Abstract
Notch signaling is involved in tumorigenesis, but its role in B–chronic lymphocytic leukemia (B-CLL) pathogenesis is not completely defined. This study examined the expression and activation of Notch receptors in B-CLL cells and the role of Notch signaling in sustaining the survival of these cells. Our results show that B-CLL cells but not normal B cells constitutively express Notch1 and Notch2 proteins as well as their ligands Jagged1 and Jagged2. Notch signaling is constitutively activated in B-CLL cells, and its activation is further increased in B-CLL cells, which resist spontaneous apoptosis after 24-hour ex vivo culture. Notch stimulation by a soluble Jagged1 ligand increases B-CLL cell survival and is accompanied by increased nuclear factor–kappa B (NF-κB) activity and cellular inhibitor of apoptosis protein 2 (c-IAP2) and X-linked inhibitor of apoptosis protein (XIAP) expression. In contrast, Notch-signaling inhibition by the γ-secretase inhibitor I (GSI; z-Leu-Leu-Nle-CHO) and the specific Notch2 down-regulation by small-interfering RNA accelerate spontaneous B-CLL cell apoptosis. Apoptotic activity of GSI is accompanied by reduction of NF-κB activity and c-IAP2 and XIAP expression. Overall, our findings show that Notch signaling plays a critical role in B-CLL cell survival and apoptosis resistance and suggest that it could be a novel potential therapeutic target.
Introduction
B-chronic lymphocytic leukemia (B-CLL) is characterized by the accumulation of malignant B CD5+ cells, refractory to apoptosis.1,2 B-CLL cell apoptosis resistance depends in part on the altered activation of various survival pathways3,4 triggered by stimuli, which are usually represented by soluble factors4 and receptor-ligand interactions5,6 derived both from the microenvironment5,7 and B-CLL cells themselves.6 Therefore, the discovery of new altered signal pathways contributing to the anomalous apoptosis resistance of B-CLL cells is important for a better understanding of disease pathogenesis and development of new therapeutic strategies.
A critical role in the pathogenesis of some hematologic and solid malignancies is played by Notch receptors, which control fundamental cellular functions associated with tumorigenesis, including proliferation, differentiation, and apoptosis.8-10 The Notch receptor family consists of 4 transmembrane proteins, Notch1, 2, 3, and 4, which have an extracellular domain (N-EC) mediating ligand binding and an intracellular domain (N-IC) mediating signaling.11,12 Notch signaling initiates when ligand, from either the Serrate/Jagged or Delta family expressed on adjacent cells, binds the receptor and induces successive proteolytic cleavages, resulting in the release and nuclear translocation of the N-IC domain. Within the nucleus, N-IC assembles a transcriptional complex that interacts with the transcription factor CBF1/RBP-Jk, leading to de-repression/activation of CBF1-dependent target genes.13,14 Many potential targets of Notch signaling have been identified in various cell contexts, and several members of the hairy/enhancer of split (Hes) family of the basic helix-loop-helix proteins are direct Notch/CBF1 targets, including Hes1.15,16
Deregulation of the Notch pathway not only diverts cell fate decisions but is also tumorigenic. Whereas the oncogenic role for Notch signaling has been well defined in T-cell malignancies,17,18 the role of Notch is controversial in B-cell malignancies, where it can function as oncogenic or tumor suppressor.8 Studies suggesting an oncogenic role of Notch in multiple myeloma and Hodgkin lymphoma, based on the high expression of Notch receptors and their ligands on these tumor cells and their proliferative response to exogenous Notch ligands,19,20 are discordant with other reports showing that constitutively active forms of all 4 Notch receptors induce growth arrest and apoptosis in these B-cell malignancies,21 although Notch-induced growth arrest may also promote malignant progression by protecting tumor cells against drug-induced apoptosis.22
Regarding the role of Notch signaling in B-CLL pathogenesis, the few studies to date are controversial. Hubmann et al23 suggested a possible role for Notch2 in B-CLL cell survival. These authors found that B-CLL cells constitutively expressed high levels of Notch2 mRNA involved in regulating expression of the survival molecule CD23a. Furthermore, both Notch2 activity and CD23a expression were down-regulated in B-CLL cells during apoptosis induced by proteasome inhibitors.24 In contrast with these studies, other authors reported that Notch is not a survival factor in B-CLL cells because it is not activated in these cells.25 Indeed, similar levels of Notch1 and Notch2 mRNA, as well as mRNA of their ligands, were found in B-CLL and normal B cells, with the Notch-Hes1 axis down-regulated in B-CLL cells compared with normal B cells.25
The few and conflicting studies about the expression of Notch receptors in B-CLL cells and the role of Notch signaling in B-CLL cell survival prompted us to investigate these issues. We show that Notch1 and Notch2 proteins, as well as their ligands Jagged1 and Jagged2, are constitutively expressed in B-CLL but not in normal B cells. Notch signaling is constitutively activated in B-CLL cells and is further stimulated in B-CLL cells, which after 24-hour ex vivo culture resist spontaneous apoptosis. Notch stimulation by a soluble Jagged1 ligand favors survival of B-CLL cells, whereas Notch inhibition with the γ-secretase inhibitor I (GSI) and the specific Notch2 down-regulation by small-interfering-RNA (siRNA) accelerate their spontaneous apoptosis. Notch modulation is also accompanied by changes in some regulatory apoptosis molecules.
Methods
Patients
Twenty-five B-CLL patients and 5 healthy donors were included in this study. Diagnoses of B-CLL were based on Stanford criteria defined by the National Cancer Institute–sponsored Working Group,26 and clinical staging was based on the Binet classification.27 This study was approved by the Perugia University Institutional Review Board, and all patients signed informed consent in accordance with the Declaration of Helsinki. Table 1 summarizes the characteristics of the patients examined.
Patient characteristics
| Patients . | Binet stage . | Treatment status . | IgVH mutational status* . | ZAP-70 expression† . | CD38 expression‡ . |
|---|---|---|---|---|---|
| B-CLL 1 | A | Untreated | Mutated | 70.3 | 0.6 |
| B-CLL 2 | A | Untreated | Mutated | 10.1 | 2.6 |
| B-CLL 3 | B | Untreated | Unmutated | 41.8 | 1.7 |
| B-CLL 4 | A | Untreated | Mutated | 23.2 | 0.2 |
| B-CLL 5 | A | Untreated | Mutated | 92.0 | 1.5 |
| B-CLL 6 | C | Treated§ | Unmutated | 72.0 | 73.2 |
| B-CLL 7 | B | Treated§ | Unmutated | 4.6 | 81.0 |
| B-CLL 8 | B | Untreated | Unmutated | 86.1 | 57.2 |
| B-CLL 9 | B | Untreated | Unmutated | 32.0 | 0.5 |
| B-CLL 10 | B | Treated§ | Mutated | 95.4 | 0.2 |
| B-CLL 11 | B | Untreated | Unmutated | 73.2 | 64.4 |
| B-CLL 12 | B | Untreated | Unmutated | 92.8 | 0.4 |
| B-CLL 13 | B | Treated§ | Mutated | 61.2 | 5.0 |
| B-CLL 14 | C | Treated§ | ND | 71.5 | 96.0 |
| B-CLL 15 | B | Untreated | Mutated | 60.9 | 1.6 |
| B-CLL 16 | B | Untreated | Unmutated | 51.6 | 10.1 |
| B-CLL 17 | B | Untreated | Mutated | ND | ND |
| B-CLL 18 | A | Untreated | Unmutated | 67.4 | 13.0 |
| B-CLL 19 | B | Untreated | Unmutated | 97.6 | 1.6 |
| B-CLL 20 | B | Untreated | Unmutated | 30.5 | 3.6 |
| B-CLL 21 | B | Untreated | Unmutated | 29.1 | 1.7 |
| B-CLL 22 | C | Treated§ | Mutated | ND | ND |
| B-CLL 23 | C | Treated§ | Unmutated | ND | 15.8 |
| B-CLL 24 | B | Untreated | Unmutated | 44.5 | 2.9 |
| B-CLL 25 | B | Untreated | Unmutated | 60.1 | 10.6 |
| Patients . | Binet stage . | Treatment status . | IgVH mutational status* . | ZAP-70 expression† . | CD38 expression‡ . |
|---|---|---|---|---|---|
| B-CLL 1 | A | Untreated | Mutated | 70.3 | 0.6 |
| B-CLL 2 | A | Untreated | Mutated | 10.1 | 2.6 |
| B-CLL 3 | B | Untreated | Unmutated | 41.8 | 1.7 |
| B-CLL 4 | A | Untreated | Mutated | 23.2 | 0.2 |
| B-CLL 5 | A | Untreated | Mutated | 92.0 | 1.5 |
| B-CLL 6 | C | Treated§ | Unmutated | 72.0 | 73.2 |
| B-CLL 7 | B | Treated§ | Unmutated | 4.6 | 81.0 |
| B-CLL 8 | B | Untreated | Unmutated | 86.1 | 57.2 |
| B-CLL 9 | B | Untreated | Unmutated | 32.0 | 0.5 |
| B-CLL 10 | B | Treated§ | Mutated | 95.4 | 0.2 |
| B-CLL 11 | B | Untreated | Unmutated | 73.2 | 64.4 |
| B-CLL 12 | B | Untreated | Unmutated | 92.8 | 0.4 |
| B-CLL 13 | B | Treated§ | Mutated | 61.2 | 5.0 |
| B-CLL 14 | C | Treated§ | ND | 71.5 | 96.0 |
| B-CLL 15 | B | Untreated | Mutated | 60.9 | 1.6 |
| B-CLL 16 | B | Untreated | Unmutated | 51.6 | 10.1 |
| B-CLL 17 | B | Untreated | Mutated | ND | ND |
| B-CLL 18 | A | Untreated | Unmutated | 67.4 | 13.0 |
| B-CLL 19 | B | Untreated | Unmutated | 97.6 | 1.6 |
| B-CLL 20 | B | Untreated | Unmutated | 30.5 | 3.6 |
| B-CLL 21 | B | Untreated | Unmutated | 29.1 | 1.7 |
| B-CLL 22 | C | Treated§ | Mutated | ND | ND |
| B-CLL 23 | C | Treated§ | Unmutated | ND | 15.8 |
| B-CLL 24 | B | Untreated | Unmutated | 44.5 | 2.9 |
| B-CLL 25 | B | Untreated | Unmutated | 60.1 | 10.6 |
ND indicates not determined.
Mutated was defined as having a frequency of mutations greater than 2% from germline VH.
Values represent percentage of B-CLL cells expressing intracellular ZAP-70.
Values represent percentage of B-CLL cells expressing surface CD38.
Patients had not received treatment for at least 3 months prior to the study.
Cell purification
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood of B-CLL patients and buffy coats of healthy donors by Ficoll density-gradient centrifugation (Nycomed, Oslo, Norway). Monocytes were removed by plastic adherence. For B-CLL cell purification, the remaining lymphocytes (peripheral blood lymphocytes [PBLs]) were subjected to rosetting with sheep erythrocytes, whereas for normal B lymphocyte purification, PBLs were subjected to negative selection using the B-cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). All B-CLL samples contained more than 96% CD19+/CD5+ B-CLL cells, as assessed by flow cytometry (EPICS-XL-MCL; Beckman Coulter, Fullerton, CA). The purity of normal B cells (B-CD19+ cells) ranged from 97% to 99%, estimated by labeling with an anti-CD19 monoclonal antibody (mAb).
B-CLL clinical laboratory characteristics
To investigate VDJ rearrangement, genomic DNA was amplified using a mixture of sense primers annealing to the VH1 through VH6 FR1 and antisense primers complementary to the germline JH regions.28 For somatic hypermutation analysis, polymerase chain reaction products were gel purified (Qiagen Gel Purification Kit; Qiagen, Milan, Italy) and directly sequenced by the automatic DNA sequencer ABI PRISM 3130 (Applied Biosystems, Warrington, United Kingdom). The National Center for Biotechnology Information (NCBI) Ig Blast database and sequence alignment tool was used to identify and compare sequences with the most homologous germline VH gene family. VH gene sequences deviating more than 2% from the corresponding germline gene were defined as mutated. CD38 surface expression was examined using double-immunofluorescence fluorescein isothiocyanate (FITC)–CD19/phycoerythrin-CD38, and the percentage of CD38+ cells was measured in the CD19+ fraction. B-CLL samples were considered CD38+ when the CD38+ cell percentage exceeded 20%. To determine zeta-chain–associated protein 70 (ZAP-70) expression, B-CLL cells were labeled with anti–CD19-Cy5, fixed and permeabilized with the Fix & Perm Kit, and then incubated with anti–ZAP-70 Alexa Fluor 488–conjugated mAb (all from Caltag Laboratories, Burlingame, CA). Flow cytometric analysis was performed following published methods,29 and the cutoff value was set at 20% of ZAP-70+ B-CLL cells.
B-CLL cell culture
Freshly isolated B-CLL cells, resuspended at 2 × 106 cells/mL in complete medium consisting of RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Invitrogen, Paisley, United Kingdom), were incubated in culture tubes (3 mL/tube) at 37°C in a 5% CO2 atmosphere. In Notch inhibition experiments, B-CLL cells were cultured for 24 hours with GSI (Calbiochem, La Jolla, CA). GSI was dissolved in dimethyl sulfoxide (DMSO) and further diluted in complete medium at the final concentrations used, which did not exceed 0.02% and did not affect B-CLL cell viability or apoptosis.
Soluble Jagged1 ligand immobilization and B-CLL cell stimulation
The recombinant rat Jagged1-Fc fusion chimera (R&D Systems, Minneapolis, MN) was dissolved in phosphate-buffered saline and immobilized in flat-bottom 96-well plates for 20 hours at 4°C, at 10 μg/mL (100 μL/well). As control for Jagged1, human IgG-Fc (R&D Systems) was used. Plates coated with Jagged1 or IgG-Fc were washed and B-CLL cells were plated at 105 cells/well. After 48-hour culture, B-CLL cells were analyzed for cell viability and in Western blot and electrophoretic mobility shift assay (EMSA). IgG-Fc did not affect B-CLL cell viability.
Apoptosis assays
Apoptosis was assessed by flow cytometric analysis (EPICS-XL-MCL) of annexin V–FITC/propidium iodide (PI)–stained cells, and hypodiploid nuclei as previously reported.30 Annexin V/PI assay, which determines simultaneously cell viability and rate of apoptosis, was performed using a commercial kit (Immunotech; Beckman Coulter) according to the manufacturer's instructions.
B-CLL cell fractionation into viable/apoptotic cells
B-CLL cells were incubated for 24 hours in complete medium and viable cells isolated from apoptotic cells by magnetic cell sorting technique, using an annexin V Microbead Kit (Miltenyi Biotec) according to the manufacturer's instructions. Purity of cells was verified using annexin V–FITC.
Western blot analysis
Whole-cell lysates were extracted with radioimmunoprecipitation assay buffer. Equal amounts of proteins were separated by 7.5% to 12.0% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Protran; Schleicher & Schuell, Keene, NH), which, after blocking, were incubated with primary Abs. Rat mAbs anti-Notch1 (clone bTAN20), -Notch2 (clone C651.6DbHN), and -Jagged1 (clone TS1.15H), developed by Spiros Artavanis-Tsakonas, were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa. Rabbit polyclonal Abs to Notch2 (25-255), Hes1 (H-140), and Jagged2 (H-143), and lysates from the F-9 cell line, used as positive control for Hes1 expression, were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal Abs to caspase-3 and poly(ADP-ribose) polymerase (PARP) were from Cell Signaling Technology (Beverly, MA) and rabbit polyclonal Ab to B cell–activating factor of tumor necrosis factor family (BAFF) from Novus Biological (Littleton, CO). Mouse mAb to β-actin (clone AC-15) was from Sigma-Aldrich (St Louis, MO) and mouse mAb to TATA-binding protein (TBP; clone 1TBP18) from Abcam (Cambridge, MA). Mouse mAb to X-linked inhibitor of apoptosis protein (XIAP; clone 28) was from BD Transduction Laboratories (Lexington, KY) and the affinity–purified goat Ab to cellular inhibitor of apoptosis protein 2 (c-IAP2) from R&D Systems. Signals were detected using appropriate horseradish peroxidase–conjugated secondary Abs and the enhanced chemiluminescence system (Amersham Biosciences, Buckinghamshire, United Kingdom). Densitometric analysis of Notch1, Notch2, and Hes1 bands was performed using Quantity One software (Bio-Rad, Hercules, CA) and the relative densitometry units calculated relative to β-actin for each lane.
EMSA
Electrophoretic mobility shift assays (EMSAs) were performed to evaluate DNA-binding activity of CBF1 and nuclear factor–kappa B (NF-κB). Nuclear extracts were prepared as previously reported.4 The oligonucleotides used for CBF1 contained 2 specific binding sites for CBF1 derived from the mouse Hes1 promoter.13 Their sequence was as follows: 5′-GTTACTGTGGGAAAGAAAGTTTGGGAAGTTTCACAC-3′ (MWG Biotech, Ebersberg, Germany). To confirm binding specificity, we used oligonucleotides with mutations (italicized portions) in both CBF1-binding sites having the following sequence: 5′-GTTACTGTGCTGCAGAAAGTTTCTCGAGTTTCACAC-3′ (MWG Biotech).
The oligonucleotides used for NF-κB contained the NF-κB–binding sequence from the Ig κ light chain promoter (5′-AGTTGAGGGGACTTTCCCAGG-3′; Promega, Milan, Italy). Each double-stranded DNA oligonucleotide was end-labeled with [γ-32P] adenosine triphosphate using a T4 polynucleotide kinase (Roche Applied Sciences, Mannheim, Germany). All DNA-binding reactions were conducted in a volume of 20 μL and started by adding 10-μg nuclear proteins to a reaction mix containing 1 μg poly [d(I-C)] as nonspecific competitor, 4 μL of 5× DNA-binding buffer, and approximately 20 000 cpm of the respective radiolabeled double-stranded DNA oligonucleotide. Cold competitor oligonucleotides were added to the reaction mix at a 50-fold molar excess 10 minutes before the radiolabeled probe. The samples were then loaded on a 4% nondenaturing polyacrylamide gel in Tris/glycine/ethylenediaminetetraacetic acid buffer. After electrophoresis, protein-DNA complexes were visualized by autoradiography. For CBF1-binding blockage, 5-μg mAbs against Notch1, Notch2, or β-tubulin (clone Tub 2.1; Sigma-Aldrich) were incubated with nuclear proteins for 12 hours at 4°C before addition of the radiolabeled probe.
siRNA nucleofection
B-CLL cells (12 × 106), resuspended in 100 μL Cell Line Solution Kit V (Amaxa, Cologne, Germany) with 0.5 μM ON-TARGETplus SMARTpool Notch2 siRNA or ON-TARGETplus siCONTROL nontargeting pool as negative control (Dharmacon, Lafayette, CO), were transfected with the Amaxa Nucleofector II device (program U-013), cultured in 12-well plates in complete medium for 72 hours, and then examined for cell viability and Notch2 protein expression.
Statistical analysis
Statistical differences between mean values were evaluated using the Student t test. The minimal level of significance was P less than .05.
Results
B-CLL but not normal B cells express Notch1, Notch2, Jagged1, and Jagged2
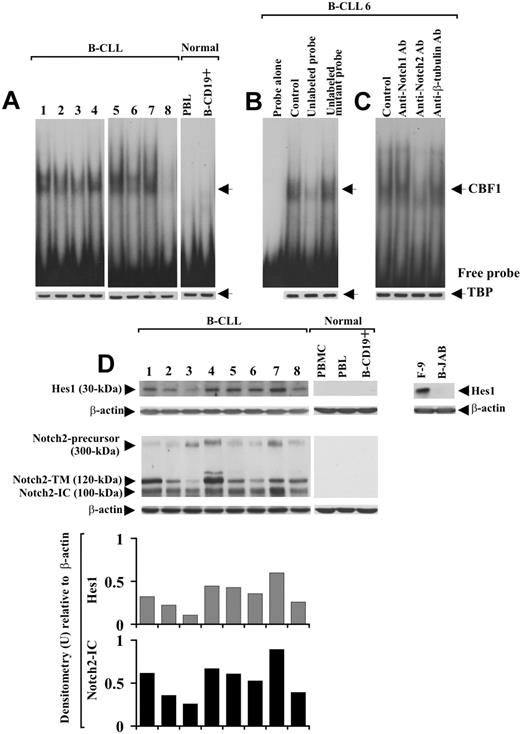
Notch1, Notch2, Jagged1, and Jagged2 protein expression was analyzed by Western blot in B-CLL cells from 25 patients and in PBMCs, PBLs, and B-CD19+ cells from 5 healthy donors. Table 1 gives the characteristics of B-CLL patients, including Binet stage, treatment, IgVH mutational status, and ZAP-70 and CD38 expression. Notch1 and Notch2 analysis, performed with mAbs directed to their N-IC domains, shows that B-CLL cells from all 25 patients examined expressed both receptors, regardless of IgVH mutational status and ZAP-70 and CD38 expression. Figure 1 shows data obtained with 8 B-CLL samples (1-8) selected to include patients with different characteristics. As shown in Figure 1A,B, each receptor appeared as 3 immunoreactive species of 300, 120, and 100 kDa, which correspond to the inactive precursor, the transmembrane/cytoplasmic portion (N-TM) and the N-IC domain, respectively.31 Notch2 expression was confirmed using an Ab raised against the N-EC domain of the receptor, able to detect a 180-kDa immunoreactive species corresponding to N-EC (Figure 1C). No Notch1 or Notch2 form was found in PBMCs, PBLs, and B-CD19+ cells from healthy donors (Figure 1A-C). Jagged1 and Jagged2 analysis shows that B-CLL cells from all 25 patients expressed both ligands, each appearing as a doublet of 150 kDa, whereas these proteins were undetectable in PBMCs, PBLs, and B-CD19+ cells from all healthy donors examined (Figure 1D,E).
Expression of Notch1, Notch2, Jagged1, and Jagged2 in B-CLL and normal B cells. The expression of Notch1 (A), Notch2 (B,C), Jagged1 (D), and Jagged2 (E) proteins was analyzed by Western blot in whole-cell lysates (25 μg for Notch1 and Notch2 using mAbs directed to N-IC domains, 70 μg for Notch2 using an Ab directed to N-EC domain, and 60 μg for Jagged1 and Jagged2) extracted from freshly isolated B-CLL cells (n = 25) and PBMCs, PBLs, and B-CD19+ cells from healthy donors (n = 5). Results from 8 B-CLL patients having different characteristics and 1 representative healthy donor are shown. Whole-cell lysates isolated from KM-H2 and B-JAB cell lines were used as positive and negative controls, respectively, for receptor expression. Whole-cell lysates isolated from IM-9 and Jurkat cell lines were used as positive controls for Jagged1 and Jagged2 expression, respectively. Whole-cell lysates isolated from NIH-3T3 cell line were used as a negative control for expression of both ligands. Protein loading was assessed by reprobing the blots with an anti–β-actin mAb.
Expression of Notch1, Notch2, Jagged1, and Jagged2 in B-CLL and normal B cells. The expression of Notch1 (A), Notch2 (B,C), Jagged1 (D), and Jagged2 (E) proteins was analyzed by Western blot in whole-cell lysates (25 μg for Notch1 and Notch2 using mAbs directed to N-IC domains, 70 μg for Notch2 using an Ab directed to N-EC domain, and 60 μg for Jagged1 and Jagged2) extracted from freshly isolated B-CLL cells (n = 25) and PBMCs, PBLs, and B-CD19+ cells from healthy donors (n = 5). Results from 8 B-CLL patients having different characteristics and 1 representative healthy donor are shown. Whole-cell lysates isolated from KM-H2 and B-JAB cell lines were used as positive and negative controls, respectively, for receptor expression. Whole-cell lysates isolated from IM-9 and Jurkat cell lines were used as positive controls for Jagged1 and Jagged2 expression, respectively. Whole-cell lysates isolated from NIH-3T3 cell line were used as a negative control for expression of both ligands. Protein loading was assessed by reprobing the blots with an anti–β-actin mAb.
Notch signaling is constitutively activated in B-CLL cells
It is known that after Notch-ligand interaction, the active N-IC is released into the cytoplasm and translocates to the nucleus, where it activates transcription of CBF1-responsive genes, including Hes1.15,16 On this basis, to investigate Notch activation and whether it could lead to Hes1 induction via CBF1, B-CLL cells from patients 1 to 8 were analyzed in EMSA using an oligonucleotide containing 2 well-characterized CBF1-binding sites derived from the murine Hes1 promoter.13 Slow-migrating DNA-protein complexes were detected in nuclear extracts from all 8 B-CLL samples examined (Figure 2A). The bands indicated by the arrow corresponded to specific CBF1-Hes1 complexes because they disappeared after incubation of the nuclear extracts with a 50-fold molar excess of unlabeled CBF1 probe but were not affected by a 50-fold molar excess of unlabeled mutant CBF1 probe (Figure 2B). The DNA-protein complexes observed in B-CLL cells were undetectable in PBLs and B-CD19+ cells from 3 healthy donors (Figure 2A).
Analysis of CBF1-Hes1 nuclear complexes and Hes1 protein expression in B-CLL cells. (A) Nuclear extracts were prepared from freshly isolated B-CLL cells (n = 8) and PBLs and B-CD19+ cells from healthy donors (n = 3). CBF1-binding activity to a 32P-labeled Hes1 probe containing 2 specific sites for CBF1 was evaluated in 10 μg nuclear proteins by EMSA. Results from all 8 B-CLL patients examined and 1 representative healthy donor are shown. (B) For competition assay, nuclear extracts from B-CLL samples were incubated with a 50-fold molar excess of unlabeled Hes1 probe or a 50-fold molar excess of unlabeled mutant Hes1 probe, 10 minutes before the addition of the 32P-labeled specific probe. (C) For blockade of CBF1-binding activity, nuclear extracts from each B-CLL sample were incubated for 12 hours at 4°C with 5 μg anti-Notch1, anti-Notch2, or anti–β-tubulin mAbs, before the addition of the 32P-labeled probe. In panels B and C, the data shown for patient 6 are representative of 8 patients. (A-C) Nuclear extracts (10 μg) used in EMSA were subjected to Western blot analysis with an anti-TBP mAb as a loading control. (D) Whole-cell lysates (25 μg) extracted from freshly isolated B-CLL cells (n = 25) and PBMCs, PBLs, and B-CD19+ cells from healthy donors (n = 5) were analyzed by Western blot for Hes1 expression. Results from 8 B-CLL patients and 1 representative healthy donor are shown. Whole-cell lysates isolated from F-9 and B-JAB cell lines were used as positive and negative controls for Hes1 expression, respectively. Protein loading was assessed by reprobing the blots with an anti–β-actin mAb. The blot of Hes1 along with the blot of Notch2, already shown in Figure 1B, were subjected to densitometric analysis and the density of the bands corresponding to Hes1 and Notch2-IC, expressed as arbitrary units, is reported for each patient. Densitometry units (U) were calculated relative to β-actin for each lane. Lane designations are identical for blots and histograms.
Analysis of CBF1-Hes1 nuclear complexes and Hes1 protein expression in B-CLL cells. (A) Nuclear extracts were prepared from freshly isolated B-CLL cells (n = 8) and PBLs and B-CD19+ cells from healthy donors (n = 3). CBF1-binding activity to a 32P-labeled Hes1 probe containing 2 specific sites for CBF1 was evaluated in 10 μg nuclear proteins by EMSA. Results from all 8 B-CLL patients examined and 1 representative healthy donor are shown. (B) For competition assay, nuclear extracts from B-CLL samples were incubated with a 50-fold molar excess of unlabeled Hes1 probe or a 50-fold molar excess of unlabeled mutant Hes1 probe, 10 minutes before the addition of the 32P-labeled specific probe. (C) For blockade of CBF1-binding activity, nuclear extracts from each B-CLL sample were incubated for 12 hours at 4°C with 5 μg anti-Notch1, anti-Notch2, or anti–β-tubulin mAbs, before the addition of the 32P-labeled probe. In panels B and C, the data shown for patient 6 are representative of 8 patients. (A-C) Nuclear extracts (10 μg) used in EMSA were subjected to Western blot analysis with an anti-TBP mAb as a loading control. (D) Whole-cell lysates (25 μg) extracted from freshly isolated B-CLL cells (n = 25) and PBMCs, PBLs, and B-CD19+ cells from healthy donors (n = 5) were analyzed by Western blot for Hes1 expression. Results from 8 B-CLL patients and 1 representative healthy donor are shown. Whole-cell lysates isolated from F-9 and B-JAB cell lines were used as positive and negative controls for Hes1 expression, respectively. Protein loading was assessed by reprobing the blots with an anti–β-actin mAb. The blot of Hes1 along with the blot of Notch2, already shown in Figure 1B, were subjected to densitometric analysis and the density of the bands corresponding to Hes1 and Notch2-IC, expressed as arbitrary units, is reported for each patient. Densitometry units (U) were calculated relative to β-actin for each lane. Lane designations are identical for blots and histograms.
To determine whether Notch1 and Notch2 were involved in CBF1-binding activity of B-CLL cells, nuclear extracts were pretreated for 12 hours with 5 μg anti-Notch1- or -Notch2-IC mAbs and then analyzed in EMSA. Anti-Notch2 mAb strongly reduced CBF1-binding activity, whereas anti-Notch1 mAb and an unrelated mAb had no effect (Figure 2C).
We next analyzed Hes1 protein expression in whole-cell lysates of B-CLL cells. B-CLL cells from all 25 patients expressed Hes1, which instead was undetectable in PBMCs, PBLs, and B-CD19+ cells from healthy donors (Figure 2D). When the expression levels of Hes1 were compared with those of Notch2-IC, we found that B-CLL samples with sustained Hes1 levels exhibited high Notch2-IC expression, and samples with lower Hes1 levels showed a low Notch2-IC expression (Figure 2D). Altogether, these results suggest that Notch signaling is constitutively activated in B-CLL cells and that Notch2 activity induces Hes1 via CBF1.
Notch-signaling activation increases in B-CLL cells that resist spontaneous apoptosis
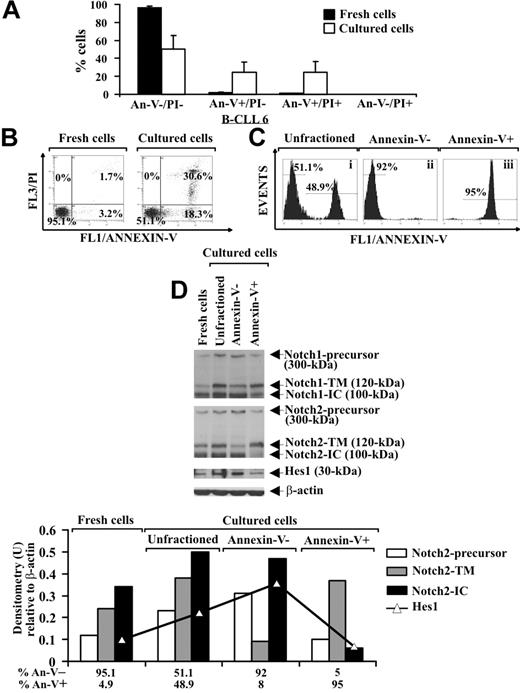
It is known that B-CLL cells undergo spontaneous apoptosis when cultured ex vivo.3,5 Analysis of apoptosis, by annexin V/PI staining, in B-CLL cells from 8 patients (2-4, 6, 16-19) shows that, after 24-hour culture in complete medium, the early and late apoptotic cells increased to 24.6% plus or minus 11.3% and 24.8% plus or minus 11.8%, respectively, compared with fresh B-CLL cells, whereas 50.6% plus or minus 14.5% of the cells remained viable. Necrotic cells were absent after 24-hour culture (Figure 3A,B). To investigate a possible correlation between Notch signaling and B-CLL cell survival, we separated cultured B-CLL cells (patients 2, 4, 6, and 16) in viable (annexin V−) and apoptotic (annexin V+) fractions using annexin V–coated magnetic microbeads and then analyzed the fractions obtained and the unfractioned population for Notch1, Notch2, and Hes1 protein expression. Two selection rounds were performed to yield more than 90% pure viable and apoptotic cells (Figure 3Cii and Ciii, respectively). Results show that, in cultured unfractioned B-CLL cells, the expression of the precursors, N-TM and N-IC forms of Notch1 and Notch2, as well as Hes1, were increased compared with fresh B-CLL cells. The N-IC forms and the precursors of both receptors were mainly expressed in viable cells, whereas apoptotic cells predominantly exhibited the N-TM forms. Hes1 was mainly expressed in viable cells (Figure 3D). These results indicate that in B-CLL cells, which had resisted spontaneous apoptosis after 24-hour culture, Notch signaling was activated more than in both fresh cells and cells that had undergone apoptosis.
Analysis of Notch1, Notch2, and Hes1 expression in B-CLL cells cultured ex vivo. (A,B) Freshly isolated B-CLL cells were cultured for 24 hours in complete medium (n = 8), and apoptosis was determined by flow cytometric analysis of annexin V/PI–stained cells. Results in panel A (mean ± SD of all 8 patients) and panel B (relative to patient 6 and representative of 8 patients) are presented as the percentage of viable (annexin V−/PI−), early apoptotic (annexin V+/PI−), late apoptotic (annexin V+/PI+), and necrotic cells (annexin V−/PI+). (C) Cultured B-CLL cells were separated into viable and apoptotic fractions (n = 4) using annexin V–coated magnetic microbeads, which bind to cells with external exposure of membrane phosphatidylserine. Flow cytometric analysis of B-CLL cells stained with annexin V–FITC shows unfractioned cells (i), negatively selected viable cells (ii), and positively selected apoptotic cells (iii). The data shown for patient 6 are representative of 4 patients. (D) Whole-cell lysates (25 μg) extracted from the indicated populations were analyzed by Western blot for Notch1, Notch2, and Hes1 expression (n = 4), and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. Densitometry was performed, and the density of the bands corresponding to Notch2-precursor, Notch2-TM, Notch2-IC, and Hes1, expressed as arbitrary units, is reported for each cell population. Densitometry units (U) were calculated relative to β-actin for each lane. The data shown for patient 6 are representative of 4 patients.
Analysis of Notch1, Notch2, and Hes1 expression in B-CLL cells cultured ex vivo. (A,B) Freshly isolated B-CLL cells were cultured for 24 hours in complete medium (n = 8), and apoptosis was determined by flow cytometric analysis of annexin V/PI–stained cells. Results in panel A (mean ± SD of all 8 patients) and panel B (relative to patient 6 and representative of 8 patients) are presented as the percentage of viable (annexin V−/PI−), early apoptotic (annexin V+/PI−), late apoptotic (annexin V+/PI+), and necrotic cells (annexin V−/PI+). (C) Cultured B-CLL cells were separated into viable and apoptotic fractions (n = 4) using annexin V–coated magnetic microbeads, which bind to cells with external exposure of membrane phosphatidylserine. Flow cytometric analysis of B-CLL cells stained with annexin V–FITC shows unfractioned cells (i), negatively selected viable cells (ii), and positively selected apoptotic cells (iii). The data shown for patient 6 are representative of 4 patients. (D) Whole-cell lysates (25 μg) extracted from the indicated populations were analyzed by Western blot for Notch1, Notch2, and Hes1 expression (n = 4), and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. Densitometry was performed, and the density of the bands corresponding to Notch2-precursor, Notch2-TM, Notch2-IC, and Hes1, expressed as arbitrary units, is reported for each cell population. Densitometry units (U) were calculated relative to β-actin for each lane. The data shown for patient 6 are representative of 4 patients.
Notch-signaling inhibition by GSI and Notch2 down-regulation by siRNA are associated with increased spontaneous B-CLL cell apoptosis
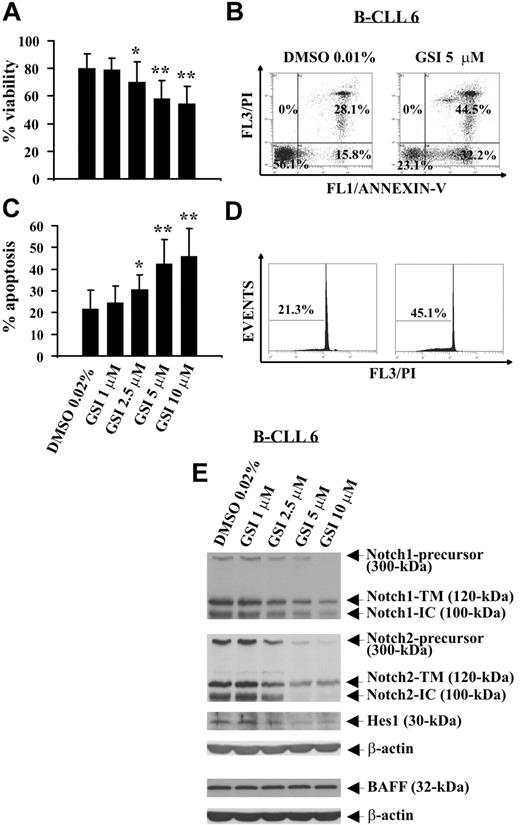
GSI, a pharmacologic inhibitor of γ-secretase able to block proteolysis of N-IC of all Notch receptors,32,33 was used to evaluate the effect of Notch inhibition on B-CLL cell survival. B-CLL cells from 12 patients (1, 2, 5, 6, 8, 14, 17, 20-24) were incubated for 24 hours with increasing GSI concentrations or DMSO, and cell viability was evaluated by trypan blue exclusion. Results show that GSI reduced B-CLL cell viability in a concentration-dependent manner, and the concentrations of 10 and 5 μM induced the greatest effects (P < .01). Significant effects were also observed with GSI 2.5 μM (P < .05), whereas GSI 1 μM had no effect (Figure 4A). These results were confirmed by annexin V/PI staining (Figure 4B; and data not shown). B-CLL cell death induced by GSI occurred by apoptosis as indicated by the analysis of annexin V+ cells (Figure 4B) and hypodiploid nuclei (Figure 4C,D). The effects of GSI on B-CLL cell apoptosis were concentration-dependent, and the concentrations 10 and 5 μM were the most effective (Figure 4C). The mean percentage of hypodiploid nuclei, which after 24-hour culture in DMSO was 21.7% plus or minus 8.6%, increased in the presence of GSI, reaching values of 45.9% plus or minus 12.8% (GSI 10 μM, P < .01), 42.4% plus or minus 11.1% (GSI 5 μM, P < .01), 30.5% plus or minus 7% (GSI 2.5 μM, P < .05), and 24.5% plus or minus 7.7% (GSI 1 μM, not significant).
Effect of GSI on spontaneous B-CLL cell apoptosis and Notch1, Notch2, and Hes1 expression. Freshly isolated B-CLL cells were cultured for 24 hours in complete medium with the indicated GSI concentrations or DMSO as control (n = 12). Cell viability was evaluated by trypan blue exclusion (A) and annexin V/PI staining (B). Apoptosis was evaluated by flow cytometric analysis of annexin V/PI–stained cells (B) and hypodiploid nuclei (C,D). Results in panels A and C are presented as the mean plus or minus SD of all 12 patients examined. *P less than .05, **P less than .01 (GSI vs DMSO) according to Student t test. Results in panel B are presented as the percentage of viable (annexin V−/PI−), early apoptotic (annexin V+/PI−), late apoptotic (annexin V+/PI+), and necrotic cells (annexin V−/PI+). Results in panel D are presented as the percentage of hypodiploid nuclei. (E) The expression of Notch1, Notch2, Hes1, and BAFF was analyzed by Western blot in whole-cell lysates (25 μg for Notch1, Notch2, and Hes1 and 50 μg for BAFF), and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. In panels B, D, and E, the results shown for patient 6 are representative of 12 patients.
Effect of GSI on spontaneous B-CLL cell apoptosis and Notch1, Notch2, and Hes1 expression. Freshly isolated B-CLL cells were cultured for 24 hours in complete medium with the indicated GSI concentrations or DMSO as control (n = 12). Cell viability was evaluated by trypan blue exclusion (A) and annexin V/PI staining (B). Apoptosis was evaluated by flow cytometric analysis of annexin V/PI–stained cells (B) and hypodiploid nuclei (C,D). Results in panels A and C are presented as the mean plus or minus SD of all 12 patients examined. *P less than .05, **P less than .01 (GSI vs DMSO) according to Student t test. Results in panel B are presented as the percentage of viable (annexin V−/PI−), early apoptotic (annexin V+/PI−), late apoptotic (annexin V+/PI+), and necrotic cells (annexin V−/PI+). Results in panel D are presented as the percentage of hypodiploid nuclei. (E) The expression of Notch1, Notch2, Hes1, and BAFF was analyzed by Western blot in whole-cell lysates (25 μg for Notch1, Notch2, and Hes1 and 50 μg for BAFF), and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. In panels B, D, and E, the results shown for patient 6 are representative of 12 patients.
To verify whether Notch signaling was inhibited by GSI, B-CLL cells tested for apoptosis were analyzed for Notch1, Notch2, and Hes1 expression. In all 12 patients examined, GSI reduced in a concentration-dependent manner Notch1- and Notch2-IC levels, as well as the expression of the precursors and N-TM forms of both receptors. Hes1 protein expression was also reduced by GSI. These results are shown in Figure 4E, which reports representative data from patient 6, and in Table 2, which summarizes the inhibitory effects of GSI on Notch1, Notch2, and Hes1 expression in all patients examined.
Percentage inhibition of Notch1, Notch2, and Hes1 expression by GSI
| Patients, GSI (μM) . | Notch1 . | Notch2 . | Hes1 . | ||||
|---|---|---|---|---|---|---|---|
| P . | TM . | IC . | P . | TM . | IC . | ||
| B-CLL 1 | |||||||
| 1 | 0 | 0 | 0 | 6 | 4 | 10 | 2 |
| 2.5 | 65 | 50 | 54 | 86 | 40 | 67 | 31 |
| 5 | 90 | 81 | 75 | 100 | 77 | 99 | 42 |
| 10 | 94 | 80 | 75 | 100 | 69 | 97 | 42 |
| B-CLL 2 | |||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.5 | ND | ND | ND | ND | ND | ND | ND |
| 5 | 18 | 10 | 15 | 21 | 15 | 18 | 10 |
| 10 | 60 | 49 | 44 | 51 | 44 | 58 | 27 |
| B-CLL 5 | |||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.5 | 51 | 40 | 46 | 60 | 35 | 44 | 29 |
| 5 | 70 | 57 | 61 | 75 | 51 | 73 | 46 |
| 10 | 71 | 66 | 70 | 96 | 89 | 96 | 49 |
| B-CLL 6 | |||||||
| 1 | 0 | 13 | 14 | 0 | 0 | 0 | 0 |
| 2.5 | 54 | 49 | 47 | 42 | 48 | 51 | 51 |
| 5 | 70 | 71 | 64 | 87 | 58 | 96 | 82 |
| 10 | 98 | 86 | 81 | 99 | 57 | 98 | 87 |
| B-CLL 8 | |||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.5 | 29 | 16 | 14 | 35 | 18 | 29 | 12 |
| 5 | 81 | 74 | 72 | 100 | 95 | 99 | 89 |
| 10 | 88 | 75 | 76 | 100 | 96 | 100 | 90 |
| B-CLL 14 | |||||||
| 1 | ND | ND | ND | ND | ND | ND | ND |
| 2.5 | 61 | 50 | 51 | 77 | 46 | 54 | 41 |
| 5 | 92 | 75 | 78 | 95 | 80 | 93 | 66 |
| 10 | 94 | 74 | 80 | 91 | 87 | 100 | 66 |
| B-CLL 17 | |||||||
| 1 | 0 | 0 | 0 | 1 | 2 | 4 | 0 |
| 2.5 | 39 | 25 | 29 | 44 | 31 | 35 | 29 |
| 5 | 93 | 79 | 82 | 100 | 81 | 87 | 59 |
| 10 | 100 | 83 | 85 | 100 | 100 | 100 | 63 |
| B-CLL 20 | |||||||
| 1 | 5 | 7 | 0 | 4 | 8 | 6 | 0 |
| 2.5 | 62 | 61 | 43 | 67 | 50 | 57 | 32 |
| 5 | 85 | 75 | 72 | 100 | 65 | 100 | 60 |
| 10 | 90 | 80 | 81 | 100 | 82 | 100 | 64 |
| B-CLL 21 | |||||||
| 1 | 13 | 15 | 16 | 17 | 10 | 21 | 15 |
| 2.5 | 21 | 18 | 24 | 18 | 11 | 20 | 15 |
| 5 | 78 | 60 | 63 | 100 | 56 | 95 | 49 |
| 10 | ND | ND | ND | ND | ND | ND | ND |
| B-CLL 22 | |||||||
| 1 | 26 | 21 | 29 | 30 | 17 | 31 | 20 |
| 2.5 | 31 | 25 | 30 | 35 | 21 | 40 | 33 |
| 5 | 81 | 73 | 69 | 99 | 99 | 98 | 70 |
| 10 | 91 | 85 | 83 | 100 | 100 | 100 | 90 |
| B-CLL 23 | |||||||
| 1 | 0 | 0 | 0 | 5 | 3 | 7 | 0 |
| 2.5 | 25 | 21 | 21 | 16 | 14 | 23 | 10 |
| 5 | 83 | 65 | 64 | 89 | 71 | 93 | 46 |
| 10 | 88 | 77 | 79 | 100 | 88 | 100 | 54 |
| B-CLL 24 | |||||||
| 1 | ND | ND | ND | ND | ND | ND | ND |
| 2.5 | ND | ND | ND | ND | ND | ND | ND |
| 5 | 92 | 80 | 77 | 100 | 81 | 96 | 39 |
| 10 | 100 | 97 | 96 | 100 | 84 | 99 | 52 |
| Patients, GSI (μM) . | Notch1 . | Notch2 . | Hes1 . | ||||
|---|---|---|---|---|---|---|---|
| P . | TM . | IC . | P . | TM . | IC . | ||
| B-CLL 1 | |||||||
| 1 | 0 | 0 | 0 | 6 | 4 | 10 | 2 |
| 2.5 | 65 | 50 | 54 | 86 | 40 | 67 | 31 |
| 5 | 90 | 81 | 75 | 100 | 77 | 99 | 42 |
| 10 | 94 | 80 | 75 | 100 | 69 | 97 | 42 |
| B-CLL 2 | |||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.5 | ND | ND | ND | ND | ND | ND | ND |
| 5 | 18 | 10 | 15 | 21 | 15 | 18 | 10 |
| 10 | 60 | 49 | 44 | 51 | 44 | 58 | 27 |
| B-CLL 5 | |||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.5 | 51 | 40 | 46 | 60 | 35 | 44 | 29 |
| 5 | 70 | 57 | 61 | 75 | 51 | 73 | 46 |
| 10 | 71 | 66 | 70 | 96 | 89 | 96 | 49 |
| B-CLL 6 | |||||||
| 1 | 0 | 13 | 14 | 0 | 0 | 0 | 0 |
| 2.5 | 54 | 49 | 47 | 42 | 48 | 51 | 51 |
| 5 | 70 | 71 | 64 | 87 | 58 | 96 | 82 |
| 10 | 98 | 86 | 81 | 99 | 57 | 98 | 87 |
| B-CLL 8 | |||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.5 | 29 | 16 | 14 | 35 | 18 | 29 | 12 |
| 5 | 81 | 74 | 72 | 100 | 95 | 99 | 89 |
| 10 | 88 | 75 | 76 | 100 | 96 | 100 | 90 |
| B-CLL 14 | |||||||
| 1 | ND | ND | ND | ND | ND | ND | ND |
| 2.5 | 61 | 50 | 51 | 77 | 46 | 54 | 41 |
| 5 | 92 | 75 | 78 | 95 | 80 | 93 | 66 |
| 10 | 94 | 74 | 80 | 91 | 87 | 100 | 66 |
| B-CLL 17 | |||||||
| 1 | 0 | 0 | 0 | 1 | 2 | 4 | 0 |
| 2.5 | 39 | 25 | 29 | 44 | 31 | 35 | 29 |
| 5 | 93 | 79 | 82 | 100 | 81 | 87 | 59 |
| 10 | 100 | 83 | 85 | 100 | 100 | 100 | 63 |
| B-CLL 20 | |||||||
| 1 | 5 | 7 | 0 | 4 | 8 | 6 | 0 |
| 2.5 | 62 | 61 | 43 | 67 | 50 | 57 | 32 |
| 5 | 85 | 75 | 72 | 100 | 65 | 100 | 60 |
| 10 | 90 | 80 | 81 | 100 | 82 | 100 | 64 |
| B-CLL 21 | |||||||
| 1 | 13 | 15 | 16 | 17 | 10 | 21 | 15 |
| 2.5 | 21 | 18 | 24 | 18 | 11 | 20 | 15 |
| 5 | 78 | 60 | 63 | 100 | 56 | 95 | 49 |
| 10 | ND | ND | ND | ND | ND | ND | ND |
| B-CLL 22 | |||||||
| 1 | 26 | 21 | 29 | 30 | 17 | 31 | 20 |
| 2.5 | 31 | 25 | 30 | 35 | 21 | 40 | 33 |
| 5 | 81 | 73 | 69 | 99 | 99 | 98 | 70 |
| 10 | 91 | 85 | 83 | 100 | 100 | 100 | 90 |
| B-CLL 23 | |||||||
| 1 | 0 | 0 | 0 | 5 | 3 | 7 | 0 |
| 2.5 | 25 | 21 | 21 | 16 | 14 | 23 | 10 |
| 5 | 83 | 65 | 64 | 89 | 71 | 93 | 46 |
| 10 | 88 | 77 | 79 | 100 | 88 | 100 | 54 |
| B-CLL 24 | |||||||
| 1 | ND | ND | ND | ND | ND | ND | ND |
| 2.5 | ND | ND | ND | ND | ND | ND | ND |
| 5 | 92 | 80 | 77 | 100 | 81 | 96 | 39 |
| 10 | 100 | 97 | 96 | 100 | 84 | 99 | 52 |
Notch1, Notch2, and Hes1 expression was analyzed by Western blot in B-CLL cells treated with GSI or DMSO for 24 hours. For each patient, the intensity of the bands corresponding to Notch1 and Notch2 (P, TM, and IC) and Hes1 was calculated by densitometric analysis and normalized to β-actin levels. Values represent the percentage inhibition of Notch1, Notch2, and Hes1 expression in GSI-treated compared with DMSO-treated B-CLL cells. P indicates precursor; ND, not determined.
To exclude that GSI can inhibit Notch expression because of a general toxicity exerted on B-CLL cells, we analyzed B-CLL cells treated with GSI (1, 2, 5, 6, 8) for BAFF receptor expression, a type II transmembrane protein constitutively expressed in B-CLL cells,6 which is neither a GSI nor γ-secretase target. Figure 4E shows that BAFF levels were not affected by GSI at all concentrations used.
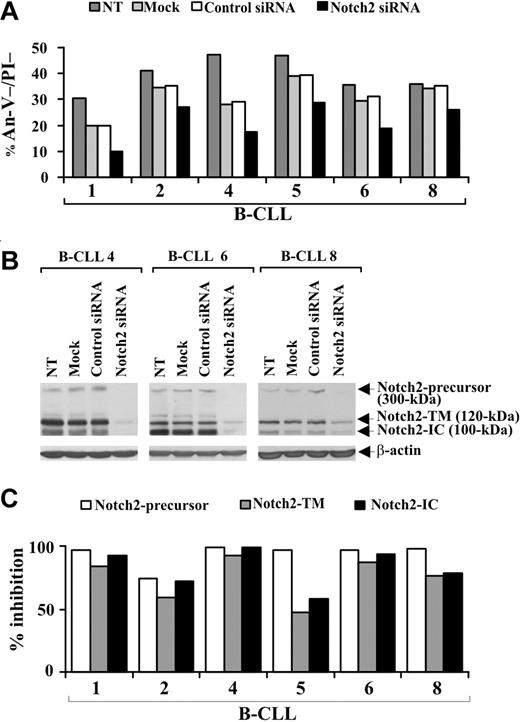
To definitely establish whether a disrupted Notch pathway limits B-CLL cell survival, we specifically down-regulated Notch2 expression by siRNA nucleofection. Preliminary experiments to optimize transfection conditions showed that the optimal siRNA dose and incubation time for inducing maximum inhibition of Notch2 protein expression were 0.5 μM for 72 hours (data not shown). B-CLL cells from 6 patients (1, 2, 4, 5, 6, 8) were nontransfected, mock-transfected, transfected with control nontargeting or Notch2 siRNA, and at 72 hours after transfection were examined for viability by annexin V/PI staining and Notch2 protein expression by Western blot. Figure 5A shows that in all samples examined the viability of mock-transfected cells decreased compared with nontransfected cells. This decrease was sample-dependent and ranged from 5% (sample 8) to 41% (sample 4). For each patient, similar values of cell viability were observed in control siRNA-transfected cells, suggesting that B-CLL cell viability is affected by the nucleofection procedure but not by nontargeting siRNA introduction. In all patients examined, cell viability was further reduced by Notch2 siRNA compared with control siRNA (Figure 5A). The percentage (mean ± SD of 6 samples) of viable B-CLL cells was 31.7% plus or minus 6.8% in control siRNA-transfected cells and decreased to 21.3% plus or minus 7.2% in Notch2 siRNA-transfected cells (P < .01). Analysis of Notch2 protein shows that, in all patients examined, the expression of all 3 forms of the receptor was inhibited in Notch2 siRNA-transfected cells with respect to control siRNA-transfected cells (Figure 5B,C), which exhibited Notch2 levels similar to those of mock-transfected and nontransfected cells (Figure 5B).
Effect of Notch2 down-regulation on B-CLL cell survival. Freshly isolated B-CLL cells were nontransfected (NT), mock-transfected (no siRNA), or transfected with 0.5 μM control nontargeting or Notch2 siRNA (n = 6) as described in “siRNA nucleofection” and then cultured in complete medium for 72 hours. (A) Cell viability was evaluated by annexin V/PI staining, and the results from each patient are presented as the percentage of annexin V−/PI− cells. (B) Notch2 protein expression was analyzed by Western blot in 25 μg whole-cell lysates from the indicated cell populations, and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. The results shown for patients 4, 6, and 8 are representative of 6 patients. (C) For each sample, blot lanes corresponding to control nontargeting and Notch2 siRNA were subjected to densitometric analysis and normalized to β-actin levels. Columns represent the percentage inhibition of Notch2-precursor, Notch2-TM, and Notch2-IC induced by Notch2 siRNA compared with control siRNA.
Effect of Notch2 down-regulation on B-CLL cell survival. Freshly isolated B-CLL cells were nontransfected (NT), mock-transfected (no siRNA), or transfected with 0.5 μM control nontargeting or Notch2 siRNA (n = 6) as described in “siRNA nucleofection” and then cultured in complete medium for 72 hours. (A) Cell viability was evaluated by annexin V/PI staining, and the results from each patient are presented as the percentage of annexin V−/PI− cells. (B) Notch2 protein expression was analyzed by Western blot in 25 μg whole-cell lysates from the indicated cell populations, and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. The results shown for patients 4, 6, and 8 are representative of 6 patients. (C) For each sample, blot lanes corresponding to control nontargeting and Notch2 siRNA were subjected to densitometric analysis and normalized to β-actin levels. Columns represent the percentage inhibition of Notch2-precursor, Notch2-TM, and Notch2-IC induced by Notch2 siRNA compared with control siRNA.
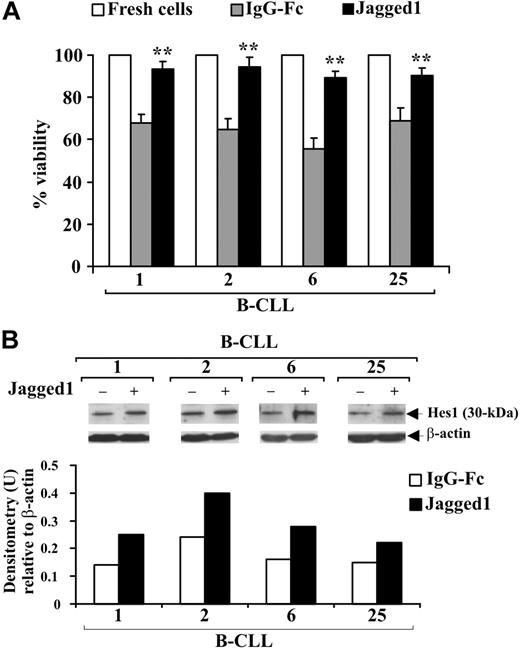
Notch-signaling stimulation with Jagged1 ligand is associated with increased B-CLL cell survival
B-CLL cells from 4 patients (1, 2, 6, 25) were cultured for 48 hours in complete medium on immobilized soluble Jagged1 ligand or IgG-Fc as control and cell viability evaluated by trypan blue exclusion. Figure 6A shows that the decrease in viability observed in B-CLL cells cultured with IgG-Fc compared with freshly isolated cells was prevented by Jagged1 ligand. To verify whether Jagged1 stimulates Notch activation, B-CLL cells were analyzed for Hes1 expression. In all patients, Hes1 levels were increased by Jagged1 compared with controls (Figure 6B).
Effect of soluble Jagged1 ligand on B-CLL cell survival. Freshly isolated B-CLL cells were cultured for 48 hours in complete medium with 10 μg/mL (100 μL/well) immobilized soluble Jagged1 ligand or IgG-Fc as control (n = 4) as described in “Soluble Jagged1 ligand immobilization and B-CLL cell stimulation.” (A) Cell viability was evaluated by trypan blue exclusion. Data for each patient are expressed as mean plus or minus SD of 4 determinations. **P less than .01, Jagged1-treated cells vs IgG-Fc–treated cells for each patient, according to the Student t test. (B) Hes1 protein expression was analyzed in 25 μg whole-cell lysates by Western blot, and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. Densitometry was performed, and the density of the bands expressed as arbitrary units is reported for each patient. Densitometry units (U) were calculated relative to β-actin for each lane. Results from all 4 patients examined are shown.
Effect of soluble Jagged1 ligand on B-CLL cell survival. Freshly isolated B-CLL cells were cultured for 48 hours in complete medium with 10 μg/mL (100 μL/well) immobilized soluble Jagged1 ligand or IgG-Fc as control (n = 4) as described in “Soluble Jagged1 ligand immobilization and B-CLL cell stimulation.” (A) Cell viability was evaluated by trypan blue exclusion. Data for each patient are expressed as mean plus or minus SD of 4 determinations. **P less than .01, Jagged1-treated cells vs IgG-Fc–treated cells for each patient, according to the Student t test. (B) Hes1 protein expression was analyzed in 25 μg whole-cell lysates by Western blot, and protein loading was assessed by reprobing the blots with an anti–β-actin mAb. Densitometry was performed, and the density of the bands expressed as arbitrary units is reported for each patient. Densitometry units (U) were calculated relative to β-actin for each lane. Results from all 4 patients examined are shown.
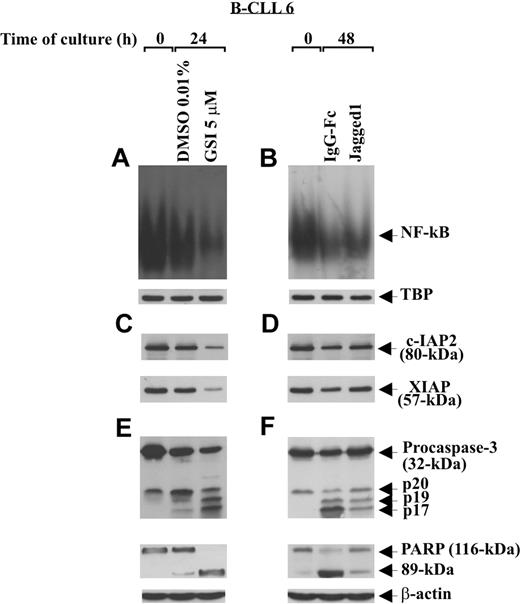
Notch-dependent survival of B-CLL cells is accompanied by increased NF-κB activity and c-IAP protein expression
To examine the effect of Notch stimulation on some regulatory and effector apoptosis molecules, B-CLL cells (patients 1, 2, 6) were cultured for 48 hours on immobilized Jagged1 ligand or IgG-Fc as control and then analyzed for NF-κB activity, c-IAP2 and XIAP expression, and caspase-3 activation. Parallel experiments were performed to examine the effect of Notch inhibition, induced by GSI 5 μM after 24-hour culture, on the same apoptosis regulators. Figure 7A,B shows that the constitutive NF-κB activity of B-CLL cells decreased progressively after 24-hour culture in DMSO and 48-hour culture on IgG-Fc. Stimulation of B-CLL cells with Jagged1 ligand partially prevented the decrease in NF-κB activity occurring at 48-hour culture (Figure 7B), whereas GSI treatment almost completely abrogated NF-κB activity observed at 24 hours (Figure 7A). The specificity of NF-κB–binding activity to the consensus sequence was confirmed in competition assays, where NF-κB complexes disappeared when nuclear extracts were incubated with a 50-fold molar excess of unlabeled probe (data not shown).
Effect of soluble Jagged1 ligand and GSI treatment on NF-κB activity, c-IAP protein expression, and caspase activation. Freshly isolated B-CLL cells (n = 3) were cultured for different times in 2 different conditions: for 24 hours in culture tubes at a density of 2 × 106/mL (3 mL/tube) with GSI 5 μM or DMSO as control (A,C,E), or for 48 hours in 96-well plates at a density of 106/mL (100 μL/well) on immobilized soluble Jagged1 ligand or IgG-Fc as control (B,D,F). NF-κB–binding activity to a 32P-labeled specific probe was evaluated by EMSA in 5 μg nuclear proteins extracted from the indicated cell populations (A,B). Nuclear extracts (5 μg) used in EMSA were subjected to Western blot analysis with an anti-TBP mAb, as a loading control (A,B). c-IAP2 and XIAP expression (C,D), caspase-3 processing, and PARP degradation (E,F) were analyzed by Western blot in 30 μg whole-cell lysates. Protein loading was assessed by reprobing the blots with an anti–β-actin mAb. The data shown for patient 6 are representative of 3 patients.
Effect of soluble Jagged1 ligand and GSI treatment on NF-κB activity, c-IAP protein expression, and caspase activation. Freshly isolated B-CLL cells (n = 3) were cultured for different times in 2 different conditions: for 24 hours in culture tubes at a density of 2 × 106/mL (3 mL/tube) with GSI 5 μM or DMSO as control (A,C,E), or for 48 hours in 96-well plates at a density of 106/mL (100 μL/well) on immobilized soluble Jagged1 ligand or IgG-Fc as control (B,D,F). NF-κB–binding activity to a 32P-labeled specific probe was evaluated by EMSA in 5 μg nuclear proteins extracted from the indicated cell populations (A,B). Nuclear extracts (5 μg) used in EMSA were subjected to Western blot analysis with an anti-TBP mAb, as a loading control (A,B). c-IAP2 and XIAP expression (C,D), caspase-3 processing, and PARP degradation (E,F) were analyzed by Western blot in 30 μg whole-cell lysates. Protein loading was assessed by reprobing the blots with an anti–β-actin mAb. The data shown for patient 6 are representative of 3 patients.
c-IAP2 and XIAP analysis shows that the constitutive levels of these proteins decreased after 24- and 48-hour culture (Figure 7C,D). Jagged1 ligand partially prevented this decrease (Figure 7D), whereas GSI further reduced c-IAP2 and XIAP levels (Figure 7C). Analysis of caspase-3 and PARP shows that, after 24- and 48-hour culture, caspase-3 was progressively cleaved in its intermediate (p20) and active fragments (p19 and p17) and PARP progressively degraded to its 89-kDa inactive form (Figure 7E,F). Caspase-3 cleavage and PARP degradation occurring at 48 hours were partially prevented by Jagged1 ligand (Figure 7F), whereas GSI treatment strongly increased both the caspase-3 cleavage and PARP degradation at 24 hours (Figure 7E).
Discussion
In this study, we identified Notch signaling activation as a novel mechanism underlying B-CLL cell apoptosis resistance and their pathologic survival. We showed that B-CLL cells but not normal B cells constitutively express Notch1 and Notch2 proteins as well as their ligands Jagged1 and Jagged2, suggesting that the expression of these proteins is a peculiar property of these malignant cells. These findings, together with observations of other authors showing comparable levels of Notch1 and Notch2 mRNA in normal and B-CLL cells,25 suggest that the anomalous Notch protein expression in leukemic cells was not related to differences in gene transcription and/or mRNA stability. A possible explanation for the different Notch protein expression between B-CLL and normal B cells could rely on a deregulation in protein turnover. In this context, B-CLL cell–defective apoptosis has been related to a deregulation of the ubiquitin-proteasome pathway,34 which with the lysosomal pathway, has been shown to control several components of the Notch-signaling cascade.35,36
We showed that Notch signaling is also constitutively activated in B-CLL cells. We found that B-CLL cells express, in addition to the precursors and TM subunits of both Notch1 and Notch2, their active N-IC domains, which are known to result from a ligand-induced cleavage of the TM subunit mediated by a membrane-associated γ-secretase complex.37 We also demonstrated that B-CLL cells have a nuclear activity that binds to the CBF1 recognition sites in the Hes1 promoter and is composed of Notch2-IC but not Notch1-IC, suggesting that the Hes1 gene may be a target of Notch2 activity in B-CLL cells. Confirming the activation of the Notch-Hes1 axis, we found that Hes1 protein is expressed in B-CLL cells. In contrast, Hes1 was undetectable in normal B cells because of an inactive Notch signaling given the absence of Notch receptors in these cells. These findings seem discordant with other studies suggesting that the Notch-Hes1 pathway is down-regulated in B-CLL cells compared with normal B cells, on the basis of higher levels of Hes1 mRNA in normal B than in B-CLL cells.25 However, given our evidence that Notch proteins are absent in normal B cells, it is possible that Hes1 mRNA found in normal B cells by those authors might result from Notch-independent mechanisms. Indeed, although the Hes1 gene is the best-characterized target of Notch signaling,13,16 its transcription can also be regulated by other transduction pathways, as observed in other systems.38 However, Notch signaling could activate targets other than Hes1,16,39 and this may explain our finding that Notch1 is activated in B-CLL cells but is not involved in Hes1 induction. The molecular targets of Notch signaling in B-CLL cells and whether Notch1 and Notch2 have redundant or distinct effects in these cells remain to be determined.
Together with Notch receptors, we have shown, for the first time, that B-CLL cells constitutively also express the Notch ligands, Jagged1 and Jagged2. These findings suggest the existence of autocrine/paracrine mechanisms responsible for the constitutive activation of Notch signaling, also reported for other hematologic malignancies, including Hodgkin and anaplastic large-cell lymphoma20 and multiple myeloma.19 It has been shown that activated Notch signaling plays a pivot role in the pathogenesis of these tumors, promoting tumor cell proliferation and survival. Therefore, an autocrine loop of Notch stimulation might also be operative in B-CLL pathogenesis, thus contributing to the defective apoptosis of leukemic cells and their prolonged survival. Consistent with this hypothesis, we demonstrated that, in B-CLL cells that have resisted spontaneous apoptosis after 24-hour ex vivo culture, Notch signaling is activated more than in both freshly isolated B-CLL cells and B-CLL cells, which have undergone apoptosis, suggesting that an increased Notch activation may be the mechanism that slows down the rate of B-CLL cell apoptosis. B-CLL cells that resist spontaneous apoptosis also show an increased Notch expression as indicated by the enhanced levels of Notch1 and Notch2 precursors in addition to their N-IC domains, suggesting that, in these cells, Notch signaling up-regulates Notch expression in a positive feedback loop. Consistent with this hypothesis, it has been shown that Notch activation drives new Notch transcription in several other systems, including T cells.40,41
The role for Notch in B-CLL cell survival was further studied modulating Notch signaling. To this end, we used GSI, a tripeptide aldehyde selected from a group of compounds originally used in Alzheimer disease, which blocks processing and activation of all 4 Notch receptors by inhibiting γ-secretase activity.32,33 Our results show that GSI increases B-CLL cell spontaneous apoptosis and concomitantly inhibits Notch1, Notch2, and Hes1 expression. The evidence that inhibition of N-IC expression by GSI is accompanied by reduced levels of the precursors and TM subunits once again suggests that Notch signaling results in its own expression in B-CLL cells. The effects of GSI are concentration-dependent, and the concentrations inducing the highest increase in spontaneous B-CLL cell apoptosis are the most effective in inhibiting Notch activation, suggesting that GSI-induced apoptosis may be associated with down-regulated Notch signaling and therefore that Notch contributes to B-CLL cell survival. GSI induces apoptosis also in other types of tumors where Notch signaling is deregulated, including melanomas,42 Kaposi sarcoma,43 and meningiomas.44 In these malignancies, Notch has been proposed as a potential target for therapeutic intervention. However, because GSI has additional targets besides γ-secretases45 and γ-secretases mediate proteolysis of several transmembrane proteins besides Notch receptors,39 the possibility that mechanisms other than Notch signaling inhibition mediate GSI-induced apoptosis cannot be excluded. Therefore, to verify whether blocking the Notch pathway contributes to increased B-CLL cell apoptosis, siRNA targeting Notch2 was used. Specific Notch2 down-regulation reduces B-CLL cell viability in all samples examined, confirming the antiapoptotic role of Notch in B-CLL cells.
The involvement of Notch in B-CLL cell survival is further supported by the evidence that Notch-signaling stimulation by a soluble Jagged1 ligand increases B-CLL cell viability. These data suggest that, in B-CLL cells, Notch-activation might be increased also by interactions with other potential ligand-bearing cells in tissue microenvironment, as also observed in other hematologic malignancies.19,20,22 Interestingly, Notch-signaling stimulation by Jagged1 is accompanied by increased NF-κB activity and c-IAP2 and XIAP expression, whereas GSI strongly down-regulates these important apoptosis regulators. It has been shown, in other cell types, that Notch signaling controls NF-κB activity17,46,47 and that NF-κB regulates the expression of several antiapoptotic molecules, including the c-IAP proteins.48 Recent studies have reported a role for Notch in regulating c-IAP1 and c-IAP2 expression49 and increasing the XIAP stability via a direct interaction between Notch and XIAP.50 At present, we do not know whether Notch signaling is directly involved in regulating NF-κB activity and c-IAP2 and XIAP expression in B-CLL cells and further studies will be necessary to clarify these points.
Overall, our study suggests that an altered Notch activation plays a critical role in B-CLL cell apoptosis resistance, thus contributing to the maintenance of the neoplastic clone. These findings might have important implications for the development of new therapeutic strategies aimed at blocking Notch activation in B-CLL cells.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Catherine Bennett Gillies for the excellent assistance in preparing the manuscript.
This work was supported by grants from Progetti di Ricerca di Interesse Nazionale (PRIN 2005) 2005068971_002 (P.M.), Fondazione Cassa di Risparmio di Perugia, Italy (2007) (P.M.), and Polo Didattico e Scientifico di Terni, Italy (2007) (P.M.).
Authorship
Contribution: E.R. conceived research, analyzed and interpreted data, and wrote the manuscript; R.S. and G.R. performed research; A.T. provided patient samples and critical suggestions; A.T. and M.D.I. provided patient clinical and laboratory data; K.F. assisted in cytofluorimetric analysis; A.B. and S.C. collected data and assisted in statistical analysis; I.S. helped in designing research and supervised the study; and P.M. conceptualized the investigation and supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pierfrancesco Marconi, Department of Clinical and Experimental Medicine, General Pathology and Immunology Section, Via Brunamonti, Policlinico-Monteluce, 06122 Perugia, Italy; e-mail: marimmun@unipg.it.
References
Author notes
*R.S. and G.R. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal