Abstract
MicroRNA (miRNA) deregulation contributes to cancer pathogenesis. However, analysis of miRNAs in diffuse large B-cell lymphoma (DLBCL) has been hindered by a focus on cell lines, limited number of miRNAs examined, and lack of copy number data. To address these restrictions, we investigated genomewide miRNA expression and copy number data in 86 DLBCLs. Permutation analysis showed that 63 miRNAs were recurrently disrupted in DLBCL, including highly expressed oncomirs not previously linked to chromosomal abnormalities. Further, using training and validation tumor groups, we defined a collection of miRNAs that robustly segregates DLBCLs into 3 subsets, which are independent of the cell-of-origin classification, extent of T-cell infiltrate, and tumor site. Instead, these unique miRNA-driven DLBCL subgroups showed markedly different MYC transcriptional activity, which explained the dominance of miRNAs regulated by MYC in their expression signatures. In addition, analysis of miRNA expression patterns of normal B cells and integration of copy number and expression data showed that genomic abnormalities and the genetic fingerprint of nonmalignant cells also contribute to the miRNA profile of DLBCL. In conclusion, we created a comprehensive map of the miRNA genome in DLBCL and, in the process, have uncovered and mechanistically elucidated the basis for additional molecular heterogeneity in this tumor.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is a frequent and often fatal neoplasm. In the past few years, significant progress has been made in the characterization of the physiologic processes that need to be subverted for the development of this malignancy. These genomewide investigations have advanced our understanding of lymphoma biology because they clarified the relation between neoplastic and normal B cells, pointed to distinct mechanisms of cell transformation, and highlighted the relevance of the tumor milieu in DLBCL pathogenesis.1 However, these data derive exclusively from mRNA studies, whereas genomewide integrative investigations of the role of microRNAs (miRNAs) in DLBCL are still unavailable.
MiRNAs are central regulators of gene expression, and their relevance in cancer is now well established.2 Attempts to define the role of miRNAs in DLBCL have been hampered by the predominant use of cell lines, investigation of limited numbers of miRNAs, and lack of miRNA gene copy number data.3,4 The latter issue is important because miRNAs have been shown to map preferentially to chromosomal regions that are disrupted in cancer,5 suggesting that their contribution to the neoplastic process may be, at least in part, rooted in genomic abnormalities. To address this knowledge gap, we designed an oligonucleotide-based array comparative genomic hybridization (array CGH) microarray platform that included probes for all known miRNA loci (miRBase release 9.06 ) tiled at very high density. We used this array to investigate copy number changes in the miRNA genome of 59 cases of primary DLBCL and 27 DLBCL cell lines. These data were subsequently integrated with miRNA expression measurements and used to construct a map of the miRNA genome in DLBCL.
Herein, we show, using training and validation DLBCL cohorts, that a collection of B-cell relevant miRNAs can robustly segregate these tumors into 3 unique subsets. This novel miRNA-driven molecular substructure is independent of mRNA-based cell-of-origin classifications, extent of T-cell infiltrate, and tumor site and may impinge on disease outcome. Importantly, we have mechanistically linked these signatures to copy number changes targeting the miRNA genome, MYC activity, and, in part, to the miRNA expression pattern of normal B cells. Thus, our data uncovered additional molecular complexity in DLBCL and have established a definitive blueprint for the detailed characterization of miRNAs that are pertinent to B-cell lymphoma.
Methods
A detailed description of the methods used is included in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Sample population
Fifty-nine primary DLBCL samples obtained from the University of Texas Health Science Center at San Antonio tumor bank and 27 cell lines (26 DLBCL and 1 mediastinal B cell lymphoma) were included in this study. The clinical and pathologic features of this collection are described in Table S1 and Document S1. All tumors were reviewed for diagnostic accuracy by a hematopathologist (R.S.R.), and the categorization in germinal center B cell–like (GCB) or nongerminal center B cell–like (non-GCB) was performed according to the algorithm described by Hans et al.7 The extent of normal cell infiltration in the DLBCL was determined by semiquantitative measurement of CD3 (T cells) and CD68 (macrophages) staining. These studies were approved by the institutional review board at the University of Texas Health Science Center at San Antonio. Overall survival data were available for 30 patients in our series and were used in the determination of Kaplan-Meier survival probability curves.
Array CGH
Platform design.
An miRNA-specific array CGH platform (termed miRTile) was designed with the use of the eArray interface (Agilent Technologies, Palo Alto, CA). These high-density tiling arrays contained 41631 oligo oligonucleotide probes that specifically covered 309 unique chromosomal regions (11.6 Mb), including the loci for 471 miRNAs and 17 mRNA genes involved in microRNA biogenesis. Importantly, the miRTile platform is highly flexible, and new versions, including the most recently identified miRNAs, can be readily implemented (Document S1).
Hybridization and data retrieval.
High molecular weight DNA from DLBCLs and sex-matched normal control DNAs (Promega, Madison, WI) were differentially labeled with Cy3 and Cy5 and cohybridized to the arrays. After array scanning, the intensity of the hybridization signals was obtained with the use of Feature Extraction Software (Agilent Technologies).
Data analysis.
The array CGH data were analyzed with the use of dChip software (www.dchip.org, version 6/7/08). Quality control resulted in exclusion of 13 samples and 4158 underperforming probes from the final analyses. To obtain the miRNA gene copy number data, we first organized the probes into miRNA regions. The log2 ratio values of all the probes in a region were averaged to obtain the miRNA region-to-level log2 ratios, and copy number summary plots were used to determine the proportion of the samples that had copy number gains (copy ≥ 2.5) or losses (copy ≤ 1.5) for all miRNA regions. To find statistically significant altered regions, we used a permutation analysis with a genomewide P value threshold of .05. A detailed description of the design of this miRNA-specific array CGH platform and data analyses are available in Document S1.
Microarray-based miRNA expression studies
Genomewide determination of miRNA expression was performed with the use of the Human miRNA Microarray Kit version 1 (Agilent Technologies), with probe sets for 470 human microRNA genes,6 according to the manufacturer's guidelines. The hybridization signal values for the multiple probes for each miRNA were obtained with the use of Feature Extraction Software (Agilent Technologies) and analyzed using dChip software (www.dchip.org, version 6/7/08). Thereafter, filters were applied to identify the miRNA genes whose normalized signal values were 50 or more in greater than 50% of the samples. The collection of miRNA genes that survived this filtering process were subjected to hierarchical clustering, identifying 3 novel DLBCL subgroups, MiRNA Groups A, B, and C (MG-A, MG-B, and MG-C). To integrate the copy number and expression data, we considered the miRNA genes with signal values of 50 or more in greater than 30% of the samples. Thereafter, each surviving miRNA gene was divided by copy number into 3 bins (loss, diploid, gain), and the significance of their differential expression was determined by analysis of variance (ANOVA) testing; a P value threshold of .05 was considered statistically significant. A detailed description of the data analyses is available in Document S1. The microarray data are available in Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession number GSE15225.
Stem-loop quantitative real-time RT-PCR
Quantitative expression measurements of selected mature miRNAs were performed with the use of stem-loop quantitative TaqMan real-time reverse transcription–polymerase chain reaction (RT-PCR) assays (MicroRNA Assays; Applied Biosystems, Foster City, CA). All reactions were performed in triplicate and normalized to the expression of the small nucleolar (sno) RNA, U6. Relative expression was determined with the ΔΔCT method and reported as 2−ΔΔCT, as we previously described.8 Subsequent hierarchical clustering analysis was performed using dChip.
Real-time RT-PCR
Quantitative real-time RT-PCR was used to define the expression of 3 MYC target genes, CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), PGK1 (phosphoglycerate kinase 1), and TFAM (transcription factor A, mitochondrial) in 24 primary DLBCL samples. Values were normalized to the expression of Cyclophilin A. All reactions were run in triplicate, and relative expression was determined and defined as described in the previous paragraph. The significance of the differences in expression between tumors assigned to MG-A and MG-C was determined with the Mann-Whitney test.
Results
Validation of the miRTile platform
To validate our array CGH platform design and aberration calling algorithm, we used 3 independent approaches. First, we defined our ability to detect previously described genomic abnormalities targeting the miR-17-92 cluster locus on chromosome 13q31 in DLBCL cell lines.9,10 We readily detected amplification (∼ 8 copies) of this chromosome region in the OCI-Ly4 and OCI-Ly7 lines, as reported, as well as in 12 additional DLBCL cell lines (Figure S1). Second, we found that miR-650 maps within the λ light chain locus (λ) on chromosome 22q11. Therefore, this miRNA is predicted to be hemizygously deleted as a consequence of the physiologic rearrangements that occur in this region in λ-expressing B cells. In agreement with this hypothesis, we encountered a significant association between loss of miR-650 and λ-expressing (P < .01, Fisher exact test; Figure S2) DLBCLs. These data validated the ability of our platform and the dChip algorithm to detect single copy loss in a complex genetic background. Interestingly, extensive analysis showed that miR-650 is not expressed in DLBCLs (data not shown), suggesting that λ rearrangement does not affect the miRNA expression profile of normal and malignant B cells. Finally, we used fluorescence in situ hybridization (FISH) to provide independent confirmation of our copy number aberration findings. In these assays, BAC probes mapping to 2 miRNA loci (miR-26a-2 and miR-15a/16-1) found to be frequently abnormal in our sample set were hybridized to relevant DLBCL cell lines and primary tumors, and gain or loss of chromosome material was confirmed in every instance (Figure S3). Taken together, these studies show that the miRTile platform and the aberration-calling algorithm will yield robust information about the copy number status of the miRNA genome in DLBCL.
Genomic integrity of the miRNA genome in DLBCL
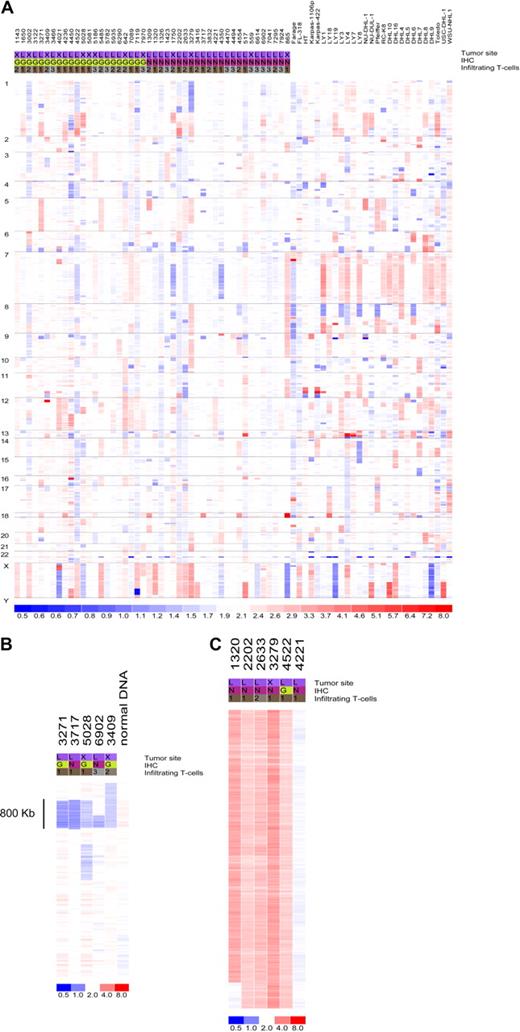
Data from 73 primary DLBCL and cell lines were used in the copy number analysis. Genomic abnormalities targeting the miRNA genes are common in DLBCL. Only 7 miRNA loci were found to be intact in all samples analyzed (Figure 1; Table S2): miR-33, let-7a-3, let-7b, miR-637, miR-657, miR-338, miR-210, and miR-202. In addition, 65 (89%) of the 73 tumors analyzed had copy number changes targeting at least one miRNA locus (Table S3). However, considering the large amount of data points derived from genomewide studies, it is critical to distinguish biologically relevant abnormalities (driver aberrations) from random changes without a functional benefit for the observed phenotype (passenger aberrations). To that end, we corrected our findings for a null hypothesis testing with the use of a false discovery rate (FDR) calculation. This led to the identification of recurrent aberrations (FDR = 0.1) targeting 63 individual miRNAs that were more likely to contribute to DLBCL pathogenesis (Table 1). These aberrations were focal, such as loss of 2 miRNAs within an 0.8-Mb region of chromosome 9p, or broad (whole or chromosome-arm size), such as those found on chromosome X (Figure 1B,C; Tables S2 and S3). Importantly, although not reaching significance with our FDR statistics, several additional miRNAs were abnormal in close to 10% of the samples analyzed (Table S2), suggesting that these genes may also play a role in lymphomagenesis. Conversely, copy number changes targeting the loci for genes involved in miRNA biogenesis were infrequent (Table S2), indicating that the proposed global disruption of this process in cancer11 is likely to result from epigenetic or regulatory defects. Finally, some of the miRNA loci commonly disrupted in our series map to regions previously shown to be abnormal in DLBCL (eg, 6q, 8p, 9p, 12q, and X chromosome),12-14 indicating that miRNAs mapping to these areas should also be considered relevant targets in DLBCL pathogenesis.
Copy number integrity of the microRNA genome in DLBCL. (A) Genomewide display of loss (blue) and gain (red) of chromosome material targeting the miRNA loci in 73 DLBCL samples. (B) Focal loss of genomic material encompassing miR-491 and miR-31 on chromosome 9 in 5 primary tumors; normal sample DNA also analyzed is displayed alongside. (C) Broad gain of chromosome material targeting the X chromosome in 5 DLBCLs with a diploid tumor is also shown. For all displays, each column represents a different sample (numerical ID for primary tumors), and each row represents an individual oligonucleotide probe. Chromosome numbers are listed on the left side of the figure. The site designation indicates the primary tumor location (L indicates lymph node; X, extranodal); immunohistochemistry (IHC), was performed as described by Hans et al7 to classify the tumors according to germinal center (G) or non-GC (N) categories. The T-cell infiltrate was quantified as described in Table S1: < 5% (1), 5%-15% (2), and 15%-25% (3).
Copy number integrity of the microRNA genome in DLBCL. (A) Genomewide display of loss (blue) and gain (red) of chromosome material targeting the miRNA loci in 73 DLBCL samples. (B) Focal loss of genomic material encompassing miR-491 and miR-31 on chromosome 9 in 5 primary tumors; normal sample DNA also analyzed is displayed alongside. (C) Broad gain of chromosome material targeting the X chromosome in 5 DLBCLs with a diploid tumor is also shown. For all displays, each column represents a different sample (numerical ID for primary tumors), and each row represents an individual oligonucleotide probe. Chromosome numbers are listed on the left side of the figure. The site designation indicates the primary tumor location (L indicates lymph node; X, extranodal); immunohistochemistry (IHC), was performed as described by Hans et al7 to classify the tumors according to germinal center (G) or non-GC (N) categories. The T-cell infiltrate was quantified as described in Table S1: < 5% (1), 5%-15% (2), and 15%-25% (3).
Recurrent copy number changes targeting the miRNA genome in DLBCL
| MicroRNA ID . | Chromosome . | Size, kb . | No. of probes . | Abnormal patients, % . |
|---|---|---|---|---|
| Copy number gain | ||||
| miR-513-1,* miR-513-2,* miR-506, miR-507, miR-508, miR-509, miR-510, miR-514-1, miR-514 | X | 105 | 194 | 24.7 |
| miR-105-1, miR-767, miR-105-2 | X | 36 | 90 | 24.7 |
| miR-17,* miR-18a,* miR-19a,* miR-20a,* miR-19b-1,* miR-92-1* | 13 | 34 | 133 | 24.7 |
| miR-224, miR-452 | X | 35 | 129 | 23.3 |
| miR-504 | X | 34 | 130 | 21.9 |
| miR-363, miR-92-2,* miR-19b-2,* miR-20b,*miR-18b, miR-106a* | X | 34 | 130 | 21.9 |
| miR-221, miR-222* | X | 34 | 106 | 20.5 |
| miR-450-1, miR-450-2, miR-542, miR-503, miR-424* | X | 40 | 132 | 20.5 |
| miR-505 | X | 34 | 91 | 20.5 |
| miR-532, miR-188,* miR-500, miR-362, miR-501, miR-660, miR-502 | X | 45 | 114 | 20.5 |
| miR-98, let-7f-2* | X | 35 | 139 | 20.5 |
| miR-653, miR-489 | 7 | 35 | 160 | 20.5 |
| miR-148b | 12 | 34 | 118 | 20.5 |
| miR-570 | 3 | 34 | 38 | 19.2 |
| miR-421 | X | 34 | 98 | 19.2 |
| miR-545, miR-374* | X | 34 | 123 | 19.2 |
| miR-223* | X | 34 | 128 | 19.2 |
| Copy number loss | ||||
| miR-650 | 22 | 34 | 78 | 31.5 |
| miR-31 | 9 | 34 | 118 | 17.8 |
| miR-588 | 6 | 34 | 61 | 17.8 |
| miR-548a-2 | 6 | 34 | 127 | 17.8 |
| miR-548b | 6 | 34 | 113 | 15.1 |
| miR-570 | 3 | 34 | 38 | 13.7 |
| miR-16-1,* miR-15a* | 13 | 34 | 121 | 13.7 |
| miR-596 | 8 | 34 | 69 | 13.7 |
| miR-587 | 6 | 34 | 110 | 12.3 |
| miR-491 | 9 | 34 | 134 | 11 |
| miR-124a-1 | 8 | 34 | 112 | 11 |
| MicroRNA ID . | Chromosome . | Size, kb . | No. of probes . | Abnormal patients, % . |
|---|---|---|---|---|
| Copy number gain | ||||
| miR-513-1,* miR-513-2,* miR-506, miR-507, miR-508, miR-509, miR-510, miR-514-1, miR-514 | X | 105 | 194 | 24.7 |
| miR-105-1, miR-767, miR-105-2 | X | 36 | 90 | 24.7 |
| miR-17,* miR-18a,* miR-19a,* miR-20a,* miR-19b-1,* miR-92-1* | 13 | 34 | 133 | 24.7 |
| miR-224, miR-452 | X | 35 | 129 | 23.3 |
| miR-504 | X | 34 | 130 | 21.9 |
| miR-363, miR-92-2,* miR-19b-2,* miR-20b,*miR-18b, miR-106a* | X | 34 | 130 | 21.9 |
| miR-221, miR-222* | X | 34 | 106 | 20.5 |
| miR-450-1, miR-450-2, miR-542, miR-503, miR-424* | X | 40 | 132 | 20.5 |
| miR-505 | X | 34 | 91 | 20.5 |
| miR-532, miR-188,* miR-500, miR-362, miR-501, miR-660, miR-502 | X | 45 | 114 | 20.5 |
| miR-98, let-7f-2* | X | 35 | 139 | 20.5 |
| miR-653, miR-489 | 7 | 35 | 160 | 20.5 |
| miR-148b | 12 | 34 | 118 | 20.5 |
| miR-570 | 3 | 34 | 38 | 19.2 |
| miR-421 | X | 34 | 98 | 19.2 |
| miR-545, miR-374* | X | 34 | 123 | 19.2 |
| miR-223* | X | 34 | 128 | 19.2 |
| Copy number loss | ||||
| miR-650 | 22 | 34 | 78 | 31.5 |
| miR-31 | 9 | 34 | 118 | 17.8 |
| miR-588 | 6 | 34 | 61 | 17.8 |
| miR-548a-2 | 6 | 34 | 127 | 17.8 |
| miR-548b | 6 | 34 | 113 | 15.1 |
| miR-570 | 3 | 34 | 38 | 13.7 |
| miR-16-1,* miR-15a* | 13 | 34 | 121 | 13.7 |
| miR-596 | 8 | 34 | 69 | 13.7 |
| miR-587 | 6 | 34 | 110 | 12.3 |
| miR-491 | 9 | 34 | 134 | 11 |
| miR-124a-1 | 8 | 34 | 112 | 11 |
Chr indicates chromosome ID; size, area (in kilobases) surrounding the miRNA precursor sequence that was tiled with the oligonucleotide probes; no. of probes, total number of probes tiling each locus; abnormal patients, percentage of cases, of 73 samples, with disruption of the indicated miRNA locus.
miRNAs also included in the expression signatures displayed in Figure 2.
Copy number changes in specific subsets of DLBCL
To better understand the contribution of the genomic aberrations targeting miRNA genes to lymphomagenesis, we also analyzed these changes within discrete DLBCL subgroups, including: GCB versus non-GCB tumors, nodal versus extranodal lymphomas, and primary tumors versus DLBCL cell lines. These analyses highlighted several statistically significant (P < .05, Fisher exact test) subgroup-specific abnormalities (Tables S4Table S5. Fisher’s Exact Test of LN vs. XN samples (PDF, 24 KB)–S6; Figures S4Figure S5. Heat map of copy number data for nodal vs. extranodal DLBCL (JPG, 122 KB)–S6). Of relevance, GCB-DLBCLs had a higher frequency of copy number gains targeting 7 miRNAs on chromosome 12q, a region previously found to exhibit abnormalities in DLBCL molecularly classified as GCB.12-14 These findings validated our immunohistochemistry (IHC)–based tumor classification and indicated the putative involvement of these miRNAs in the pathogenesis of GCB-type DLBCL. In addition, a B-cell relevant miRNA cluster on chromosome 9 (miR-23b, miR-27b, miR-24-1) was predominantly gained in extranodal DLBCL, whereas gains of the miR-17-92 cluster were far more common in cell lines than in primary tumors. Indeed, 14 (∼ 50%) of the 27 DLBCL cell lines analyzed displayed gains of miR-17-92 locus, whereas only 6 (∼ 13%; 3 GCB and 3 non-GCB) of 46 primary DLBCLs studied had similar changes (P < .01, Fisher exact test). This difference remained significant even when 8 primary tumors with more than 25% infiltrating normal cells were removed from the analysis; this is an important consideration given that samples with high percentage of infiltrating cells tended to cluster together because of their attenuated copy changes (Figure S7). These findings suggested that gain of miR-17-92 may be a later event in lymphoma progression or important for in vitro immortalization of malignant B cells or both. Importantly, although the distinctions between GCB and non-GCB or nodal and extranodal tumors were relatively modest, chromosomal gains and losses targeting miRNA loci were markedly overrepresented in DLBCL cell lines in comparison to primary tumors (Figure S6). These findings indicate that cell lines may not represent an authentic model to study the role of miRNAs in DLBCL. For this reason, we decided to focus our miRNA expression studies on primary DLBCLs.
miRNA expression profiling defines unique subclasses of DLBCL
Initially, we used a microarray platform to determine the expression profiles of 470 cellular miRNAs in 21 well-characterized DLBCLs (training set). Hierarchical clustering analysis (Document S1) showed the presence of 3 robust DLBCL clusters, MG-A, -B, and -C, defined by the expression of 98 miRNAs (Figure 2A; Table S7). To identify the miRNAs that more effectively distinguished these clusters, we used 2 rounds of one-way ANOVA testing. The expression of specific groups of 38 and 16 miRNA genes were found to effectively discriminate DLBCL into the same 3 unique groups (P < .01 and < .001, respectively; Figure 2B,C; Table S7). The ability of these small miRNA sets to identify discrete subgroups of DLBCL was tested with a stem-loop real-time RT-PCR approach in a second cohort of 42 primary tumors (validation set). Unsupervised hierarchical clustering confirmed the presence of the same substructure identified in our training set and correctly assigned all the reanalyzed tumors (n = 10) to their original MG clusters (Figure 2D). Thus, the miRNA expression profiles of a total of 53 primary DLBCLs defined the presence of 3 unique subgroups, which could be identified with as few as 16 individual miRNAs. These novel subgroups were unrelated to GCB and non-GCB classifications, tumor site, and the percentage of infiltrating cells (Figure 2D; Figure S8). These data indicate that miRNA expression could define additional molecular heterogeneity in DLBCL, beyond that derived from analysis of mRNA genes.15,16 Support for the uniqueness of this novel miRNA-driven substructure and for our suggestion that cell lines may make subpar models for the study of miRNA in DLBCL, was obtained when we determined that a 9-miRNA signature reported to distinguish GCB from non-GCB DLBCL cell lines was unable to cluster our primary tumors in these categories (Figure S9). In addition, initial evaluation of the clinical effect of this molecular substructure suggests a worse overall survival probability (Kaplan-Meier estimation) for patients assigned to the MG-A subgroup (P < .05, MG-A vs MG-B/-C, n = 30, log-rank and permutation tests; Figure S10).
MicroRNA expression profiling in DLBCL, training and validation sets. (A) Unsupervised hierarchical clustering analysis of 21 DLBCLs (training set) defined 3 unique subsets of tumors based on the differential expression of 98 miRNA genes (MiRNA Groups A-C). (B,C) Two rounds of one-way ANOVA testing identified subsets of 38 and 16 miRNAs whose expression could effectively discriminate DLBCLs into these 3 subsets. (D) Validation of the miRNA-defined molecular substructure in an extended cohort of DLBCLs. Expression of 17 mature miRNAs was defined by stem-loop real-time RT-PCR, and unsupervised hierarchical clustering of 42 DLBCLs was performed with the use of dChip. All tumors reanalyzed by RT-PCR clustered into their originally assigned groups. In each heat-map, a column represents a DLBCL sample and a row represents a miRNA. Tumor features listed at the top of the figure include the following: site, GC or non-GC origin (IHC), extent of T-cell infiltrate, and copy number of the miR-17-92 cluster for samples in which both array CGH and expression analyses were performed; G indicates gain; D, diploid, L, loss. Original MG cluster was determined by microarray analyses.
MicroRNA expression profiling in DLBCL, training and validation sets. (A) Unsupervised hierarchical clustering analysis of 21 DLBCLs (training set) defined 3 unique subsets of tumors based on the differential expression of 98 miRNA genes (MiRNA Groups A-C). (B,C) Two rounds of one-way ANOVA testing identified subsets of 38 and 16 miRNAs whose expression could effectively discriminate DLBCLs into these 3 subsets. (D) Validation of the miRNA-defined molecular substructure in an extended cohort of DLBCLs. Expression of 17 mature miRNAs was defined by stem-loop real-time RT-PCR, and unsupervised hierarchical clustering of 42 DLBCLs was performed with the use of dChip. All tumors reanalyzed by RT-PCR clustered into their originally assigned groups. In each heat-map, a column represents a DLBCL sample and a row represents a miRNA. Tumor features listed at the top of the figure include the following: site, GC or non-GC origin (IHC), extent of T-cell infiltrate, and copy number of the miR-17-92 cluster for samples in which both array CGH and expression analyses were performed; G indicates gain; D, diploid, L, loss. Original MG cluster was determined by microarray analyses.
Mechanistic basis for the miRNA-driven DLBCL substructure
To gain insight into the basis for these newly discovered subgroups of DLBCL, we examined the identity of the smallest set of miRNAs that defined this distinction. MG-A is primarily comprised of the oncogenic miR-17-92 cluster and the paralog miR-106a-363 of chromosome X (Figure 2C,D). Considering the presence of genomic abnormalities targeting these regions, we integrated copy number and expression data for 40 DLBCLs that were subjected to both array CGH analysis and miR-17-92/miR-106a expression measurements (Figures 1A, 2A,D). These investigations showed that a significantly larger fraction of DLBCLs assigned to the MG-A subgroup (5 of 15) had gains of the miR-17-92 locus, whereas only 1 tumor of the combined MG-B and MG-C subgroups (1 of 25) showed these structural changes (P < .05, 2-tailed Fisher exact test). No association was found between MG assignment and copy number at the miR-106a-363 locus. These data suggest that genomic abnormalities contribute, at least in part, to the miRNA-driven DLBCL subgroupings. However, a sizable fraction of tumors assigned to the MG-A subgroup was diploid for the miR-17-92 locus, indicating that additional mechanisms may account for the overexpression of these miRNAs. The MYC oncogene has been recently shown to up-regulate the expression of the miR-17-92 cluster and to also directly suppress the expression of a large number of miRNAs.17,18 Remarkably, the majority (∼ 95%) of these miRNAs were found to be down-regulated in the MG-A subgroup and overexpressed in the MG-C subgroup (Table S8), suggesting that MYC may play an important role in the miRNA expression signature of DLBCL. To test this hypothesis, we measured the expression of 3 independent MYC target genes (CAD, PGK1, and TFAM) that are functionally related to cell metabolism and mitochondrial function but unconnected to known lymphoma biology.19-21 These analyses showed a significantly higher MYC transcriptional activity in tumors assigned to the MG-A subgroup than to the MG-C subgroup (P < .05, Mann-Whitney test; Figure 3), suggesting that this transcription factor markedly influences the miRNA-driven molecular substructure of DLBCL.
Differential expression of MYC targets in miRNA-driven subsets of DLBCL. Box plot display of the real-time RT-PCR analyses of 3 independent MYC target genes, CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), PGK1 (phosphoglycerate kinase 1), and TFAM (transcription factor A, mitochondrial) in 24 primary DLBCL samples (16 assigned to MG-A and 8 to MG-C). As hypothesized, MYC activity, defined by the expression of these target genes, was significantly higher in MG-A tumors (P < .01, Mann-Whitney test [CAD and PGK1] and P < .05 [TFAM]).
Differential expression of MYC targets in miRNA-driven subsets of DLBCL. Box plot display of the real-time RT-PCR analyses of 3 independent MYC target genes, CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), PGK1 (phosphoglycerate kinase 1), and TFAM (transcription factor A, mitochondrial) in 24 primary DLBCL samples (16 assigned to MG-A and 8 to MG-C). As hypothesized, MYC activity, defined by the expression of these target genes, was significantly higher in MG-A tumors (P < .01, Mann-Whitney test [CAD and PGK1] and P < .05 [TFAM]).
Finally, additional genomewide integration of our copy number and expression data (Document S1; Figure S11) showed that miR-100, miR-125b-1, and miR-130a (all mapping to chromosome 11 and members of the MG-B cluster) were overexpressed as a consequence of chromosomal gain or amplification, pointing to additional contributions of genomic abnormalities to the expression profile of miRNAs in DLBCL. Interestingly, in these integrative analyses we did not find that copy number loss at chromosome 13q14 influenced the expression of miR-15a/16-1. Because previous reports in related mature B-cell malignancies suggested the presence of such association,22 we decided to evaluate this issue more extensively. Thus, we used quantitative RT-PCR strategies to specifically measure the expression of mature miR-15a and miR-16-1 expression in a larger number of DLBCLs (n = 30) for which we had also performed array CGH investigations. These analyses confirmed the lack of correlation between copy number and expression of miR-15a/16-1 locus (Figure S12). It should be noted that, given the differences between miR-15a (chromosome 13) and miR-15b (chromosome 3), the coregulation of miR-16 and miR-15,22 and the specificity of this RT-PCR strategy,23 we could confidently interpret these findings. Thus, it is likely that mechanisms other than copy number variation, such as direct MYC regulation,18 may account for the differential expression of these miRNAs in DLBCL.
miRNA expression signatures of normal B-cells in DLBCL
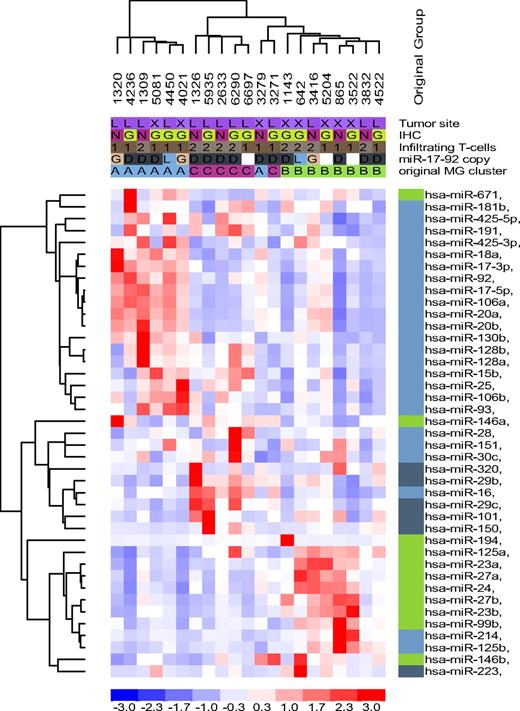
In DLBCL, the genetic composition of developmentally regulated B cells is of critical importance.15 However, a recent report has indicted that a robust miRNA signature of developmentally regulated normal B cells was not recapitulated in biologically distinct subtypes of DLBCL cell lines.3 Because we found that DLBCL cell lines made a poor model for studying miRNAs in DLBCL, we decided to test whether the recently described miRNA signature capable of distinguishing naive, centroblasts (CBs) and memory B cells,3 could also capture the substructure that we had identified in primary DLBCLs. An early indication that this could indeed be the case was the significant overlap noted between the individual miRNAs that comprise the normal B-cell miRNA signature and those that we found to define unique subsets of DLBCL (Table S9). In fact, using an unsupervised hierarchical clustering approach, we found that the miRNA signature derived from normal B cells clustered most of the tumors in our collection into their original MG subgroups, with the exception of 2 samples originally assigned to the MG-A and MG-C subgroups that now were branching from MG-B (Figure 4). In general, miRNAs expressed at highest levels in CB, memory, and “non-CB” cells (naive and memory cells combined) were also overexpressed in the MG-A, -B, and -C subgroups, respectively (Figure 4; Table S9). However, our analysis also showed that tumor-associated dysfunctions superseded the coordinated miRNA expression of normal B cells, as evident in the MG-B and MG-C clusters, which despite being related to memory and non-CB signatures, respectively, also exhibited a significant component (25%-50%) derived from other B-cell populations (Figure 4).
Normal B-cell miRNA expression signature in DLBCL. Unsupervised hierarchical clustering analysis of 21 DLBCL with 39 miRNA genes related to normal B cells recapitulated the subsets defined by the DLBCL-relevant miRNAs in the most of the samples; except for 2 tumors originally assigned to MG-A and MG-C and now branching from MG-B. However, the malignant transformation process also disrupted the coordinated miRNA expression of normal B cells as illustrated by a mix of miRNAs related to centroblast (light blue), noncentroblast (memory/naive; dark blue), and memory cells (green) clustering in MG-B and MG-C. MG-A was primarily composed of centroblast-related miRNAs, although miR-671 (memory cells) was also clustered with this group.
Normal B-cell miRNA expression signature in DLBCL. Unsupervised hierarchical clustering analysis of 21 DLBCL with 39 miRNA genes related to normal B cells recapitulated the subsets defined by the DLBCL-relevant miRNAs in the most of the samples; except for 2 tumors originally assigned to MG-A and MG-C and now branching from MG-B. However, the malignant transformation process also disrupted the coordinated miRNA expression of normal B cells as illustrated by a mix of miRNAs related to centroblast (light blue), noncentroblast (memory/naive; dark blue), and memory cells (green) clustering in MG-B and MG-C. MG-A was primarily composed of centroblast-related miRNAs, although miR-671 (memory cells) was also clustered with this group.
Discussion
Genomewide examinations of mRNA genes in DLBCL have advanced our understanding of lymphoma biology and have become a beacon for the development of novel therapeutic interventions.1,24-26 Here, we described the construction of a map of the miRNA genome in DLBCL that fully integrates miRNA gene copy number and expression data. Considering the critical role of miRNAs in various physiologic processes and their common disruption in cancer, these findings may spearhead advances in DLBCL biology similar to those that followed the initial mRNA-based expression profiling studies.27
MiRNAs have been reported to be frequently located at fragile sites and genomic regions involved in cancers.5 In agreement with this suggestion, genomic abnormalities have been firmly linked to miRNA deregulation in cancer,22,28,29 including lymphomas.9 Therefore, we reasoned that for our study to reach more meaningful conclusions, it would be important to also comprehensively investigate the copy number changes targeting the miRNA genome in DLBCL. To achieve this goal, we designed a highly specialized oligonucleotide-based array CGH platform that tiled the loci for all human miRNAs at a high density. This tool gave us an unprecedented view of the integrity of these genes in DLBCL. With the use of a stringent permutation testing to determine significance, we identified 63 individual miRNAs, including components of 28 unique miRNA clusters, with recurrent copy number change in DLBCL (Table 1). Importantly, a third of these miRNAs were also highly expressed in DLBCL and were included in our newly identified miRNA-driven tumor classifier. These initial observations suggested that structural abnormalities do play an important role in the dysfunctional expression of miRNAs in lymphomas. The miRNAs that were both highly expressed and common targets for copy number changes included those previously linked to mature B-cell malignancies (miR-17-92 and miR-15a/16-1 clusters), as well as others in which the putative contribution to DLBCL pathogenesis is novel, such as miR-222 and let-7f, which have been associated with other malignancies,30-33 miR-51334 and miR-223,3,35 linked to immune regulation and related B-cell tumors, miR-424, which plays a role in hematopoiesis,36 and miR-188 and miR-374, with no known physiologic or pathologic functions. It should be noted that several of these miRNA loci map to chromosome X. Although copy number changes targeting this chromosome are common in DLBCL,13,14 no candidate target genes have been consistently ascribed to these abnormalities. Thus, our data suggest that miRNA deregulation may be the underlying mechanism that confers biologic advantage to tumors with chromosome X disruption. Finally, although not surviving our permutation analysis, an additional 70 miRNAs exhibited gains, and 26 exhibited losses in a significant percentage of the DLBCL cases (11%-17.8% of cases for gain and 8.2%-9.6% for loss). These data suggest that the effect of structural abnormalities on the miRNA deregulation found in DLBCL may be more pronounced than presently appreciated.
In agreement with previous reports, we found that gains of chromosomes 12q was more commonly associated with tumors classified as the GCB type.12-14 These findings gave us confidence in our IHC-based tumor assignment, despite the recognized limitation of this strategy in recapitulating the molecular classification of DLBCLs.7 Among the multiple miRNA loci on 12q that were frequently (> 10% of the tumors) targeted by copy number gains, miRNAs 26a-2 and let-7i, were also highly expressed in the primary tumors. These data suggest that these are the chromosome 12q miRNAs that are likely to contribute to the pathogenesis of GCB-type DLBCL. A recent report has suggested that amplification of the miR-17-92 cluster was limited to GCB tumors.12 However, we found that this chromosome abnormality did not segregate with either GCB or non-GCB tumors. Although this discrepancy may be related to the limitation of the IHC-based classification used in our work, it is more important to consider miR-17-92 expression, which is also modulated by MYC activity and is independent of the GCB and non-GCB distinction.4,12 One of the most notable features of our array CGH investigation was the marked increase in the frequency of copy number alterations observed at miRNA loci in DLBCL cell lines compared with primary tumors. This difference certainly reflects the well-known karyotypic complexity of DLBCL cell lines,37 but it also validates the biologic relevance of the preferential localization of miRNAs to unstable chromosomal regions. With that in mind, we considered DLBCL cell lines an inappropriate model to define the role of miRNA expression in DLBCL biology and, thus, focused our analyses on primary tumors.
With the use of an unsupervised hierarchical clustering algorithm in training and validation tumor sets, we defined a miRNA signature that robustly segregated DLBCL in 3 novel subgroups. The uniqueness of this molecular classification was confirmed by its lack of correlation to the GCB and non-GCB classifications, tumor site, or the extent of T-cell infiltrate. The latter is of interest because T cells and the tumor microenvironment significantly influence the mRNA-driven expression signatures of DLBCL.16,38 However, differently from mRNAs, there is a high degree of correlation in the expression patterns of miRNAs in distinct immune cell lineages at comparable stages of differentiation.39 Thus, we propose that this particular property of miRNAs provides a strong physiologic basis for our observations and indicates that the similarities in the miRNAs expressed by mature B cells and Th1 and Th2 lymphocytes may attenuate the effect of T cells in the miRNA signature of DLBCL.
Independence from a mRNA-based cell-of-origin classification15 is also an important concept to address in light of a recent study that suggested that the expression of 9 miRNAs was consistently higher in non-GCB DLBCL cell lines and could distinguish them from GCB cell lines.3 However, when we tested this cell line–derived miRNA signature in our dataset, we did not find that it could segregate primary DLBCLs in the described clusters.3 Thus, our findings, along with those of Roehle et al,4 support the idea that a comprehensive miRNA expression signature of DLBCL is unrelated to the cell-of-origin classification.4 These data further indicate that meaningful information about the role of miRNAs in DLBCL is unlikely to emerge from studies centered in cell lines.
Preliminary assessment of the clinical effect of this newly identified molecular signature in DLBCL suggested that patients assigned to the MG-A subgroup have a less favorable outcome than patients assigned to the MG-B or -C categories. These data certainly need to be validated in larger cohorts, but investigations with single miRNAs indicated that these small regulatory elements can indeed determine outcome in DLBCL.3 Importantly, as each MG cluster is composed of multiple miRNAs, insights into commonly disrupted pathways with clinical translational potential may emerge. For example, because the MG-A subgroup is enriched for the miR-17-92 cluster, which directly targets the tumor suppressor PTEN,40 it is possible that these tumors would be particularly responsive to therapeutic maneuvers that target the PI3K/AKT/mTOR pathway.41
After the identification of this novel miRNA-driven substructure in DLBCL, we sought to define its mechanistic basis. As addressed in “Results,” copy number changes contribute to the MG-A and, probably, MG-B subgroups. However, it quickly became apparent that additional factors were also associated with this molecular distinction, which led us to consider molecules that are relevant for lymphoma biology and miRNA regulation. In this realm, the transcriptional factor MYC stands out. MYC has been recently shown to negatively regulate the expression of a host of miRNAs and to transcriptionally activate the miR-17-92 cluster.17,18 Remarkably, most of the miRNAs known to be down-regulated by MYC were underexpressed in the MG-A subgroup and overexpressed in the MG-C subgroup. Conversely, miR-17-92 was overexpressed in the MG-A subgroup (independently of copy number gain) and down-regulated in the MG-C subgroup. These results suggested that MYC played a key role in determining the miRNA expression signature of DLBCL. We confirmed this hypothesis by showing that MYC transcriptional activity, defined by the expression of 3 independent target genes, was significantly higher in the MG-A subgroup than in the MG-C subgroup. With the use of this well-established approach to determine MYC activity,20,21 we limited the confounding effects derived from MYC's posttranslational modifications42,43 and avoided the imprecise conclusions derived from directly measuring MYC expression. Thus, as MYC emerges as a key regulator of the miRNA profile in DLBCL, it highlights a new mechanism by which it may contribute to lymphomagenesis. Future investigation on the role of other B-cell–relevant transcriptional factors should further delineate the interaction between mRNA and miRNAs in DLBCL biology.
Finally, we have shown that the genetic signature of normal B cells influences the miRNA expression profile of DLBCL. Thirty-nine miRNAs that could efficiently segregate 3 subpopulations of mature B cells3 also clustered our DLBCL collection to largely the same MG-A, -B, and -C subgroups. However, it should be noted that the correlation between the miRNA signatures of normal B cells and these newly defined subsets of DLBCL was not absolute. Accordingly, although the miRNA signature of the MG-B subgroup was closely related to that of memory B cells, a significant fraction of the miRNAs defining this subgroup were derived from centroblasts. Likewise, the signature of the MG-C subgroup was influenced by the expression signatures of both centroblast and non–centroblast-derived miRNAs. These findings suggest that tumor-associated events disrupt the coordinated miRNA expression of normal B cells and yield a unique miRNA expression signature that typifies DLBCL. Indeed, these features are reminiscent of those observed in mRNA-based studies in which both cell-of-origin and tumor-associated defects contribute to DLBCL expression profiles.15,16
In conclusion, we have described a comprehensive map of the miRNA genome in DLBCL. In particular, we uncovered novel, miRNA-driven, molecular heterogeneity in DLBCL that is mechanistically explained by copy number changes targeting the miRNA loci, MYC activity, and the genetic fingerprint of normal B cells. Furthermore, the identification of a small number of miRNAs that can robustly recapitulate this substructure will facilitate studies aimed at confirming its effect on clinical outcome and the use of miRNAs as biomarkers for serum-based DLBCL diagnosis.44-46 Finally, untangling the functional repercussions of the differential expression of these miRNAs with global genomic and proteomics approaches will advance our understanding of lymphoma biology and may also improve our ability to more effectively treat this disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Richard Meier for help in collecting clinical data.
This work was supported by grants from the AT&T Research Foundation (San Antonio, TX) and the Concern Foundation (Beverly Hills, CA). Ricardo Aguiar is a scholar of the American Society of Hematology.
Authorship
Contribution: C.L. analyzed array CGH and expression data and wrote part of the manuscript; S.-W.K., D.R., and A.R.B. performed research; S.A. performed FISH studies; M.C.K. and R.S.R. identified and characterized tumor specimens; and R.C.T.A. designed the study, analyzed and interpreted the results, and wrote the manuscript. All authors read the manuscript and agreed with its contents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ricardo Aguiar, Division of Hematology and Medical Oncology, University of Texas Health Science Center at San Antonio, 7703 Floyd Curl Dr, MC7880, San Antonio, TX 782229; e-mail: aguiarr@uthscsa.edu.
References
Supplemental data
Additional supplemental tables and supplemental figures can be found here.


![Figure 3. Differential expression of MYC targets in miRNA-driven subsets of DLBCL. Box plot display of the real-time RT-PCR analyses of 3 independent MYC target genes, CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), PGK1 (phosphoglycerate kinase 1), and TFAM (transcription factor A, mitochondrial) in 24 primary DLBCL samples (16 assigned to MG-A and 8 to MG-C). As hypothesized, MYC activity, defined by the expression of these target genes, was significantly higher in MG-A tumors (P < .01, Mann-Whitney test [CAD and PGK1] and P < .05 [TFAM]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/26/10.1182_blood-2009-01-202028/4/m_zh89990935560003.jpeg?Expires=1769165921&Signature=4IJXmUYBNS2eCP~n2ZXSLn1cpXJYDreLsLben8nwsESc9KWyn4eO-NeVr9~lA9Wy5cvhHhtKZDrWrfOhMJcU6WAB5-XGr8XhRFc~BL0juw~z5~IvD2My8G68gjxAwC8a92Er3LBJs4nIS94QM54FUSIELKIJHnCIV~RxIP1fYJCXOXrpctRkVuIYLBK1Jq6LLKoWLavC4c1Pw4LXAEEkrzk0xjOq2D6p-a6SfViDP2njPqa54OXtH~9EYF2J6CunP6xUwHF~fgHw6xAVD-lVuVOi3xzsnJD36xZL9O3nnrNj97vx2xWDInKdDbnYwDggjZokSJoX2IexGxjXP5pM7A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)