 in monocytes triggers inflammatory cytokine production
Several studies have demonstrated that the ligation of CD163 on monocytes, either by monoclonal antibodies or Hp-Hb complexes, results in the secretion of inflammatory cytokines.7,8,22 To investigate whether recognition of bacteria by CD163 also induces a cytokine response in monocytic cells we expressed the full-length CD163 in THP-1 cells, which express little or no endogenous CD163 (Figure 4A). The CD163-expressing THP-1 cells (THP-CD163) showed a moderately but significantly enhanced binding of bacteria as compared with the empty vector cells (THP-EV) (Figure 4B). The relatively low level of bacterial binding of the CD163-expressing THP-1 cells, in comparison to that of the CD163-expressing CHO cells, is probably related to the considerably lower CD163 expression level in THP-1 cells (Figure S3). At the same time, the THP-EV cells, which do not express any detectable CD163, already displayed a considerable bacterial-binding capacity by themselves, which is presumably due to the activity of unrelated pattern recognition receptors endogenously expressed on these cells. To determine whether binding of bacteria by CD163 resulted in cytokine production the cells were incubated with bacteria, including S mutans and E coli, for 4 hours, and the production of TNF-α was measured. A 2-fold increase in TNF-α production was observed upon CD163 expression on THP-1 cells (Figure 4C), suggesting that binding of bacteria by CD163 triggered, or at least supported, cytokine production in monocytic cells.
Bacterial binding and bacteria-induced cytokine production in CD163-expressing THP-1 cells. (A) Human CD163 expression on THP-1 cells expressing empty vector (THP EV; filled histogram) or CD163 (THP CD163; open histogram) as analyzed by flow cytometry using the anti-CD163 mAb EDhu1. (B) Binding of DsRed-transfected E coli to THP EV cells and THP CD163 cells as described in “Cell binding assays.” Means plus or minus SD from 3 independent experiments are shown. (C) TNF-α production by THP EV and THP CD163 cells in response to S mutans and E coli bacteria. Bacteria were added in a concentration of 100 bacteria per cell and incubated for 4 hours. Means plus or minus SD from 2 independent experiments are shown. Statistics: 2-tailed Student t test.
Bacterial binding and bacteria-induced cytokine production in CD163-expressing THP-1 cells. (A) Human CD163 expression on THP-1 cells expressing empty vector (THP EV; filled histogram) or CD163 (THP CD163; open histogram) as analyzed by flow cytometry using the anti-CD163 mAb EDhu1. (B) Binding of DsRed-transfected E coli to THP EV cells and THP CD163 cells as described in “Cell binding assays.” Means plus or minus SD from 3 independent experiments are shown. (C) TNF-α production by THP EV and THP CD163 cells in response to S mutans and E coli bacteria. Bacteria were added in a concentration of 100 bacteria per cell and incubated for 4 hours. Means plus or minus SD from 2 independent experiments are shown. Statistics: 2-tailed Student t test.
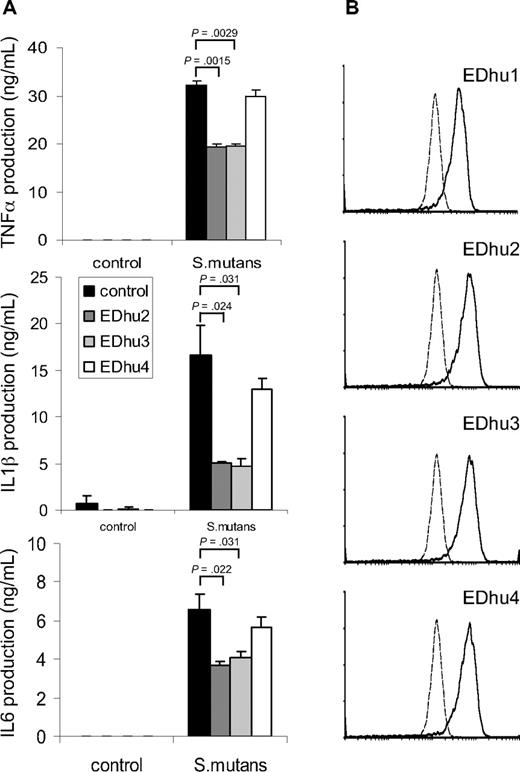
To investigate whether CD163 on freshly isolated monocytes contributed to cytokine production induced by intact bacteria we wanted to identify monoclonal antibodies against CD163 that antagonized bacterial binding. First, several previously reported monoclonal antibodies directed against different domains of CD163,14 including Mac2-158 (d1), EDhu1 (d3), KiM8 (d3), GHI/61 (d7) and RM3/1 (d9), were tested, but none of these showed significant blocking of bacterial binding (data not shown). Therefore, we generated a novel set of monoclonal antibodies against human CD163. Hybridomas producing antibodies against CD163 were first identified by specific staining of CD163-CHO by flow cytometry and by immunohistochemical staining of CD163-expressing splenic red pulp macrophages and liver Kupffer cells (not shown), followed by testing for inhibition of bacterial binding and cytokine production. Several antibodies, including clones EDhu2 and EDhu3, were shown to significantly reduce bacterial interaction with CD163-coated beads (data not shown), binding of bacteria to THP-CD163 cells (Figure S2A), and bacteria-induced TNF-α production by THP-CD163 cells (Figure S2B). Instead, a nonblocking EDhu4 mAb of the same isotype did not affect these parameters. Next, we investigated the effect of these mAb on cytokine production triggered by bacteria in freshly isolated human monocytes. A significant (30%-70%) inhibition of the production of all measured cytokines, including TNF-α, IL-1β, and IL-6, was observed with both antagonistic mAb at saturating antibody concentrations, but there was little or no effect with the nonblocking antibody (Figure 5A). The effects of the 2 antibodies were dose-dependent and nonadditive under conditions of maximal inhibition (data not shown). Collectively, when taking into account that freshly isolated monocytes express relatively low levels of CD163 (Figure 5B), particularly in comparison to, for example, mature tissue macrophages,1,3,7 these results suggests that CD163 harbors a potent intrinsic signaling capacity with respect to cytokine production elicited by intact bacteria in macrophages. Finally, none of the antibodies (EDhu1-4) significantly inhibited the binding and/or phagocytosis of bacteria by monocytes (Figure S4), suggesting that CD163 primarily functions as a sensor for bacterial infection that triggers monocyte/macrophage cytokine responses, rather than acting as a phagocytic receptor.
Suppression of bacteria-induced cytokine production by freshly isolated monocytes by blocking antibodies against CD163. (A) Human monocytes were incubated with S mutans bacteria (100 bacteria/cell) in the presence of blocking (EDhu2 or EDhu3) or nonblocking (EDhu4) antibodies against CD163, and cytokine production was measured in supernatants after 4 hours (TNF-α) or 18 hours (IL-1β and IL-6). Means plus or minus SD from triplicate incubations of a representative experiment of 4 are shown. Statistics: 2-tailed Student t test. (B) Flow cytometric analysis of CD163 expression on monocytes using EDhu1, EDhu2, EDhu3 and EDhu4 mAb. Histograms show control cells stained without primary antibody (dotted line) or with the indicated anti-CD163 mAb. Note that monocytes express only moderate levels of surface CD163.
Suppression of bacteria-induced cytokine production by freshly isolated monocytes by blocking antibodies against CD163. (A) Human monocytes were incubated with S mutans bacteria (100 bacteria/cell) in the presence of blocking (EDhu2 or EDhu3) or nonblocking (EDhu4) antibodies against CD163, and cytokine production was measured in supernatants after 4 hours (TNF-α) or 18 hours (IL-1β and IL-6). Means plus or minus SD from triplicate incubations of a representative experiment of 4 are shown. Statistics: 2-tailed Student t test. (B) Flow cytometric analysis of CD163 expression on monocytes using EDhu1, EDhu2, EDhu3 and EDhu4 mAb. Histograms show control cells stained without primary antibody (dotted line) or with the indicated anti-CD163 mAb. Note that monocytes express only moderate levels of surface CD163.
Discussion
Our current findings identify the human CD163 as a macrophage surface receptor for recognition of intact Gram-positive and Gram-negative bacteria. Furthermore, we demonstrate that the binding of bacteria to CD163 triggers, or at least contributes to, macrophage cytokine production. This suggests for the first time, that CD163, in addition to its formerly implicated homeostatic roles in erythroblast adhesion and Hb-Hp clearance, may also be involved in host defense. The role of CD163 as a bacterial receptor is in line with that of other SCRC family members, such as gp340/SAG/DMBT1,10,11 Spα,12 CD613 and MARCO,20 which have recently also been implicated in bacterial recognition, and clearly provides further support for the concept that bacterial and cellular recognition constitute unifying and perhaps also primordial functions of the scavenger domain.23 With respect to the role in host defense of CD163, which is broadly and constitutively expressed by resident macrophages in tissues, we propose that it acts as a typical innate immune sensor for the detection of bacterial infection. Cytokines and other inflammatory mediators produced as the consequence of CD163-mediated bacterial recognition and signaling are anticipated to trigger, or at least contribute to, the initiation of a local inflammatory response that leads to the elimination of the infection. The high levels of proteolytically shed CD163 observed in plasma during bacterial sepsis24-26 and a variety of other inflammatory conditions27 may reflect a shut-down mechanism attempting to prevent excessive cytokine production at a later stage during infection.
While the current results support the idea that bacterial recognition by CD163 generates signals that trigger macrophage cytokine production, they argue against a prominent role for CD163 in phagocytosis. In this sense CD163 may fulfill a similar function to Toll-like receptors (TLR), which appear dedicated to the regulation of the production of cytokines and other mediators. It will be of interest to determine whether and to what extent the CD163 and TLR signaling pathways collaborate and/or converge. The observation that antibodies against CD163, such as EDhu1, can induce cytokine production by themselves suggests at least that signaling via CD163 is sufficient to trigger cytokine generation.7-9 One notable difference between CD163 and TLRs is that most, if not all, TLRs constitute receptors for soluble, dissociated microbial components and do not mediate recognition of intact bacteria, whereas CD163 is apparently able to do so. Clearly, future studies will have to establish the nature of the relevant bacterial ligands, the relevant intracellular signaling pathways, and the actual role(s) of CD163 during bacterial infection in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Taco W. Kuijpers and Dirk Roos are gratefully acknowledged for critically reading the manuscript and for their useful comments.
Authorship
Contribution: B.O.F. performed experiments, analyzed data, and wrote the paper; R.v.B. performed experiments and analyzed data; D.M.D. performed experiments and analyzed data; A.J.M.L. provided expertise and vital reagents; K.N. performed experiments; K.S. perfomed experiments and analyzed data; R.V. performed experiments; C.D.D. provided expertise and management; and T.K.v.d.B. designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timo K. van den Berg, Phagocyte Laboratory, Sanquin Research and Landsteiner Laboratory, Academic Medical Center, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail: t.k.vandenberg@sanquin.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal