Abstract
Hematopoietic stem cells (HSCs) are generally defined by their dual properties of pluripotency and extensive self-renewal capacity. However, a lack of experimental clarity as to what constitutes extensive self-renewal capacity coupled with an absence of methods to prospectively isolate long-term repopulating cells with defined self-renewal activities has made it difficult to identify the essential components of the self-renewal machinery and investigate their regulation. We now show that cells capable of repopulating irradiated congenic hosts for 4 months and producing clones of cells that can be serially transplanted are selectively and highly enriched in the CD150+ subset of the EPCR+CD48−CD45+ fraction of mouse fetal liver and adult bone marrow cells. In contrast, cells that repopulate primary hosts for the same period but show more limited self-renewal activity are enriched in the CD150− subset. Comparative transcriptome analyses of these 2 subsets with each other and with HSCs whose self-renewal activity has been rapidly extinguished in vitro revealed 3 new genes (VWF, Rhob, Pld3) whose elevated expression is a consistent and selective feature of the long-term repopulating cells with durable self-renewal capacity. These findings establish the identity of a phenotypically and molecularly distinct class of pluripotent hematopoietic cells with lifelong self-renewal capacity.
Introduction
Each day, the hematopoietic system of the adult mouse produces billions of mature blood cells. The multistep process that underlies the production of these cells is controlled by complex mechanisms that enable changing physiologic demands to be met without overwhelming the system. It is now clear that a small population of cells known as hematopoietic stem cells (HSCs) are ultimately responsible for maintaining the lifelong output of new blood cells.1 Nevertheless, a molecular understanding of what constitutes an HSC and how its key functions are maintained are still poorly understood.
The existence of hematopoietic cells with the individual potential to produce large numbers of multiple blood cell types in vivo for prolonged periods was first suggested by retrospective clonal tracking experiments that used unique chromosomal,2 and later, retroviral marking approaches.3,4 The relatively short lifespan of many mature blood cell types and the contrasting longevity of some of the multilineage clones identified in these experiments implied an origin of the clones from undifferentiated cells with an extensive ability to divide and maintain a derivative population of similarly undifferentiated cells. Serial transplantation experiments demonstrated that such cell divisions did occur and thus provided the first definitive evidence of HSC self-renewal.3,4 These findings prompted a search for functional end points that would allow HSCs to be specifically quantified independent of the presence of other cell types when assayed in limiting dilution transplantation strategies using suitably irradiated congenic hosts.5,6 A sustained output of at least 1% of all the circulating white blood cells (WBCs) for at least 4 months is now widely assumed to be suitable for this purpose.7
Interestingly, most analyses of individual pluripotent hematopoietic cells proliferating either in vivo or in vitro have generally found the self-renewal activity actually displayed to be highly variable.3,4,8,9 How such a variable behavior is related to the molecular state of the initial cells is not well defined and remains a subject of intense interest and investigation.10 Variations in external cues may be one contributing parameter, at least in vivo, because it is known that self-renewal responses can be directly and rapidly modulated in this way in vitro.11,12 In addition, there is some evidence of predetermined heterogeneity in HSC self-renewal potential. This is exemplified by the differences in regenerative activity of HSCs from fetal and adult sources13,14 and the finding of a consistent association of short-term and long-term multilineage WBC outputs with distinct phenotypes of adult bone marrow (BM) cells (eg, according to their expression of CD49b and CD34).15,16 Taken together, these results suggest that some hematopoietic cells can remain pluripotent for several divisions even though they are already destined to undergo terminal differentiation within a finite period. This is the basis of the concept of durable versus finite self-renewal potential that is the subject of the studies presented here.
Recently, we and others have provided evidence that even among cells with longer term repopulating activity, there is predetermined heterogeneity in HSC self-renewal durability.17-20 Specifically, from analyses of more than 90 single cell or clonal transplantations, we resolved 4 subtypes of adult BM HSCs, only 2 of which consistently produced significant numbers of daughter HSCs for extended periods (3 cycles of clone expansion in vivo), whereas the other 2 failed to generate detectable HSCs after 6 months of clonal expansion in primary recipients.17 Interestingly, the 2 repopulation patterns that do not have secondary repopulation ability showed relatively stronger contributions to the lymphoid lineages and extend the findings of other groups that reported robust myelopoiesis at 4 months after transplantation to be associated with the presence of cogenerated HSCs with repopulating activity in secondary hosts.17-20
We now present further evidence that this difference in HSC self-renewal durability is a reflection of intrinsic differences in the starting populations that allow their separate isolation in highly purified form. From a comparative analysis of the gene expression profiles of these purified populations, we show that they are also molecularly distinct and, among a subset of genes that are expressed at higher levels in the HSCs with durable self-renewal activity, we have identified 3 that have not been previously implicated as having a role in regulating HSC self-renewal activity.
Methods
Mice
C57Bl/6J (B6)-Ly5.1 or -Ly5.2 mice and congenic B6-W41/W41-Ly5.1 and -Ly5.2 (W41-5.1 and W41-5.2) mice were bred and maintained in the British Columbia Cancer Agency Animal Resource Center in microisolator cages and provided continuously with sterile food, water, and bedding. All procedures involving these mice were carried out in accordance with Canadian Council on Animal Care guidelines with approval from the University of British Columbia Animal Care Committee.
Isolation of subsets of fetal liver and adult BM cells
Fetal livers (FLs) from E14.5 mice were suspended in 1 mL 2% (vol/vol) fetal bovine serum (FBS) supplemented-Hanks balanced salt solution (collectively called HF) by forcing the tissue through a sieve and then through a 40-μm filter. Erythroid precursors were depleted by immunomagnetic removal of biotin-conjugated anti-Ter119–labeled cells using EasySep reagents (StemCell Technologies, Vancouver, BC) as recommended by the manufacturer. The unlabeled fraction of cells was then centrifuged and suspended at 107-108 cells/mL of Hanks containing 100 μL/mL rat serum (Sigma-Aldrich, St Louis, MO) and 2.5 μL/mL 2.4G2 anti–mouse FcR antibody (ATCC, Rockville, MD; blocking solution) and incubated on ice for 10 minutes before further labeling. Suspensions of BM cells from adult (8-12-week-old) B6 donors were first depleted of red blood cells (RBCs) by a 10-minute incubation on ice in the presence of NH4Cl and were then resuspended at 107-108 cells/mL in the blocking solution and incubated for 10 minutes on ice. To isolate lineage marker-negative (lin−) populations from BM or FL cells, the cells were stained for 30 minutes with biotin-conjugated anti-Ly1, anti-Gr1, anti-CD5, and anti-B220 (StemCell Technologies), washed, resuspended in HF, stained for 15 minutes with phycoerythrin (PE)–conjugated streptavidin (BD Biosciences, San Jose, CA), washed, and suspended in HF containing 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) to mark dead cells.
Highly HSC-enriched populations used for the construction of long serial analysis of gene expression (LongSAGE) libraries were isolated by sorting for lin−CD43+Mac1+Sca1+ E14.5 FL cells and CD45midlin−Rhodamine-123− side population (SP) adult BM cells as previously described.14,17 To isolate CD45+ endothelial protein C receptor (EPCR)+CD48−CD150+ (E-SLAM) and/or CD45+EPCR+CD48−CD150− FL or adult BM cells, the initial populations were first stained for 30 minutes with fluorescein isothiocyanate (FITC)–conjugated anti-CD45 (purified and conjugated in the Terry Fox Laboratory), PE-conjugated anti-EPCR (StemCell Technologies), biotin-conjugated or PE-Cyanin7–conjugated anti-CD150 and allophycocyanin (APC)–conjugated anti-CD48 (Biolegend, San Diego, CA). When biotin-conjugated anti-CD150 was used, cells were secondarily stained with streptavidin-PE-Texas Red (BD Biosciences).
All cell sorting was performed using either a FACSVantage (BD), FACSDiVa (BD), or Influx Cell Sorter (Cytopeia, Seattle, WA). Cells were first sorted at a high rate (10 000-15 000 cells/s) using a EPCR+CD48− gate that captured approximately 0.5% of all the viable cells and were then resorted at a slower rate (1-200 cells/s) to improve the purity. When single E-SLAM or CD45+EPCR+CD48−CD150− cells were required, the single cell deposition unit of the sorter was used to place these individually into the wells of round-bottom 96-well plates, each well having been preloaded with 100 to 200 μL serum-free medium (SFM; Iscove modified Dulbecco medium supplemented with 10 mg/mL bovine serum albumin, 10 μg/mL insulin, and 200 μg/mL transferrin, 100 U/mL penicillin, 100 μg/mL streptomycin [all from StemCell Technologies], and 10−4 M β-mercaptoethanol [Sigma-Aldrich]). Each well was then visually inspected to identify those that contained 1 and only 1 viable cell.
Cell cultures
Sixteen-hour cultures of E-SLAM cells were incubated in SFM plus 20 ng/mL interleukin-11 (IL-11; Genetics Institute, Cambridge, MA) and either 1, 10, or 300 ng/mL Steel factor (SF; StemCell Technologies). Long-term culture-initiating cell (LTC-IC) assays were performed by plating visually confirmed single sorted cells onto irradiated AFT024 feeder cells21 in flat-bottomed wells of a 96-well plate. The cocultures were maintained for 6 weeks at 37°C in Myelocult (StemCell Technologies) supplemented with 10−6 M hydrocortisone (Sigma-Aldrich) with weekly half-medium changes as previously described.22 The contents of each well were then harvested individually and dissociated into a single-cell suspension that was assayed in methylcellulose cultures (MethoCult GF M3434; StemCell Technologies) to determine whether colony-forming cells (CFCs) were present or not. The input cell was inferred to be a LTC-IC if one or more colonies were produced after 14 days of incubation in the methylcellulose assay.
Transplantation of cells and analysis of mice that received transplants
All transplantations were performed by standard intravenous tail vein injection of sublethally irradiated Ly5-congenic adult W41/W41 mice as previously described.23 Peripheral blood samples were collected from the tail vein of some mice at 4 weeks and all mice at 8, 16, and 24 weeks after transplantation and RBCs lysed using NH4Cl. All samples were stained with both anti–CD45.1-APC (eBiosciences, San Diego, CA) and anti–CD45.2-FITC (Terry Fox Laboratory) to enable donor and recipient WBCs to be distinguished and to ensure that any double positive CD45.1 and CD45.2 events were excluded from the analysis of donor contributions to specific WBC subsets. The latter were identified by staining with anti–Ly6g-PE and anti–Mac1-PE to detect myeloid (GM) cells, anti–B220-PE for B cells, and anti–CD5-PE for T cells (all antibodies from BD). Cells that contributed at least 1% of the WBCs at 4 and 8 weeks, but not later, were defined as short-term repopulating cells (STRCs). Those in which at least 1% of the WBCs were donor-derived at 16 and/or 24 weeks after transplantation were considered to be repopulated with HSCs. HSCs (α, β, γ, and δ) were further discriminated according to previously described high (α and β) or low (γ and δ) ratios of their proportional contributions to the GM and B- and T-cell subsets of the circulating WBCs assessed at 16 weeks after transplantation.17 The α and β subtypes of HSC consistently show continuing lifelong self-renewal activity in contrast to the γ and δ subtypes of HSC, from which progeny HSCs are generally not detectable after 6 to 7 months. In the present studies, secondary transplantations of single CD45+EPCR+CD48−CD150+ (E-SLAM) or bulk (∼ 2 × 107) BM cells harvested from 4 primary mice repopulated with α or β HSCs into secondary irradiated W41/W41 hosts.
Construction and sequencing of SAGE libraries
RNA (5 ng) was collected for cDNA synthesis and then amplified by polymerase chain reaction (PCR) to generate 200 ng,24 and then LongSAGE libraries were constructed as previously described.25 In brief, RNA was reverse-transcribed, and the cDNA obtained was amplified using the switching mechanism at the 5′ end of RNA transcripts (SMART) cDNA amplification kit (BD) following the manufacturer's protocol, but using a modified template switching primer containing an AscI digestion site. The first strand cDNA was purified with a NucleoSpin column (Clontech Laboratories, Mountain View, CA) and then amplified using a modified PCR primer that contained a biotin molecule at its 5′ end and the Advantage II PCR kit (BD). The biotinylated 5′ ends of the amplified cDNAs were then removed by digestion of the initial amplified product with AscI (New England BioLabs, Beverly, MA). The cDNA was purified on a Chroma-Spin 200 Column (BD), and its concentration determined using a spectrophotometer (GeneQuant Pro; Biochrom, Cambridge, United Kingdom). The amplified cDNA was then processed according to a modified version of the standard LongSAGE protocol using the I-SAGE Long kit (Invitrogen, Carlsbad, CA) as described in detail in Khattra et al.25 After analysis of data quality from a first 384-well sequencing plate, each library was sequenced to a sampling depth of 44 506 (adult BM) and 200 319 (FL) raw tags (GEO Series accession no. GSE13243; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE13243).
Quantitative real-time-PCR analysis
A PicoPure kit (Arcturus Bioscience, Mountain View, CA) was used to extract RNA. The extracted RNA was then reverse-transcribed into cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Burlington, ON), and quantitative real-time PCR analyses were performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). The primers are listed in Table 1.
List of primers used to assess transcript levels in murine HSCs and various downstream cell types
| Gene . | GenBank no. . | Forward primer . | Reverse primer . |
|---|---|---|---|
| Gapdh | NM_008084 | 5′-AACTTTGGCATTGTGGAAGG-3′ | 5′-ATGCAGGGATGATGTTCTGG-3′ |
| Cdkn2c | NM_007671 | 5′-GAACTGCGCTGCAGGTTAT-3′ | 5′-TCAAATTGGGATTAGCACCTC-3′ |
| Stat3 | NM_213659 | 5′-GGCACCTTGGATTGAGAGTC-3′ | 5′-ACTCTTGCAGGAATCGGCTA-3′ |
| MEF | NM_019680 | 5′-TCTGTGGATGAGGAGGTTCC-3′ | 5′-GGGTGCTGGAGAAGAACTCA-3′ |
| ATM | NM_007499 | 5′-GCAGAGTGTCTGAGGGTTTGT-3′ | 5′-AACTTCCAGCAACCTTCACC-3′ |
| Gata2 | NM_008090 | 5′-GCAGAGTGTCTGAGGGTTTGT-3′ | 5′-AACTTCCAGCAACCTTCACC-3′ |
| Lyl1 | NM_008535.2 | 5′-GACCCTTCAGCATCTTCCCTAACA-3′ | 5′-AGCCACCTTCTGGGGTTGGT-3′ |
| G3bp1 | NM_013716.2 | 5′-GGGAAGCCAGCAGATGCAGT-3′ | 5′-ATCCACGTGGCGGATCTTG-3′ |
| Eif1a | NM_010120.3 | 5′-CCGCTGCGTTTTGGTCACTA-3′ | 5′-TCGGTTCTGGCCTGGTTCTC-3′ |
| Tubb5 | NM_011655.3 | 5′-CAGCTGGACCGAATCTCTGTGT-3′ | 5′-GGACCTGAGCGAACGGAGTC-3′ |
| Hmga2 | NM_010441.1 | 5′-GGTGCCACAGAAGCGAGGAC-3′ | 5′-GGGCTCACAGGTTGGCTCTT-3′ |
| Psap | NM_011179.2 | 5′-ACTGTGGGGCCGTGAAGC-3′ | 5′-GTCGCAAGGAAGGGATTTCG-3′ |
| Prnp | NM_011170.1 | 5′-TCCAATTTAGGAGAGCCAAGCA-3′ | 5′-GCCGACATCAGTCCACATAGTCA-3′ |
| Pld3 | NM_0111161.1 | 5′-GCTGAGGAACCGGAAGCTGT-3′ | 5′-GGGAAAGGGGTGGTCCTGAG-3′ |
| Rhob | NM_007483.2 | 5′-GGGGCACGCAGAGTGGTT-3′ | 5′-GCAACAGTAGTGGCTTGCTGGTT-3′ |
| Vwf | NM_011708.3 | 5′-GGCGAGGATGGAGTTCGACA-3′ | 5′-TGACAGGGCTGATGGTCTGG-3′ |
| Car3 | NM_007606.3 | 5′-TGGCTAAGGAGTGGGGCTAC-3′ | 5′-GTCCCCTTTGGCAATTGGAT-3′ |
| Plp1 | NM_011123.2 | 5′-TCAGTCTATTGCCTTCCCTAGCAA-3′ | 5′-GCATTCCATGGGAGAACACCA-3′ |
| Hdac3 | NM_010411 | 5′-TCAACGTGGGTGATGACTGC-3′ | 5′-GCAGAGATGCGCCTGTGTA-3′ |
| Chd4 | NM_145979 | 5′-GCTGCCAGAGATCCCAAACG-3′ | 5′-TTGCCCTTAAGAGCTGGACAA-3′ |
| Cul4a | NM_146207 | 5′-GGAGAACATTTGACAGCAATTCTACA-3′ | 5′-GTGAGGTCGGGCACCCTGT-3′ |
| Trim27 | NM_009054 | 5′-GAAGAGACGGCGGGCACA-3′ | 5′-GCTGCTCAAACTCCCAGACAA-3′ |
| Fus | NM_139149 | 5′-CACTGGTTGCATTCATTTCTCCA-3′ | 5′-CCAGTGGAGGTGGTGGAGGT-3′ |
| Cebpa | NM_007678 | 5′-CCAGTCAGACCAGAAAGCTGA-3′ | 5′-CCACAAAGCCCAGAAACCTA-3′ |
| Mlx | NM_011550 | 5′-TGTGTCTTCAGCTGGATTGAGGA-3′ | 5′-GGACACCGATCACAATCTCTCG-3′ |
| Smarcc1 | NM_009211 | 5′-TGGGAGAGCCCGGACACG-3′ | 5′-TTGGTAGGAGCATCTGCATGAAC-3′ |
| Smarcc2 | NM_198160 | 5′-TCTTCAGCCGAAGCCTCCAC-3′ | 5′-CCCTTCTCAGGGAAGTTCAGCA-3′ |
| Cdkn1a | NM_007669 | 5′-GTACTTCCTCTGCCCTGCTG-3′ | 5′-TCTGCGCTTGGAGTGATAGA-3′ |
| 1200009F10Rik | NM_026166 | 5′-GAAAAACAATACCGGTTACTGCAAA-3′ | 5′-CCACGAGAGCTTCACATTCCTG-3′ |
| Ezh2 | NM_007971.1 | 5′-CGCTCTTCTGTCGACGATGTTTT-3′ | 5′-GTTGGGTGTTGCATGGAAGG-3′ |
| Gata3 | NM_008091.2 | 5′-GGTATCCTCCGACCCACCAC-3′ | 5′-CCAGCCAGGGCAGAGATCC-3′ |
| Bmi1 | NM_007552.3 | 5′-AAACCAGACCACTCCTGAACA-3′ | 5′-TCTTCTTCTCTTCATCTCATTTTTGA-3′ |
| Sh2b3 | NM_008507 | 5′-CAACACACACAAGGCTGTCA-3′ | 5′-CCTGTGCACAAGAACTACATCTG-3′ |
| Rae28 | NM_001042623 | 5′-CGCACATCATTGAAGGCTTTGTT-3′ | 5′-TTCTTTCAGGAACTGAGAACATCC-3′ |
| Gene . | GenBank no. . | Forward primer . | Reverse primer . |
|---|---|---|---|
| Gapdh | NM_008084 | 5′-AACTTTGGCATTGTGGAAGG-3′ | 5′-ATGCAGGGATGATGTTCTGG-3′ |
| Cdkn2c | NM_007671 | 5′-GAACTGCGCTGCAGGTTAT-3′ | 5′-TCAAATTGGGATTAGCACCTC-3′ |
| Stat3 | NM_213659 | 5′-GGCACCTTGGATTGAGAGTC-3′ | 5′-ACTCTTGCAGGAATCGGCTA-3′ |
| MEF | NM_019680 | 5′-TCTGTGGATGAGGAGGTTCC-3′ | 5′-GGGTGCTGGAGAAGAACTCA-3′ |
| ATM | NM_007499 | 5′-GCAGAGTGTCTGAGGGTTTGT-3′ | 5′-AACTTCCAGCAACCTTCACC-3′ |
| Gata2 | NM_008090 | 5′-GCAGAGTGTCTGAGGGTTTGT-3′ | 5′-AACTTCCAGCAACCTTCACC-3′ |
| Lyl1 | NM_008535.2 | 5′-GACCCTTCAGCATCTTCCCTAACA-3′ | 5′-AGCCACCTTCTGGGGTTGGT-3′ |
| G3bp1 | NM_013716.2 | 5′-GGGAAGCCAGCAGATGCAGT-3′ | 5′-ATCCACGTGGCGGATCTTG-3′ |
| Eif1a | NM_010120.3 | 5′-CCGCTGCGTTTTGGTCACTA-3′ | 5′-TCGGTTCTGGCCTGGTTCTC-3′ |
| Tubb5 | NM_011655.3 | 5′-CAGCTGGACCGAATCTCTGTGT-3′ | 5′-GGACCTGAGCGAACGGAGTC-3′ |
| Hmga2 | NM_010441.1 | 5′-GGTGCCACAGAAGCGAGGAC-3′ | 5′-GGGCTCACAGGTTGGCTCTT-3′ |
| Psap | NM_011179.2 | 5′-ACTGTGGGGCCGTGAAGC-3′ | 5′-GTCGCAAGGAAGGGATTTCG-3′ |
| Prnp | NM_011170.1 | 5′-TCCAATTTAGGAGAGCCAAGCA-3′ | 5′-GCCGACATCAGTCCACATAGTCA-3′ |
| Pld3 | NM_0111161.1 | 5′-GCTGAGGAACCGGAAGCTGT-3′ | 5′-GGGAAAGGGGTGGTCCTGAG-3′ |
| Rhob | NM_007483.2 | 5′-GGGGCACGCAGAGTGGTT-3′ | 5′-GCAACAGTAGTGGCTTGCTGGTT-3′ |
| Vwf | NM_011708.3 | 5′-GGCGAGGATGGAGTTCGACA-3′ | 5′-TGACAGGGCTGATGGTCTGG-3′ |
| Car3 | NM_007606.3 | 5′-TGGCTAAGGAGTGGGGCTAC-3′ | 5′-GTCCCCTTTGGCAATTGGAT-3′ |
| Plp1 | NM_011123.2 | 5′-TCAGTCTATTGCCTTCCCTAGCAA-3′ | 5′-GCATTCCATGGGAGAACACCA-3′ |
| Hdac3 | NM_010411 | 5′-TCAACGTGGGTGATGACTGC-3′ | 5′-GCAGAGATGCGCCTGTGTA-3′ |
| Chd4 | NM_145979 | 5′-GCTGCCAGAGATCCCAAACG-3′ | 5′-TTGCCCTTAAGAGCTGGACAA-3′ |
| Cul4a | NM_146207 | 5′-GGAGAACATTTGACAGCAATTCTACA-3′ | 5′-GTGAGGTCGGGCACCCTGT-3′ |
| Trim27 | NM_009054 | 5′-GAAGAGACGGCGGGCACA-3′ | 5′-GCTGCTCAAACTCCCAGACAA-3′ |
| Fus | NM_139149 | 5′-CACTGGTTGCATTCATTTCTCCA-3′ | 5′-CCAGTGGAGGTGGTGGAGGT-3′ |
| Cebpa | NM_007678 | 5′-CCAGTCAGACCAGAAAGCTGA-3′ | 5′-CCACAAAGCCCAGAAACCTA-3′ |
| Mlx | NM_011550 | 5′-TGTGTCTTCAGCTGGATTGAGGA-3′ | 5′-GGACACCGATCACAATCTCTCG-3′ |
| Smarcc1 | NM_009211 | 5′-TGGGAGAGCCCGGACACG-3′ | 5′-TTGGTAGGAGCATCTGCATGAAC-3′ |
| Smarcc2 | NM_198160 | 5′-TCTTCAGCCGAAGCCTCCAC-3′ | 5′-CCCTTCTCAGGGAAGTTCAGCA-3′ |
| Cdkn1a | NM_007669 | 5′-GTACTTCCTCTGCCCTGCTG-3′ | 5′-TCTGCGCTTGGAGTGATAGA-3′ |
| 1200009F10Rik | NM_026166 | 5′-GAAAAACAATACCGGTTACTGCAAA-3′ | 5′-CCACGAGAGCTTCACATTCCTG-3′ |
| Ezh2 | NM_007971.1 | 5′-CGCTCTTCTGTCGACGATGTTTT-3′ | 5′-GTTGGGTGTTGCATGGAAGG-3′ |
| Gata3 | NM_008091.2 | 5′-GGTATCCTCCGACCCACCAC-3′ | 5′-CCAGCCAGGGCAGAGATCC-3′ |
| Bmi1 | NM_007552.3 | 5′-AAACCAGACCACTCCTGAACA-3′ | 5′-TCTTCTTCTCTTCATCTCATTTTTGA-3′ |
| Sh2b3 | NM_008507 | 5′-CAACACACACAAGGCTGTCA-3′ | 5′-CCTGTGCACAAGAACTACATCTG-3′ |
| Rae28 | NM_001042623 | 5′-CGCACATCATTGAAGGCTTTGTT-3′ | 5′-TTCTTTCAGGAACTGAGAACATCC-3′ |
Statistical analysis
DiscoverySpace software version 4.01,26 which uses Audic Claverie statistics,27 was used to identify tags that were over- or under-represented in a comparison of the LongSAGE libraries. Quantitative real-time PCR data were collected from 3 to 6 biologic replicates, and each sample was assigned a δ-cycle threshold (Ct) value with respect to the levels of Gapdh transcripts present in the same sample. The SEM of the resulting data were then calculated and plotted, and significant differences (P < .05) were established using the Student t test. L-Calc software (StemCell Technologies) was used to quantify the number of HSCs present in cells that had been stimulated for 16 hours in vitro with 20 ng/mL IL-11 plus 1, 10, or 300 ng/mL SF by limiting dilution analysis of transplantation recipients that showed less than a 1% donor-derived contribution to their circulating WBCs 16 to 24 weeks later.
Results
Selective purification of HSCs with durable self-renewal from fetal and adult sources
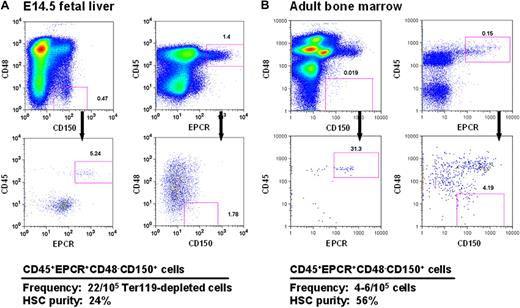
Current strategies for obtaining mouse HSCs at high purities from both FL and adult BM exploit combinations of stable features (eg, lin−kit+Sca1+5,28 and CD150+CD48−29 ) that distinguish these cells from their more mature derivatives. However, to obtain high HSC purities, additional elements are required, and these typically differ to accommodate the differing phenotypic characteristics of fetal and adult HSCs14,30,31 caused by their different cycling status.14,31 In preliminary studies, we found that a high expression of EPCR, a powerful discriminator of adult BM HSCs,11 was stable on HSCs that had been proliferating in vitro for 5 to 7 days (data not shown). This finding suggested that selection of EPCR+ cells within the CD48−CD150+ fraction (E-SLAM cells) might allow fetal and adult HSCs to be purified using the same simple protocol. Representative FACS profiles of these cells from both E14.5 FL and adult BM are shown in Figure 1.
CD45+EPCR+CD48−CD150+ (E-SLAM) cells from E14.5 FL cells and adult BM are highly enriched for HSCs. Representative profiles of viable cells after being stained with antibodies to CD45-FITC, EPCR-PE, CD48-APC, and one of CD150-biotin/streptavidin-PE-TexasRed or CD150-PECy7 from E14.5 FL (A) and adult mouse BM (B). The E-SLAM fraction of cells represents approximately 0.02% of Ter119-depleted FL cells and approximately 0.004% of the adult BM population. The HSC purities in these suspensions were determined from monitoring mice injected with single E-SLAM cells for at least 16 weeks after transplantation. Plots were generated using FlowJo software from TreeStar (Ashland, OR).
CD45+EPCR+CD48−CD150+ (E-SLAM) cells from E14.5 FL cells and adult BM are highly enriched for HSCs. Representative profiles of viable cells after being stained with antibodies to CD45-FITC, EPCR-PE, CD48-APC, and one of CD150-biotin/streptavidin-PE-TexasRed or CD150-PECy7 from E14.5 FL (A) and adult mouse BM (B). The E-SLAM fraction of cells represents approximately 0.02% of Ter119-depleted FL cells and approximately 0.004% of the adult BM population. The HSC purities in these suspensions were determined from monitoring mice injected with single E-SLAM cells for at least 16 weeks after transplantation. Plots were generated using FlowJo software from TreeStar (Ashland, OR).
FL and adult BM E-SLAM cells were then assessed for their repopulating activity in single-cell transplanted irradiated hosts that were then monitored over a period of 4 to 6 months to determine the contribution of each separately transplanted E-SLAM cell to the peripheral blood lymphoid and myeloid compartments. The pooled results of 3 experiments with 49 single E14.5 FL E-SLAM cells and 4 experiments with 62 single adult BM E-SLAM cells are shown in Figures 1 and 2. The purity of HSCs in the E-SLAM FL cells was 24% (Figure 1A), of which 50% showed a WBC output pattern characteristic of HSCs with durable self-renewal (data not shown). Because all FL HSCs are proliferating and only those in the G1 phase of the cell cycle are detectable by conventional transplantation strategies,32,33 this frequency is almost certainly an underestimate. If one assumes that half of the cells are in the S/G2/M phases of the cell cycle and cannot be detected, this would imply an actual FL HSC purity of approximately 48% (24% HSCs with durable self-renewal potential).
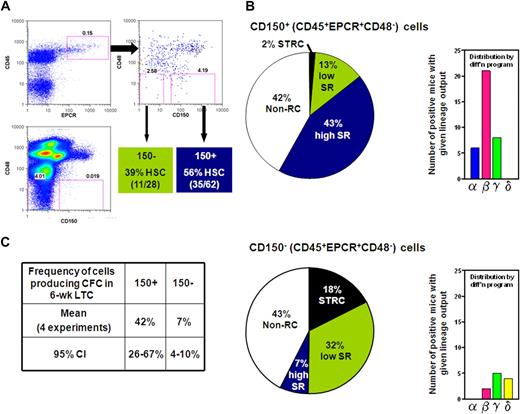
CD150 expression divides the CD45+EPCR+CD48− adult BM cell population into fractions differentially enriched in HSCs with differing abilities to sustain further self-renewal divisions. (A) Gates used to subdivide CD45+EPCR+CD48− adult BM cells into CD150+ and CD150− fractions as generated by FlowJo. (B) Distribution of different types of repopulating cells according to their self-renewal capacity (pie chart on the left) or differentiation (diff'n) programs displayed (bar chart on the right) from analyses of 62 mice that received a single transplantation of E-SLAM cells (top) and 28 mice that received a single translplantationof CD45+EPCR+CD48−CD150− cells (bottom). (C) Pooled results of single-cell 6-week LTC-IC assays of E-SLAM (CD150+) and CD45+EPCR+CD48−CD150− adult BM cells. (High and low SR HSCs = HSCs with durable and finite self-renewal potential, respectively; STRCs = short-term repopulating cells; and CI = confidence interval.)
CD150 expression divides the CD45+EPCR+CD48− adult BM cell population into fractions differentially enriched in HSCs with differing abilities to sustain further self-renewal divisions. (A) Gates used to subdivide CD45+EPCR+CD48− adult BM cells into CD150+ and CD150− fractions as generated by FlowJo. (B) Distribution of different types of repopulating cells according to their self-renewal capacity (pie chart on the left) or differentiation (diff'n) programs displayed (bar chart on the right) from analyses of 62 mice that received a single transplantation of E-SLAM cells (top) and 28 mice that received a single translplantationof CD45+EPCR+CD48−CD150− cells (bottom). (C) Pooled results of single-cell 6-week LTC-IC assays of E-SLAM (CD150+) and CD45+EPCR+CD48−CD150− adult BM cells. (High and low SR HSCs = HSCs with durable and finite self-renewal potential, respectively; STRCs = short-term repopulating cells; and CI = confidence interval.)
The experiments with the adult BM E-SLAM cells showed these to be 56% pure HSCs (Figure 1B) and, of these, approximately 75% had WBC output features associated with durable self-renewal potential. Hence the purity of this type of HSC in the E-SLAM fraction of adult BM was 43% (Figure 2B). These values are much higher than we previously reported for other powerful HSC isolation strategies (eg, CD45midRho−SP cells12,17 and E-Rho− cells34 ). Secondary mice transplanted with BM cells harvested individually from 4 primary mice that had each been repopulated with a single adult BM E-SLAM cell more than 26 weeks before confirmed that, in each case, progeny HSCs had been generated from the original input E-SLAM cell (data not shown). A small minority of the remaining cells in the E-SLAM fraction of adult BM were found to be STRCs (2%, defined here as cells that generate clones containing greater than 1% of the circulating WBC population between 4 and 8 weeks after transplantation but not later). The rest did not show any repopulating activity at or beyond 8 weeks after transplantation (earlier times were not assessed in these experiments). Thus, isolation of E-SLAM cells offers a simple method for obtaining HSCs with durable self-renewal activity at very high purities and is applicable to HSCs from both fetal and adult sources.
Prospective isolation of a phenotypically distinct population of adult BM HSCs that lack durable self-renewal activity
Because of the apparent depletion of HSCs lacking durable self-renewal activity from the CD150+ fraction of CD45+EPCR+CD48− adult BM cells, we examined the repopulating activity of cells in the matching CD150− subset. Analysis of 28 mice (2 experiments) that each received a transplantion of a single cell from the CD150− subset (see Figure 2A) showed that 39% of these cells could also be classified as HSCs, but most of these showed WBC outputs associated with a time-limited self-renewal activity (Figure 2B). Thus, within the total CD45+EPCR+CD48− population, approximately 90% of the HSCs with durable self-renewal were in the CD150+ subset at a purity of 43%, and approximately 50% of the HSCs that do not show evidence of continuing self-renewal for 4 months were in the corresponding CD150− subset at a purity of 32%. These results indicate that increasing expression of CD150 distinguishes adult BM HSCs with durable and finite self-renewal activity suggesting that these represent 2 biologically distinct cell types.
Similarities in the phenotype and frequency of 6-week long-term culture-initiating cells and HSCs with durable self-renewal potential
We then asked whether the CD150+ and CD150− subsets of CD45+EPCR+CD48− BM cells might also display differences in their ability to produce CFCs in LTC-IC assays. A total of 213 LTCs were set up, each with a single E-SLAM or CD45+EPCR+CD48−CD150− cell, and these cultures were then assessed 6 weeks later for the presence or absence of 1 or more CFCs in 4 independent experiments. As summarized in Figure 2C, we found the frequencies of LTC-ICs in both populations analyzed to be almost identical to the frequencies of HSCs expected to have durable self-renewal that were measured in the same fractions (42% vs 43% and 7% vs 7%), supporting the possibility that both assays actually detect the same cells with 100% efficiency.
Prnp, Gata3, and Bmi1 transcripts are elevated in adult BM E-SLAM cells that are highly enriched in HSCs with durable self-renewal potential
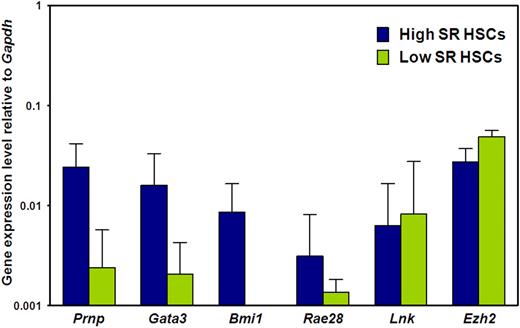
We then asked whether the biologic differences between E-SLAM and CD45+EPCR+CD48−CD150− adult BM cells are associated with differences in expression of 6 genes previously implicated in the regulation of HSC self-renewal, (ie, Prnp, Bmi1, Gata3, Rae28, Lnk, and Ezh2).35-41 Figure 3 shows the results of a comparison of the transcript levels measured in these 2 populations of adult BM cells by quantitative real-time PCR. Transcripts for all 6 genes were consistently detected in the CD150+ subset (selectively enriched in HSCs with durable self-renewal potential), albeit at low levels relative to Gapdh, as expected. Moreover, transcript levels for 3 of these 6 genes (Prnp, Gata3, and Bmi1) were significantly higher in the CD150+ subset (P < .05), suggesting a possible functional role for these molecules in maintaining a high self-renewal state in HSCs.
Prnp, Gata3, and Bmi1 transcripts are differentially expressed by E-SLAM and CD45+EPCR+CD48−CD150− adult BM cells. Quantitative real-time PCR analyses of transcript levels in extracts of 300 to 400 E-SLAM cells (blue bars) or CD45+EPCR+CD48−CD150− cells (green bars). All values are normalized to Gapdh transcript levels. Values shown are the mean ± SEM of values obtained in 3 to 6 independent experiments, with each measurement being derived from triplicate assays. Results for Prnp, Gata3, and Bmi1 in the 2 fractions are significantly different (P < .05).
Prnp, Gata3, and Bmi1 transcripts are differentially expressed by E-SLAM and CD45+EPCR+CD48−CD150− adult BM cells. Quantitative real-time PCR analyses of transcript levels in extracts of 300 to 400 E-SLAM cells (blue bars) or CD45+EPCR+CD48−CD150− cells (green bars). All values are normalized to Gapdh transcript levels. Values shown are the mean ± SEM of values obtained in 3 to 6 independent experiments, with each measurement being derived from triplicate assays. Results for Prnp, Gata3, and Bmi1 in the 2 fractions are significantly different (P < .05).
Generation of a list of new candidate genes regulating HSC self-renewal durability
To search for new candidate regulators of HSC self-renewal durability, we first compared the tag counts of LongSAGE libraries generated from highly purified HSC populations isolated from E14.5 FL and adult BM (Figure S1 and Tables S1,S2, available on the Blood website; see the Supplemental Materials link at the top of the online article). The HSCs from these 2 sources are known to differ in their content of HSC subtypes defined both in terms of their lineage outputs and in their self-renewal properties.14,17 To reduce the number of candidate genes for follow-up quantitative real-time PCR analysis, a further comparison was made with differentially expressed genes identified in a previously published microarray-based comparison of E14.5 FL HSCs, adult BM HSCs pre- and 5 days after treatment of the mice with 5-fluorouracil (5-FU).42 These microarrays were analyzed using the GEO microarray analysis tool with one-tailed Student t tests and a 0.100 significance stringency. When we compared all gene lists (SAGE, GEO FL versus adult BM HSCs, and GEO FL vs 5-FU–treated BM HSCs), we identified 13 candidate genes that appeared to be up-regulated in either the FL HSCs (eg, Hmga2, Lyl1, G3bp1, Eif1a, Fus, and Tubb5) or the BM HSCs (eg, Psap, Pld3, Rhob, VWF, Car3, and Plp1). Additional genes were selected from the SAGE library comparison based on previous studies from our group and others,14,43,44 or because they had potentially interesting roles in chromatin remodeling (Chd4, Hdac3, Trim27, Smarcc1, and Smarcc2).
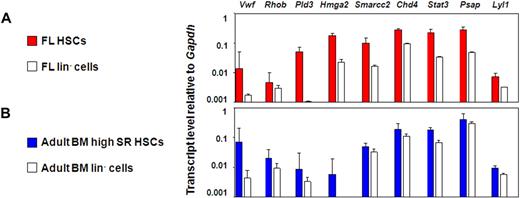
This approach yielded 27 candidate genes whose expression could be anticipated to be specifically up-regulated in HSCs with durable self-renewal potential. To test this hypothesis, we measured the transcript levels of these 27 genes in different HSC-enriched and closely related populations by quantitative real-time PCR. The 27 genes selected were: Rhob, Hmga2, Trim27, Mef, Pld3, Gata2, Cdkn2c, VWF, Atm, Cdkn1a, Lyl1, Car3, Plp1, Smarcc1, Cul4a, Stat3, G3bp1, Chd4, Eif1a, Cebpa, Smarcc2, Fus, Psap, Tubb5, Mlx, Hdac3, and Rik1200009F10. An initial comparison of the E14.5 FL and adult BM E-SLAM cells with their corresponding lin− fractions showed that 9 of these 27 genes were consistently expressed at higher levels in the E-SLAM cell fractions (Chd4, Pld3, Lyl1, Psap, Stat3, VWF, Rhob, Smarcc2, and Hmga2; Figure 4A,B). Results for the other genes tested are found in Figure S2A.
Nine candidate genes are consistently expressed in the E-SLAM fraction versus the lineage negative fraction of BM and FL blood cells. (A) Quantitative real-time PCR analyses of transcript levels in extracts of 300 to 400 E-SLAM cells (red bars) or 104 lin− (white bars) from FL. (B) Quantitative real-time PCR analyses of transcript levels in extracts of 300 to 400 E-SLAM cells (blue bars) or 104 lin− (white bars) from adult BM. All values are normalized to Gapdh. Values shown are the mean ± SEM of values obtained in 3 to 6 independent experiments, with each measurement being derived from triplicate assays. N.D. indicates not done.
Nine candidate genes are consistently expressed in the E-SLAM fraction versus the lineage negative fraction of BM and FL blood cells. (A) Quantitative real-time PCR analyses of transcript levels in extracts of 300 to 400 E-SLAM cells (red bars) or 104 lin− (white bars) from FL. (B) Quantitative real-time PCR analyses of transcript levels in extracts of 300 to 400 E-SLAM cells (blue bars) or 104 lin− (white bars) from adult BM. All values are normalized to Gapdh. Values shown are the mean ± SEM of values obtained in 3 to 6 independent experiments, with each measurement being derived from triplicate assays. N.D. indicates not done.
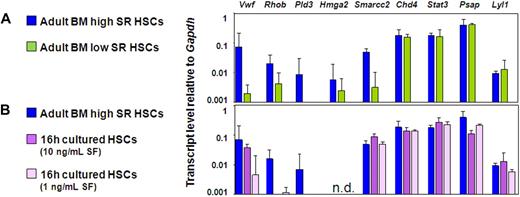
VWF, Rhob, and Pld3 expression is associated with HSCs that have durable self-renewal potential
We then compared the gene expression profiles of the CD150+ and CD150− subsets of CD45+EPCR+CD48− adult BM cells. These cells showed no difference in Lyl1, Stat3, Hmga2, Psap, and Chd4 transcript levels, but the CD150+ fraction contained significantly higher levels of Smarcc2, VWF, Rhob, and Pld3 transcripts (Figure 5A). We then took advantage of the finding that freshly isolated adult BM HSCs retain or lose (by 75%) their repopulating ability in the absence of any change in their quiescent status, rate of subsequent entry into the cell cycle, or viability when exposed for 16 hours in vitro to a glycoprotein 130-activating ligand (IL-11) in combination with an adequate (300 ng/mL) or a limiting concentration (1-10 ng/mL) of SF, respectively.34 Previous measurements showed that Bmi-1 and Ezh-2 transcript levels in such cultured HSC populations are markedly lower in the cells cultured in limiting concentrations of SF.34 These findings were confirmed in the experiments described here. In addition, the present set of experiments identified transcripts for VWF, Rhob, and Pld3 to be present at reduced levels in the cells cultured in concentrations of SF that selectively and rapidly eliminate their repopulating activity (Figure 5B). Additional quantitative real-time PCR data from these experiments can be found in Figure S2A through C.
Elevated VWF, Rhob, and Pld3 expression is consistently associated with HSCs possessing durable self-renewal potential. (A,B) Quantitative real-time PCR analyses of transcript levels from adult BM E-SLAM cells (blue bars) compared with CD45+EPCR+CD48−CD150− cells (A, green bars) and E-SLAM cells cultured in 20 ng/mL of IL-11 supplemented with 10 ng/mL SF (B, dark purple bars) or 1 ng/mL SF (B, light purple bars). All values are normalized to Gapdh. Values shown are the mean ± SEM of values obtained in 3 to 6 independent experiments, with each measurement being derived from triplicate assays. N.D. indicates not done.
Elevated VWF, Rhob, and Pld3 expression is consistently associated with HSCs possessing durable self-renewal potential. (A,B) Quantitative real-time PCR analyses of transcript levels from adult BM E-SLAM cells (blue bars) compared with CD45+EPCR+CD48−CD150− cells (A, green bars) and E-SLAM cells cultured in 20 ng/mL of IL-11 supplemented with 10 ng/mL SF (B, dark purple bars) or 1 ng/mL SF (B, light purple bars). All values are normalized to Gapdh. Values shown are the mean ± SEM of values obtained in 3 to 6 independent experiments, with each measurement being derived from triplicate assays. N.D. indicates not done.
Discussion
Understanding HSC self-renewal control remains a priority in medicine, as this knowledge is likely to provide insights into methods for expanding HSCs and producing blood products in vitro in addition to identifying key pathways that are perturbed or hijacked by leukemic stem cells. Furthermore, it is possible that parts of this machinery are used by other stem cell populations to maintain their undifferentiated status. Already, a surprisingly large number of genes have been identified from overexpression or knock-out experiments as able to influence HSC function to varying degrees in assays designed to test the ability of these cells to amplify their numbers either in vitro or in vivo. These include genes like Bmi138 and Cdkn1a (p21),43 whose expression appears to be required for optimal HSC self-renewal, as well as Lnk45 and Cdkn2c (p18),46 whose expression appears to suppress HSC self-renewal. However, whether these genes constitute the complete self-renewal circuitry of HSCs is unclear, and there is a large, understudied role of noncoding RNAs that is beginning to emerge. It also remains a mystery as to how cells are molecularly configured to be able to execute sufficient self-renewal divisions to sustain large clonal outputs for up to 4 to 6 months but not longer, in contrast to others that do not exhaust within the lifetime of the mouse. Indeed, evidence of a clear distinction between the latter 2 types of HSCs has only recently become available from studies that have assessed the in vivo self-renewal properties of a large number of individual HSCs at a clonal level.17 Thus, it has not been possible to implicate any of the genes thus far identified as to whether they might be particularly important in keeping HSCs in a state where the probability of executing a self-renewal division when stimulated in vivo remains high.
As a first step toward addressing this issue, we sought to identify a method of obtaining different highly purified populations of HSCs with either durable or limited self-renewal properties. We and others have shown that a 4-month output of myeloid or balanced output of progeny in a primary animal is a signature property of HSCs with unlimited self-renewal ability and, conversely, that a lymphoid biased output is a signature property of HSCs with limited self-renewal ability.17 Here we show that this change in self-renewal durability is accompanied by a decline in cell surface CD150 expression. This, in turn, allows HSCs with durable versus finite self-renewal abilities to be separately isolated at very high purities within the CD150+ and CD150− fractions of CD45+EPCR+CD48− adult BM cells. Interestingly, CD150 expression on HSCs has been the subject of much recent discussion in the literature, with conflicting reports regarding the presence and frequency of HSCs in CD150− fraction of adult mouse BM.47,48 Our results provide definitive evidence that some HSCs with durable self-renewal potential are present in the CD150− fraction as demonstrated by single-cell transplantation of CD45+EPCR+CD48−CD150− cells. However, these constitute a small fraction of all HSCs with this activity. Our studies also show that at least one in every million adult mouse BM cells is a CD150− HSC with durable self-renewal potential. We did not, however, test CD150− cells that were either EPCR− or CD48+, so we may be underestimating the total frequency of HSCs in the CD150− fraction.
Interestingly, LTC-ICs defined by a 6-week CFC output are also selectively enriched in the E-SLAM fraction. In fact, the similarity of their absolute frequencies in both subsets to the corresponding frequencies of the HSCs with durable self-renewal potential is strikingly suggestive of their possible identity. It will therefore be of interest to determine whether 6-week LTC-ICs and/or CD150 expression can be used as direct monitors of an unlimited self-renewal HSC state in the mouse system. We also found that Prnp, Bmi1, and Gata3 were selectively up-regulated in the E-SLAM population (enriched in HSCs with durable self-renewal potential), consistent with previous evidence that these genes regulate the maintenance of HSC self-renewal ability. We now show from pairwise comparisons of several other genes that another 3 (Rhob, Pld3, and VWF) may have similar roles given their selective but consistently elevated expression in populations that are most enriched in HSCs with durable self-renewal potential.
Prion protein (Prnp), which appeared in the LongSAGE comparison as being highly up-regulated in adult BM, and doubles as a positive control for this comparison, has a very interesting role in HSCs. In 2005, Zhang et al36 showed that PRNP was expressed on adult BM HSCs both by phenotypic overlap studies and by affinity purification followed by competitive repopulation experiments. The primary transplantations of Prnp−/− BM cells were found to compete equally with wild-type cells, but the secondary and tertiary transplantations of Prnp−/− cells showed a 2- to 3-fold reduction in repopulating activity compared with wild-type HSCs. This is in strong agreement with Prnp having a functional role in maintaining a high self-renewal capability in HSCs.36
Rhob encodes a small GTPase that has been described to have a role in the inhibition of malignant cell growth and is the putative target of farnesyltransferase inhibitors.49 It has been shown to interact with mDia1,50 which in both heterozygous and homozygous knockout animals leads to age-dependent myeloproliferative defects that include splenomegaly, a hypercellular BM, and extramedullary hematopoiesis.51
Phospholipase D3, on the other hand, is a gene about which relatively little is known. Pld3, also known as Sam-9 is strongly expressed in neural tissue,52 and transcripts have also been identified in T-cell lines,53 T-regulatory cells, and Foxp3-transduced CD25−CD4+ T cells.54 Notably, higher levels of Pld3 transcripts have been previously detected in primitive hematopoietic cell populations expanded in culture after transduction with 2 different Nucleoporin98-Hox fusion genes that induce HSC expansion compared with cells transduced with a control vector.55 Pld3 has 30% to 40% homology to Pld1 and Pld2, whose products are stimulated by PIP2, G proteins, ARF, and RHOA. PLD1 is directly regulated by PKC in most mammalian cells.56 Most interesting is the strong connection between Phospholipases and Rho family members, as 2 of the 3 new candidate regulators identified here are RHOB and PLD3.
Absence of VWF is primarily associated with defective hemostasis and thrombosis. VWF knockout animals are normal at birth, but have a highly prolonged bleeding time and reduced levels of Factor VIII.57 However, a role of VWF in the control of HSC self-renewal has not been previously indicated.
Lyl1 (an Scl family member) is a gene that remains high in the HSCs that do not have durable self-renewal activity and show poor maintenance of myelopoietic potential. Recently, Capron et al showed that Lyl1−/− BM cells are 1.6- to 5.7-fold less competitive than wild-type BM cells with the vast majority of the long-term impairment being attributable to poor B- and T-cell production.58 Furthermore, overexpression of Lyl1 has been clearly implicated in B- and T-cell lymphoma, with 30% of Lyl1 transgenic mice developing malignant lymphoma.59
In summary, we have now shown that HSCs with long-term multilineage potential in vivo, but markedly different self-renewal properties, can be prospectively isolated as separate, highly purified cell types and found to display unique gene expression signatures. These findings strongly support a model in which the most primitive hematopoietic cells can postpone indefinitely the activation of programs associated with expression of their multilineage competence, which may then proceed more or less rapidly according to the signals the cell receives. The present identification of 3 genes not previously associated with HSC self-renewal activity offers an exciting approach to further deciphering the mechanisms that regulate the relevant molecular machinery.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Flow Cytometry Facility of the Terry Fox Laboratory, the Animal Resource Center of the BC Cancer Agency, StemCell Technologies, and Genetics Institute for gifts of reagents.
This work was supported by grants from the National Cancer Institute of Canada (NCIC, Toronto, ON, with funds from the Terry Fox Foundation), the Canadian Institutes of Health Research (CIHR, Toronto, ON) and the Natural Science and Engineering Research Council (Ottawa, ON). D.K. received Studentships from the Stem Cell Network (Ottawa, ON), the CIHR, and the Michael Smith Foundation for Health Research (MSFHR, Vancouver, BC). M.C. received a studentship from the MSFHR. C.B. received a Fellowship from the Deutsche Forschungsgemeinschaft (Bonn, Germany). S.W. received an Erwin Schroedinger Auslands Fellowship from the Austrian Science Fund (Vienna, Austria). B.D. received Studentships from NCIC and the Stem Cell Network.
Authorship
Contribution: Experiments were designed by D.G.K. and C.J.E. with input from M.R.C., C.B., S.W., and B.J.D.; HSC isolation, transplantations, and flow cytometric analysis were carried out primarily by D.G.K. with assistance from M.R.C., C.B., B.J.D., and M.G.; transcript analysis was performed by D.G.K., M.R.C., E.M., and J.C.; peripheral blood collection and analysis was performed by D.G.K., C.B., E.M., and J.C.; long-term culture experiments were performed by S.W.; material for LongSAGE libraries was isolated by D.G.K, B.J.D., and M.B.B. with isolation and amplification performed by D.G.K. with assistance from Y.Z.; SAGE libraries were made at the Michael Smith Genome Sciences Center under the direction of M.M. with construction by Yongjun Zhao and statistical assessment by D.G.K., A.D., and C.J.E.; and D.G.K. wrote the paper with input from M.R.C., C.B., S.W., B.J.D., and C.S.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Connie J. Eaves, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail: ceaves@bccrc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal