Abstract
In most human somatic cells, telomeres shorten as a function of DNA replication. Telomere length is therefore an indirect measure of the replicative history of cells. We measured the telomere lengths at XpYp chromosomes in purified human hematopoietic populations enriched for stem cells (Lin−CD34+CD38−Rho−) and successively more mature cells. The average telomere length showed expected length changes, pointing to the utility of this method for classifying novel differentiation markers. Interestingly, the frequency of abruptly shortened telomeres increased in terminally differentiated adult populations, suggesting that damage to telomeric DNA occurs or is not repaired upon hematopoietic differentiation. When Lin−CD34+CD38−Rho− cord blood cells were transplanted into immunodeficient mice, the telomeres of the most primitive regenerated human hematopoietic cells lost approximately 3 kb, indicative of more than 30 cell divisions. Further losses in differentiating cells were similar to those observed in pretransplantation cell populations. These results indicate extensive self-renewal divisions of human hematopoietic stem cells are the primary cause of telomere erosion upon transplantation rather than added cell divisions in downstream progenitors.
Introduction
The extreme terminus of linear chromosomes is organized into nucleoprotein structures known as telomeres. In humans, telomeres are composed of heterogeneous arrays of up to 20 kb of TTAGGG1 that are in equilibrium with a complex of associated proteins.2 The G-rich strand is orientated 5′ to 3′ and ends in an overhang of 130 to 210 bp in length3 that is vital for telomere function.4 Telomeres serve a protective function, preventing chromosome end-to-end fusions and protecting against the loss of coding DNA during normal cellular turnover.5 In most somatic cells, telomeres lose 50 to 100 bp at every cell division6 due to incomplete end-replication.7 When a critical length is reached, the capping function of the telomere is abated, and cells either die by apoptosis or enter senescence.8 Thus, telomeres have been described as “mitotic clocks,” acting as indicators of the remaining replicative capacity in cells.9 Consequently, as telomere repeats are lost as a function of cell division, overall telomere length also provides an indirect measure of cell replicative history and may be useful to derive cellular hierarchies. Under particular circumstances, such as is provided by conditions of high oxidative stress, cells senesce more rapidly even in the absence of significant loss of mean telomere length.10 These and other data have prompted a revised model of telomere shortening where it is the shortest, rather than the average telomere length that is important to cell viability and the triggering of senescence.11 An accurate measurement of these “outlying” ultrashort telomeres is therefore important in understanding the role of cell division and senescence in stem cell responses to mitogenic stimuli.
Telomeres are maintained in the germ line by a specialized ribonucleoprotein reverse transcriptase called telomerase, which adds telomeric repeats de novo onto the 3′ overhang, counteracting the attrition of telomeric repeats from incomplete end-replication.12,13 In addition to the germ line, telomerase is present at detectable levels in many types of somatic cells including hematopoietic stem cells (HSCs), albeit in limited amounts.14 HSCs have extensive replicative potential as highlighted by their ability to permanently regenerate blood formation in myeloablated recipients. Nevertheless, there is a consistent decline in telomere length in their progeny as humans age15 ; thus telomerase levels in human HSCs are not sufficient to maintain replication indefinitely.16,17 The telomere loss in most somatic cells reflects limiting levels of telomerase, although activated B and T lymphocytes as well as CD34+ progenitor cells express telomerase at detectable levels.18,19 It has been proposed that although the low telomerase level present in somatic cells is not sufficient to maintain constant telomere length, it is required to repair rare critically short telomeres arising from DNA damage.20 The replicative limit that telomeres impose on the proliferation of most somatic cells appears to act as a tumor suppressor mechanism.21 Because cells must bypass senescence or apoptosis to become cancerous, the majority of cancer cells are telomerase positive.22
As the average life expectancy of human recipients of HSC transplants increases, so too does the frequency of posttransplantation age-related hematologic disorders.23 It has been proposed that the increased frequency of these disorders may be due to an accelerated “aging” phenotype in the transplanted donor cells manifested by short telomeres.24,25 Previous studies have shown that telomere length in terminally differentiated subpopulations rapidly decreases after successful engraftment and hematopoietic reconstitution.24,26,27 This decrease can range from 0.4 to more than 3 kb,24,26 although other studies suggest that the extent of telomere loss by the regenerated cells can be very small.28 To gain more precise information about this issue, we used a modified single-molecule polymerase chain reaction (PCR)–based technique called STELA29 (single telomere length amplification) to assess the telomere length of small numbers of phenotypically defined subsets of primitive and differentiating cells from normal human cord blood (CB) and granulocyte colony-stimulating factor (G-CSF)–mobilized adult peripheral blood (MPB) samples and their progeny before and after transplantation into immunodeficient mice.
Methods
Cells
CB cells were obtained from deliveries of normal, full-term infants. MPB cells were obtained from healthy volunteer donors who had been given a 5-day course of subcutaneous G-CSF injections. These were all obtained as anonymized samples and used according to procedures approved by the Review Ethics Board of the University of British Columbia. From each sample, the low-density (< 1.077 g/mL) cells were isolated using Ficoll Hypaque density separation and then used either directly or after freezing and thawing when required.
Cell staining and flow cytometry
Suspensions enriched in CD34+ cells were obtained by EasySep immunomagnetic removal of cells expressing various lineage markers as described by the supplier (StemCell Technologies, Vancouver, BC). Where indicated, the low-density, Lin− cells obtained were stained with rhodamine-123 (Molecular Probes, Eugene, OR) with and without 0.5mM Verapamil (Sigma-Aldrich, St Louis, MO) for 20 minutes at 37°C. A final incubation of 20 minutes at 37°C to allow the cells to efflux the rhodamine dye was performed. For flow cytometry and cell sorting, cells were incubated with 10% human serum and an anti–mouse FcR antibody (2.4G2; ATCC, Rockville, MD) for 10 minutes at 4°C and then for 30 minutes at 4°C with antibodies against human CD34-phycoerythrin (PE; StemCell Technologies), human anti–CD38-fluorescein isothiocyanate (FITC; Becton Dickinson, San José, CA), and donkey-anti–rat-allophycocyanin (APC; Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were washed twice in 2% FCS in HBSS with 0.1% propidium iodide (PI; Sigma-Aldrich) added to the final wash. Analyses were performed on a FACSCalibur and cells were isolated using either a FACSVantage or a FACSDiva (Becton Dickinson) with gates chosen to exclude at least 99.95% of cells incubated with either verapamil and/or isotype control antibodies labeled with the same fluorochromes. For cell sorting of subpopulations, cells were first incubated with 10% human serum and an anti–mouse FcR antibody (2.4G2; ATCC) for 10 minutes at 4°C and then for 30 minutes at 4°C with antibodies against one or more of the following human antigens: CD45, CD19, CD20, CD15, CD66b, CD56, CD3, CD4, CD8 (Becton Dickinson), and CD34 (StemCell Technologies). The cells were then washed twice and sorted using either a FACSVantage or FACSDiva (Becton Dickinson).
Xenotransplantation of immunodeficient mice
All animal studies were performed in accordance with institutional guidelines at the University of British Columbia. Primary test cells were injected into the tail vein together with 106 irradiated (15 Gy) human bone marrow cells into 6- to 12-week-old NOD.Cg-PrkdcscidIL2rgtmlWjl/Sz (NOD/SCID/IL2Rγcnull) mice within 12 hours after administration of 275 cGy x-rays to maximize immediate engraftment. Six weeks later, the mice were killed and bone marrow cells were removed from femurs and tibiae. After lysis of red blood cells with 0.8% NH4Cl (StemCell Technologies), the cells were stained with monoclonal antibodies against CD45, CD19, CD20, CD15, CD66b, CD56, CD3, CD4, CD8, and CD34 and analyzed on a FACSCalibur (Becton Dickinson) or isolated using either a FACSVantage or a FACSAria (Becton Dickinson).
STELA
STELA was conducted as described previously.29 Briefly, cells were sorted directly into tubes containing 1 × SSC and 1 × 105 SF9 insect cells. Because sorted populations were routinely fewer than 100 cells, inclusion of SF9 cells acted as both a cellular and genomic carrier. DNA was extracted from sorted populations by standard proteinase K, RNase A, and phenol/chloroform protocols. This DNA was ligated at 35°C for 12 hours with 0.001 μM telorette linker 806, which anneals to the telomeric 3′ G-rich overhang and ligates directly to the 5′ end of the telomere. DNA was diluted to 50 genome equivalents per microliter based on cell numbers obtained from sorting and added to a PCR reaction containing oligonucleotides 800 and 810, specific for the XpYp subtelomeric region and a sequence on the linker 806, respectively, Extensor Hi-fidelity PCR enzyme (ABgene, Epsom, United Kingdom) and PCR buffer. After 24 rounds of amplification, samples were resolved on 0.7% LE agarose, Southern blotted, and hybridized to a probe specific to the XpYp subtelomeric region (generated by PCR using primers 800 and 801).
STELA calculations and statistics
Amplicons were allocated into bins based on length, and the frequency within each bin was used to calculate the mean telomere length, SEM, and SD. Data are presented here as the mean plus or minus SEM. Because SD is sensitive to the presence of statistical outliers, a more robust scale estimator, the mean absolute deviation (MAD) was used to identify ultrashort telomeres. Outliers were classified as any amplicon falling outside of 1.4826 MAD from the median under the assumption that telomere length variation is normally distributed. For analyses where absolute measurements are given, outlying telomeres have been included in calculations for mean lengths. For analyses where the average number of cell divisions has been estimated, outlying telomeres have been excluded as it is likely that ultrashort telomeres have been generated via a mechanism other than normal replication, and will therefore lead to the overestimation of the number of replication events. When ultrashort telomeres were compared in different groups, significance was determined by logistical regression analysis.
Oligonucleotides
The oligonucleotide sequences were as follows: 800, 5′-TTGTCTCAGGGTCCTAGTG-3′; 801, 5′-TCTGAAAGTGGACCWATCAG-3′; 806, 5′-TGCTCCGTGCATCTGGCATCCCTAACC-3′, 810, 5′-TGCTCCGTGCATCTGGCATC-3′.
Results
STELA allows telomere length analysis from highly limiting starting material
STELA has been shown previously to be an accurate and reliable way to measure telomere lengths in a variety of cell types,29-31 including hematopoietic cells.32,33 However, there are several important caveats to consider when interpreting STELA results. First, the efficiencies of flow sorting, DNA extraction, ligation, and PCR need to be considered when working with limiting cell numbers. Second, as with any PCR-based technique, amplification is restricted to a maximum size limit, and we found that amplicons more than 18 kb were not amplified reproducibly, similar to previously reported data.30 Third, PCR techniques are inherently biased toward shorter amplicons, and fourth, the XpYp telomere might not be representative of telomere length as a whole in the cell. To address the first caveat, the efficiency of STELA was measured by sorting exact numbers of cells from whole MPB (donor MPB-1). Only with the addition of 105 SF9 insect carrier cells directly to the populations did STELA reproducibly generate amplicons. Three tubes each containing 100 cells generated 38, 40, and 40 STELA products, and a further 3 tubes each containing 50 cells each generated 17, 19, and 20 STELA products. Assuming all cells are diploid, using these data the overall efficiency was calculated as approximately 20%. This efficiency was used to calculate the number of amplifiable molecules from available XpYp substrates in a genomic DNA preparation. Samples in which the number of amplicons generated was significantly lower than the calculated overall efficiency and in which the mean telomere length was high were excluded from analysis. The bias for preferential amplification of smaller products was reduced by ensuring all STELA reactions were performed at subvisible levels, requiring Southern blot analysis for detection. The final caveat has been addressed previously,31 and was confirmed here as identical patterns of telomere loss were observed when STELA analysis was conducted on the 12q telomere (data not shown).
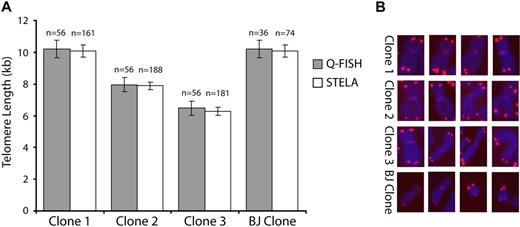
The accuracy of STELA was first assessed by comparing telomere lengths obtained by STELA and quantitative fluorescent in situ hybridization (Q-FISH) specifically at the XpYp telomere. Four cloned primary fibroblasts lines (3 female, 1 male) were analyzed and a total of 56 XpYp ends from 28 Q-FISH metaphases were compared with STELA conducted on 10 ng extracted DNA. The results showed that the mean telomere lengths obtained by both techniques were not significantly different (Figure 1), even in one clone that had telomeres ranging up to 21 kb in length (clone 1, Figure 1). At least 3 separate repeats of STELA experiments were undertaken for each subpopulation of each sample studied and results were not significantly different between each repeat. Depending on the yield of primitive cells, results from some populations isolated from particular MPBs and CBs were omitted due to low yields. However, overall, we found telomere length analysis using STELA gave reliable and reproducible data from very little starting material, allowing telomere length analysis of very small subpopulations of cells.
Comparison between STELA and Q-FISH. (A) Telomere lengths were determined in 4 cloned fibroblast lines by STELA and Q-FISH. The graph shows mean telomere length (± SEM) for both techniques. (B) Representative Q-FISH images of the allosomes in each clone analyzed. The X chromosome was identified with the addition of an X-specific centromeric PNA probe,34 whereas the Y chromosome (BJ clone only) was identified by G-banding patterns.
Comparison between STELA and Q-FISH. (A) Telomere lengths were determined in 4 cloned fibroblast lines by STELA and Q-FISH. The graph shows mean telomere length (± SEM) for both techniques. (B) Representative Q-FISH images of the allosomes in each clone analyzed. The X chromosome was identified with the addition of an X-specific centromeric PNA probe,34 whereas the Y chromosome (BJ clone only) was identified by G-banding patterns.
Telomere lengths in CB and MPB subpopulations show similar decreases with increasing differentiation
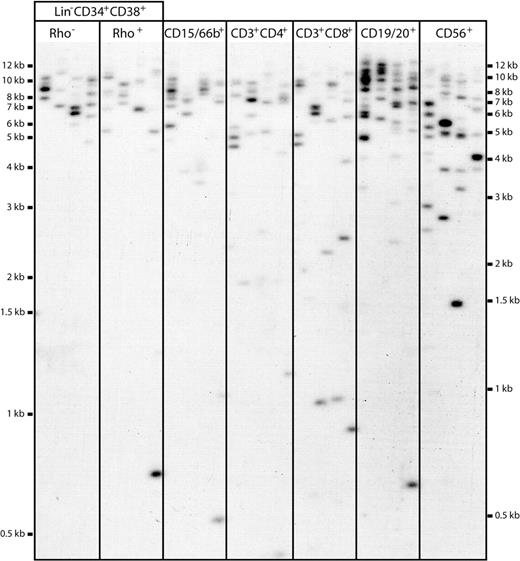
Different subpopulations of hematopoietic cells were isolated by flow cytometry from a total of 3 MPB and 5 CB samples and then analyzed by STELA. A representative blot from an analysis of various subsets of MPB cells is shown in Figure 2. These included different types of primitive cells: that is, Lin−CD34+CD38−Rho−, Lin−CD34+CD38−Rho+, CD34+CD38−, and CD34+CD38+ cells,35-37 as well as different types of fully differentiated granulocytes, T cells, B cells, and natural killer (NK) cells: that is, CD15/66b+, CD3+CD4+, CD3+CD8+, CD19/20+, and CD56+ cells.
Representative STELA blot. DNA extracted from sorted subpopulations of cells was taken through STELA PCR to amplify XpYp telomeres. An estimated concentration of 50 amplifiable molecules was added to each master mix, and aliquoted into 5 separate PCR tubes to ensure clear identification of resolved products. Amplicons were resolved on 0.7% agarose, Southern blotted, and detected with an XpYp-specific probe. Each amplicon represents the product from a single telomere end from a single cell, allowing telomere measurements of highly limiting cell populations. Amplicons were binned into size windows to determine mean telomere lengths, and products falling outside 1.4826 MAD from the median were counted and classified as ultrashort “outlier” telomeres.
Representative STELA blot. DNA extracted from sorted subpopulations of cells was taken through STELA PCR to amplify XpYp telomeres. An estimated concentration of 50 amplifiable molecules was added to each master mix, and aliquoted into 5 separate PCR tubes to ensure clear identification of resolved products. Amplicons were resolved on 0.7% agarose, Southern blotted, and detected with an XpYp-specific probe. Each amplicon represents the product from a single telomere end from a single cell, allowing telomere measurements of highly limiting cell populations. Amplicons were binned into size windows to determine mean telomere lengths, and products falling outside 1.4826 MAD from the median were counted and classified as ultrashort “outlier” telomeres.
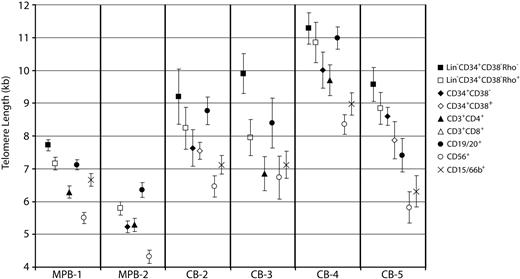
Despite marked variation between individual samples, the average telomere length determined from STELA amplicon measurements was higher in CB than in MPB populations (Table 1). In addition, telomere length heterogeneity within each subpopulation was observed (representative data are shown in Figure 3). Within each MPB and CB sample, telomere length from the various purified subsets correlated with their reported differentiation status. The most primitive (Lin−CD34+CD38−Rho−) cells had the longest telomeres, with modest but consistent progressive losses in Lin−CD34+CD38−Rho+ and CD34+CD38+ cells. In addition, the trends seen in terminally differentiated cells were consistent among all samples, with CD56+ cells having the shortest telomeres of all the sorted subpopulations, and CD19/20+ cells showing on average long telomeres (Figures 2–3). These findings establish the validity of using XpYp telomere measurements to obtain insight into the relationship of differentiation status to hematopoietic cell replication history.
Mean telomere lengths and ultrashort telomeres in subpopulations of CB and MPB
| Subpopulation . | Mean . | n . | Outlier . | Mean, − outliers . | Mean . | n . | Outlier . | Mean, − outliers . |
|---|---|---|---|---|---|---|---|---|
| MPB-1 | MPB-2 | |||||||
| CD34+38−Rho− | 7.72 ± 0.17 | 231 | 0.09 | 8.21 ± 0.16 | — | — | — | — |
| CD34+38−Rho+ | 7.16 ± 0.20 | 245 | 0.13 | 7.93 ± 0.17 | 5.79 ± 0.21 | 173 | 0.03 | 5.96 ± 0.21 |
| CD34+CD38− | — | — | — | — | 5.23 ± 0.18 | 269 | 0.11 | 5.79 ± 0.19 |
| CD3+CD4+ | 6.29 ± 0.18 | 302 | 0.14 | 7.13 ± 0.16 | 5.29 ± 0.20 | 204 | 0.08 | 5.68 ± 0.20 |
| CD3+CD8+ | 6.13 ± 0.18 | 311 | 0.13 | 6.85 ± 0.16 | 5.14 ± 0.20 | 226 | 0.06 | 5.42 ± 0.20 |
| CD19/20+ | 7.10 ± 0.16 | 448 | 0.14 | 7.97 ± 0.15 | 6.33 ± 0.23 | 207 | 0.11 | 7.00 ± 0.21 |
| CD56+ | 5.50 ± 0.17 | 327 | 0.10 | 6.04 ± 0.16 | 4.32 ± 0.20 | 198 | 0.07 | 4.62 ± 0.20 |
| CD15/66b+ | 6.64 ± 0.19 | 264 | 0.17 | 7.71 ± 0.15 | — | — | — | — |
| MPB-3 | CB-1 | |||||||
| CD34+38−Rho− | 5.74 ± 0.28 | 118 | 0.06 | 6.07 ± 0.27 | — | — | — | — |
| CD34+38−Rho+ | 5.64 ± 0.29 | 85 | 0.07 | 5.99 ± 0.27 | 9.64 ± 0.65 | 39 | 0.10 | 10.54 ± 0.54 |
| CD34+CD38− | — | — | — | — | 8.55 ± 0.57 | 56 | 0.09 | 9.17 ± 0.55 |
| CD34+CD38+ | — | — | — | — | 8.16 ± 0.52 | 67 | 0.10 | 8.97 ± 0.48 |
| CD19/20+ | 5.53 ± 0.34 | 95 | 0.15 | 6.33 ± 0.32 | 7.66 ± 0.49 | 108 | 0.09 | 8.39 ± 0.48 |
| CD56+ | — | — | — | — | 6.51 ± 0.36 | 145 | 0.10 | 7.20 ± 0.38 |
| CD15/66b+ | 4.62 ± 0.32 | 76 | 0.20 | 5.54 ± 0.29 | — | — | — | — |
| CB-2 | CB-3 | |||||||
| CD34+38−Rho− | 9.20 ± 0.84 | 26 | 0.11 | 10.25 ± 0.68 | 9.90 ± 0.61 | 59 | 0.12 | 10.84 ± 0.58 |
| CD34+38−Rho+ | 8.24 ± 0.64 | 40 | 0.08 | 8.80 ± 0.60 | 7.95 ± 0.55 | 88 | ND | 7.95 ± 0.55 |
| CD34+CD38− | 7.63 ± 0.56 | 74 | 0.12 | 8.58 ± 0.53 | — | — | — | — |
| CD34+CD38+ | 7.55 ± 0.26 | 184 | 0.09 | 8.17 ± 0.24 | — | — | — | — |
| CD3+CD4+ | — | — | — | — | 6.85 ± 0.52 | 104 | ND | 6.85 ± 0.52 |
| CD3+CD8+ | — | — | — | — | 7.61 ± 0.50 | 87 | 0.10 | 8.41 ± 0.48 |
| CD19/20+ | 8.74 ± 0.42 | 108 | 0.14 | 9.84 ± 0.37 | 8.37 ± 0.76 | 48 | 0.17 | 9.86 ± 0.70 |
| CD56+ | 6.46 ± 0.32 | 123 | 0.11 | 7.19 ± 0.30 | 6.73 ± 0.66 | 56 | 0.02 | 6.85 ± 0.66 |
| CD15/66b+ | 7.10 ± 0.28 | 147 | 0.14 | 7.99 ± 0.23 | 7.10 ± 0.41 | 118 | 0.06 | 7.51 ± 0.40 |
| CB-4 | CB-4 after transplantation | |||||||
| CD34+38−Rho− | 11.28 ± 0.48 | 78 | 0.08 | 11.97 ± 0.43 | — | — | — | — |
| CD34+38−Rho+ | 10.85 ± 0.62 | 58 | 0.10 | 11.82 ± 0.55 | — | — | — | — |
| CD34+CD38+ | 10.01 ± 0.55 | 57 | 0.16 | 11.22 ± 0.48 | 7.32 ± 0.43 | 57 | 0.11 | 8.00 ± 0.38 |
| CD3+CD4+ | 9.70 ± 0.47 | 82 | 0.10 | 10.53 ± 0.42 | 6.15 ± 0.30 | 31 | 0.10 | 6.55 ± 0.21 |
| CD19/20+ | 10.96 ± 0.34 | 129 | 0.09 | 11.70 ± 0.29 | 7.28 ± 0.26 | 95 | 0.13 | 8.02 ± 0.19 |
| CD56+ | 8.35 ± 0.30 | 170 | 0.12 | 9.17 ± 0.28 | 5.27 ± 0.38 | 35 | 0.20 | 6.07 ± 0.32 |
| CD15/66b+ | 8.95 ± 0.34 | 113 | 0.07 | 9.46 ± 0.33 | 5.78 ± 0.34 | 59 | 0.19 | 6.85 ± 0.19 |
| CB-5 | CB-5 after transplantation | |||||||
| CD34+38−Rho− | 9.57 ± 0.52 | 50 | 0.08 | 10.11 ± 0.49 | — | — | — | — |
| CD34+38−Rho+ | 8.84 ± 0.48 | 64 | 0.09 | 9.48 ± 0.45 | — | — | — | — |
| CD34+CD38− | 8.60 ± 0.28 | 167 | 0.12 | 9.38 ± 0.27 | — | — | — | — |
| CD34+CD38+ | 7.87 ± 0.57 | 48 | 0.08 | 8.46 ± 0.54 | 6.62 ± 0.21 | 131 | 0.14 | 7.32 ± 0.16 |
| CD19/20+ | 7.39 ± 0.51 | 67 | 0.13 | 8.37 ± 0.47 | 7.58 ± 0.32 | 101 | 0.17 | 8.67 ± 0.24 |
| CD56+ | 5.82 ± 0.48 | 52 | 0.15 | 6.74 ± 0.44 | 4.78 ± 0.46 | 27 | 0.19 | 5.59 ± 0.38 |
| CD15/66b+ | 6.29 ± 0.48 | 58 | 0.12 | 7.05 ± 0.45 | 4.90 ± 0.19 | 125 | 0.15 | 5.68 ± 0.13 |
| CB-6 | CB-6 mouse 1 | |||||||
| CD34+CD38+ | 9.10 ± 0.23 | 264 | 0.10 | 9.92 ± 0.20 | 6.61 ± 0.20 | 163 | 0.15 | 7.32 ± 0.18 |
| CD3+CD4+ | 7.71 ± 0.19 | 284 | 0.07 | 8.19 ± 0.17 | 5.30 ± 0.26 | 75 | 0.16 | 6.01 ± 0.21 |
| CD15/66b+ | 6.96 ± 0.25 | 188 | 0.12 | 7.68 ± 0.24 | 4.96 ± 0.21 | 99 | 0.22 | 5.76 ± 0.14 |
| CB-6 mouse 2 | CB-6 mouse 3 | |||||||
| CD34+CD38+ | 6.58 ± 0.21 | 143 | 0.13 | 7.24 ± 0.18 | 6.35 ± 0.23 | 168 | 0.13 | 7.12 ± 0.19 |
| CD3+CD4+ | 5.23 ± 0.20 | 100 | 0.11 | 5.67 ± 0.17 | 5.35 ± 0.17 | 148 | 0.18 | 6.06 ± 0.14 |
| CD15/66b+ | 5.38 ± 0.29 | 54 | 0.13 | 5.99 ± 0.21 | 4.93 ± 0.21 | 113 | 0.16 | 5.66 ± 0.15 |
| Subpopulation . | Mean . | n . | Outlier . | Mean, − outliers . | Mean . | n . | Outlier . | Mean, − outliers . |
|---|---|---|---|---|---|---|---|---|
| MPB-1 | MPB-2 | |||||||
| CD34+38−Rho− | 7.72 ± 0.17 | 231 | 0.09 | 8.21 ± 0.16 | — | — | — | — |
| CD34+38−Rho+ | 7.16 ± 0.20 | 245 | 0.13 | 7.93 ± 0.17 | 5.79 ± 0.21 | 173 | 0.03 | 5.96 ± 0.21 |
| CD34+CD38− | — | — | — | — | 5.23 ± 0.18 | 269 | 0.11 | 5.79 ± 0.19 |
| CD3+CD4+ | 6.29 ± 0.18 | 302 | 0.14 | 7.13 ± 0.16 | 5.29 ± 0.20 | 204 | 0.08 | 5.68 ± 0.20 |
| CD3+CD8+ | 6.13 ± 0.18 | 311 | 0.13 | 6.85 ± 0.16 | 5.14 ± 0.20 | 226 | 0.06 | 5.42 ± 0.20 |
| CD19/20+ | 7.10 ± 0.16 | 448 | 0.14 | 7.97 ± 0.15 | 6.33 ± 0.23 | 207 | 0.11 | 7.00 ± 0.21 |
| CD56+ | 5.50 ± 0.17 | 327 | 0.10 | 6.04 ± 0.16 | 4.32 ± 0.20 | 198 | 0.07 | 4.62 ± 0.20 |
| CD15/66b+ | 6.64 ± 0.19 | 264 | 0.17 | 7.71 ± 0.15 | — | — | — | — |
| MPB-3 | CB-1 | |||||||
| CD34+38−Rho− | 5.74 ± 0.28 | 118 | 0.06 | 6.07 ± 0.27 | — | — | — | — |
| CD34+38−Rho+ | 5.64 ± 0.29 | 85 | 0.07 | 5.99 ± 0.27 | 9.64 ± 0.65 | 39 | 0.10 | 10.54 ± 0.54 |
| CD34+CD38− | — | — | — | — | 8.55 ± 0.57 | 56 | 0.09 | 9.17 ± 0.55 |
| CD34+CD38+ | — | — | — | — | 8.16 ± 0.52 | 67 | 0.10 | 8.97 ± 0.48 |
| CD19/20+ | 5.53 ± 0.34 | 95 | 0.15 | 6.33 ± 0.32 | 7.66 ± 0.49 | 108 | 0.09 | 8.39 ± 0.48 |
| CD56+ | — | — | — | — | 6.51 ± 0.36 | 145 | 0.10 | 7.20 ± 0.38 |
| CD15/66b+ | 4.62 ± 0.32 | 76 | 0.20 | 5.54 ± 0.29 | — | — | — | — |
| CB-2 | CB-3 | |||||||
| CD34+38−Rho− | 9.20 ± 0.84 | 26 | 0.11 | 10.25 ± 0.68 | 9.90 ± 0.61 | 59 | 0.12 | 10.84 ± 0.58 |
| CD34+38−Rho+ | 8.24 ± 0.64 | 40 | 0.08 | 8.80 ± 0.60 | 7.95 ± 0.55 | 88 | ND | 7.95 ± 0.55 |
| CD34+CD38− | 7.63 ± 0.56 | 74 | 0.12 | 8.58 ± 0.53 | — | — | — | — |
| CD34+CD38+ | 7.55 ± 0.26 | 184 | 0.09 | 8.17 ± 0.24 | — | — | — | — |
| CD3+CD4+ | — | — | — | — | 6.85 ± 0.52 | 104 | ND | 6.85 ± 0.52 |
| CD3+CD8+ | — | — | — | — | 7.61 ± 0.50 | 87 | 0.10 | 8.41 ± 0.48 |
| CD19/20+ | 8.74 ± 0.42 | 108 | 0.14 | 9.84 ± 0.37 | 8.37 ± 0.76 | 48 | 0.17 | 9.86 ± 0.70 |
| CD56+ | 6.46 ± 0.32 | 123 | 0.11 | 7.19 ± 0.30 | 6.73 ± 0.66 | 56 | 0.02 | 6.85 ± 0.66 |
| CD15/66b+ | 7.10 ± 0.28 | 147 | 0.14 | 7.99 ± 0.23 | 7.10 ± 0.41 | 118 | 0.06 | 7.51 ± 0.40 |
| CB-4 | CB-4 after transplantation | |||||||
| CD34+38−Rho− | 11.28 ± 0.48 | 78 | 0.08 | 11.97 ± 0.43 | — | — | — | — |
| CD34+38−Rho+ | 10.85 ± 0.62 | 58 | 0.10 | 11.82 ± 0.55 | — | — | — | — |
| CD34+CD38+ | 10.01 ± 0.55 | 57 | 0.16 | 11.22 ± 0.48 | 7.32 ± 0.43 | 57 | 0.11 | 8.00 ± 0.38 |
| CD3+CD4+ | 9.70 ± 0.47 | 82 | 0.10 | 10.53 ± 0.42 | 6.15 ± 0.30 | 31 | 0.10 | 6.55 ± 0.21 |
| CD19/20+ | 10.96 ± 0.34 | 129 | 0.09 | 11.70 ± 0.29 | 7.28 ± 0.26 | 95 | 0.13 | 8.02 ± 0.19 |
| CD56+ | 8.35 ± 0.30 | 170 | 0.12 | 9.17 ± 0.28 | 5.27 ± 0.38 | 35 | 0.20 | 6.07 ± 0.32 |
| CD15/66b+ | 8.95 ± 0.34 | 113 | 0.07 | 9.46 ± 0.33 | 5.78 ± 0.34 | 59 | 0.19 | 6.85 ± 0.19 |
| CB-5 | CB-5 after transplantation | |||||||
| CD34+38−Rho− | 9.57 ± 0.52 | 50 | 0.08 | 10.11 ± 0.49 | — | — | — | — |
| CD34+38−Rho+ | 8.84 ± 0.48 | 64 | 0.09 | 9.48 ± 0.45 | — | — | — | — |
| CD34+CD38− | 8.60 ± 0.28 | 167 | 0.12 | 9.38 ± 0.27 | — | — | — | — |
| CD34+CD38+ | 7.87 ± 0.57 | 48 | 0.08 | 8.46 ± 0.54 | 6.62 ± 0.21 | 131 | 0.14 | 7.32 ± 0.16 |
| CD19/20+ | 7.39 ± 0.51 | 67 | 0.13 | 8.37 ± 0.47 | 7.58 ± 0.32 | 101 | 0.17 | 8.67 ± 0.24 |
| CD56+ | 5.82 ± 0.48 | 52 | 0.15 | 6.74 ± 0.44 | 4.78 ± 0.46 | 27 | 0.19 | 5.59 ± 0.38 |
| CD15/66b+ | 6.29 ± 0.48 | 58 | 0.12 | 7.05 ± 0.45 | 4.90 ± 0.19 | 125 | 0.15 | 5.68 ± 0.13 |
| CB-6 | CB-6 mouse 1 | |||||||
| CD34+CD38+ | 9.10 ± 0.23 | 264 | 0.10 | 9.92 ± 0.20 | 6.61 ± 0.20 | 163 | 0.15 | 7.32 ± 0.18 |
| CD3+CD4+ | 7.71 ± 0.19 | 284 | 0.07 | 8.19 ± 0.17 | 5.30 ± 0.26 | 75 | 0.16 | 6.01 ± 0.21 |
| CD15/66b+ | 6.96 ± 0.25 | 188 | 0.12 | 7.68 ± 0.24 | 4.96 ± 0.21 | 99 | 0.22 | 5.76 ± 0.14 |
| CB-6 mouse 2 | CB-6 mouse 3 | |||||||
| CD34+CD38+ | 6.58 ± 0.21 | 143 | 0.13 | 7.24 ± 0.18 | 6.35 ± 0.23 | 168 | 0.13 | 7.12 ± 0.19 |
| CD3+CD4+ | 5.23 ± 0.20 | 100 | 0.11 | 5.67 ± 0.17 | 5.35 ± 0.17 | 148 | 0.18 | 6.06 ± 0.14 |
| CD15/66b+ | 5.38 ± 0.29 | 54 | 0.13 | 5.99 ± 0.21 | 4.93 ± 0.21 | 113 | 0.16 | 5.66 ± 0.15 |
Data are represented as mean telomere lengths (± SEM) of each subpopulation in the 3 MPB and 6 CB studied. The number of XpYp telomeres scored (n), and frequency of outliers, representing telomeres that are likely to have shortened by processes other than normal replication, are also shown. Measures of mean telomere length (± SEM) with the outliers excluded are also given, and are used in subsequent analyses where the number of divisions is inferred from the changes in telomere length.
— indicates no reliable data.
Mean telomere lengths in subpopulations of MPB and CB. Representative MPB and CB samples are shown with mean telomere lengths (± SEM) in all analyzed subpopulations. Primitive subpopulations show the longest mean telomere lengths within each sample. Of the terminally differentiated cell types, CD56+ cells routinely have the shortest telomeres. CD19/20+ cells have the longest mean telomere length of terminally differentiated cells, predominantly due to a subset of cells having very long telomeres. This has been observed elsewhere and is presumably due to the reactivation of telomerase during B-cell development. Full telomere length data and distribution from all samples analyzed are presented in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Mean telomere lengths in subpopulations of MPB and CB. Representative MPB and CB samples are shown with mean telomere lengths (± SEM) in all analyzed subpopulations. Primitive subpopulations show the longest mean telomere lengths within each sample. Of the terminally differentiated cell types, CD56+ cells routinely have the shortest telomeres. CD19/20+ cells have the longest mean telomere length of terminally differentiated cells, predominantly due to a subset of cells having very long telomeres. This has been observed elsewhere and is presumably due to the reactivation of telomerase during B-cell development. Full telomere length data and distribution from all samples analyzed are presented in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Changes in telomere length can be used to predict numbers of cell divisions
By measuring the change in telomere length between different cell populations and assuming a loss of approximately 100 bp telomere per cell division,6 it is possible to infer the number of divisions undertaken as cells differentiate. Telomere length is dictated both by replicative loss and atypical sporadic shortening from damage.20 As such, the length used in estimating cell divisions should exclude atypically short telomeres. Therefore, for each subpopulation analyzed, the number of statistical outliers representing abruptly shortened telomeres was calculated and excluded from the final length measurement to ensure length changes occurred predominantly via replicative loss (Table 1). However, the level of telomerase expression and degree of telomere elongation in different subpopulations is unknown, hence the calculation of cell divisions between different cell types is likely to represent an underestimate. The difference in telomere length between the Lin−CD34+CD38−Rho− and Lin−CD34+CD38−Rho+ MPB cells ranged from 80 to 280 bp, equivalent to approximately 1 to 5 cell divisions. In CB, this difference was more pronounced, with 150-bp, 630-bp, 1.45-kb, and 2.89-kb differences in the samples analyzed, equivalent to 1 to 29 cell divisions. The change in telomere length from Lin−CD34+CD38−Rho+ to CD34+CD38+ in CB ranged from 630 to 1570 bp (mean = 968 bp), equivalent to 6 to 16 cell divisions. Relative to the Lin−CD34+CD38−Rho+ cells, terminally differentiated CD56+ cells showed the greatest (and most variable) decrease in average telomere length, ranging from 1.1 to 3.37 kb (11 to 34 cell divisions). Therefore, although general changes in telomere length showed the same trend in all samples, the changes in absolute length between subpopulations were heterogeneous. Furthermore, this heterogeneity appeared more pronounced in CB samples than MPBs.
Outlier frequency increases from primitive to differentiated cell types in MPB
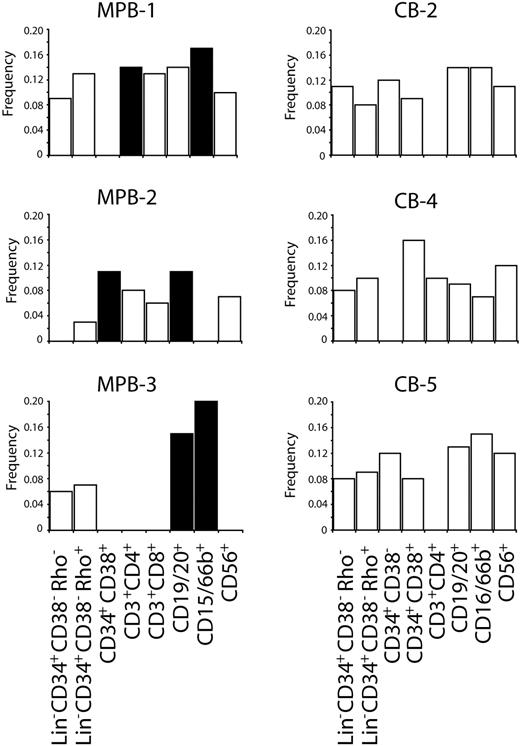
Abruptly shortened telomeres can be defined as telomeres that fall outside of the normal range of telomere length within a subpopulation. Because abruptly shortened telomeres are likely to have arisen from DNA damage rather than normal replicative loss, it is important to exclude them from determining number of cell divisions between subpopulations. However, analysis of these telomeres also offers insight into the degree of DNA damage and repair occurring at telomeres in cells. Although it remains unknown whether damage at the telomere is reflective of more widespread damage in genomic DNA, abruptly shortened telomeres are more likely to reach a critical length and initiate activation of senescence/apoptotic pathways. To assess whether some cell types have less abruptly shortened telomeres, either by replication history, prevention, or repair, the frequency of telomeres falling outside of the normal range was determined for each subpopulation and then compared. All 3 of the MPBs analyzed had significantly increased frequencies of atypically short telomeres in at least 2 of the terminally differentiated subpopulations compared with the most primitive population (P < .05, Figure 4; Table 1), whereas there was no significant increase in atypically short telomeres between the most primitive CD34+CD38−Rho− and Lin−CD34+CD38−Rho+ cells in any of the MPBs analyzed. However, a significant increase in abruptly shortened telomeres was observed between the Lin−CD34+CD38−Rho+ and CD34+CD38+ populations from one of the MPB samples (MPB-3, P = .006). Significant increases in atypically short telomeres with respect to more primitive cells were recorded in 2 of 2 samples for CD15/66b+ populations, 2 of 3 samples for CD19/20+ populations, with the remaining sample approaching significance (P = .08), and 1 of 2 samples for CD3+CD4+ populations (Figure 4). In contrast, of the 5 CB samples analyzed, no subpopulations had significantly higher atypically short telomeres with respect to the more primitive cells.
Frequency of ultrashort telomeres in subpopulations of hematopoietic cells. Bar graphs show the frequency of statistical outliers within each sorted subpopulation. Where no bar is shown, no telomere data exist for that particular subpopulation. The frequency of statistical outliers in more differentiated cells was compared with the most primitive cells available for each sample. Linear regression analyses were performed to identify significant increases in ultrashort telomeres, with significant subpopulations denoted as ■. MPB subpopulations showed significant increases in ultrashort telomeres with respect to more primitive cells; however, this trend was not observed in CB samples. An additional 2 CBs were analyzed and also showed no significant increases in ultrashort telomeres (data not shown).
Frequency of ultrashort telomeres in subpopulations of hematopoietic cells. Bar graphs show the frequency of statistical outliers within each sorted subpopulation. Where no bar is shown, no telomere data exist for that particular subpopulation. The frequency of statistical outliers in more differentiated cells was compared with the most primitive cells available for each sample. Linear regression analyses were performed to identify significant increases in ultrashort telomeres, with significant subpopulations denoted as ■. MPB subpopulations showed significant increases in ultrashort telomeres with respect to more primitive cells; however, this trend was not observed in CB samples. An additional 2 CBs were analyzed and also showed no significant increases in ultrashort telomeres (data not shown).
Transplantation experiments indicate progenitor cell expansion before differentiation
To determine whether telomere losses are the result of HSC expansion during an early repopulating phase in the recipient bone marrow or selective induced expansion in later stage progenitor cells, we used STELA to analyze hematopoietic subpopulations regenerated in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice that received a transplant 6 weeks previously with 1000 Lin−CD34+CD38−Rho− cells from 2 separate cord bloods (CB-4 and CB-5). We then compared these data with STELA measurements performed on subsets isolated from the fresh cells in the same original sample. The regenerated CD34+CD38−Rho− cells were too rare to analyze, but primitive CD34+CD38+ cells showed a 3970-bp and 2790-bp decrease in telomere length from the levels in the Lin−CD34+CD38−Rho− subset in the pretransplantation samples (Figure 5). Assuming a loss of 100 bp per cell division,6 this corresponds to 40 and 28 divisions. The difference in telomere length between CD34+CD38−Rho− and CD34+CD38+ in freshly isolated cells correspond to 7 divisions, indicating that at least an additional 21 cell divisions occur in regenerated primitive cell types 6 weeks after transplantation. The terminally differentiated CD3+, CD15/66b+, and CD56+ cells also possessed shorter telomeres than their counterparts in the initial samples, although this loss can be largely attributed to the initial decrease in telomere length seen between the Lin−CD34+CD38−Rho− and CD34+CD38+ populations. Hence, in the 2 CBs analyzed, the telomere loss seen after transplantation appears to occur primarily in the earliest hematopoietic cells. A total of 17 divisions were deduced to have occurred between the CD34+CD38+ cells and the CD15/66b+ cells present in the initial CB cells, compared with 12 divisions between the regenerated CD34+CD38+ and CD15/66b+ cells in the CB-4 transplant. Similar results were observed in CB-5, with 23 and 17 cell divisions calculated to separate the same 2 subsets in the fresh and regenerated populations. Corresponding results for the CD56+ cells were 21 and 20 cell divisions for sample CB-4, and 26 and 18 cell divisions for sample CB-5 (Figure 5B). In contrast to the other terminally differentiated populations, at least a subset of B cells was found to have long telomeres after transplantation, with longer telomere lengths than those measured in the CD34+CD38+ cells from both CBs. Thus, the process by which these cells are regenerated in the mice appears able to reactivate telomerase expression in these cells.
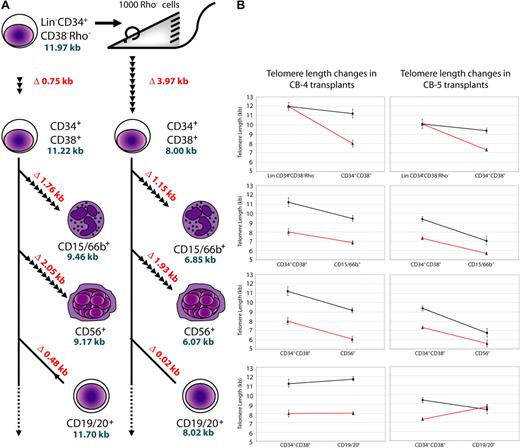
Telomere length changes in donor cells after transplantation of purified human CB cells into immune-deficient mice. (A) Schematic for transplantation, sorting, and STELA analysis is shown on the left panel. Lin−CD34+CD38−Rho− cells were transplanted into mice, and differentiated cell types were sorted 6 weeks after transplantation. STELA analysis was then performed and telomere lengths were compared. Differences in telomere lengths between differentiated and progenitor cell types in CB-4 are shown as kilobases, in red, whereas telomere lengths (excluding outliers) are shown in blue. The number of cell divisions between differentiated and progenitor cells are represented by arrowheads, each representing 2.5 cell divisions (assuming ∼ 100 bp loss per cell division). (B) Graphs showing the difference between telomere lengths from progenitor to daughter cells before and after transplantation in CB-4 and CB-5. Note the large decrease in telomere length from the Lin−CD34+CD38−Rho− to the CD34+CD38− populations in both transplants with respect to pretransplantation telomere loss.
Telomere length changes in donor cells after transplantation of purified human CB cells into immune-deficient mice. (A) Schematic for transplantation, sorting, and STELA analysis is shown on the left panel. Lin−CD34+CD38−Rho− cells were transplanted into mice, and differentiated cell types were sorted 6 weeks after transplantation. STELA analysis was then performed and telomere lengths were compared. Differences in telomere lengths between differentiated and progenitor cell types in CB-4 are shown as kilobases, in red, whereas telomere lengths (excluding outliers) are shown in blue. The number of cell divisions between differentiated and progenitor cells are represented by arrowheads, each representing 2.5 cell divisions (assuming ∼ 100 bp loss per cell division). (B) Graphs showing the difference between telomere lengths from progenitor to daughter cells before and after transplantation in CB-4 and CB-5. Note the large decrease in telomere length from the Lin−CD34+CD38−Rho− to the CD34+CD38− populations in both transplants with respect to pretransplantation telomere loss.
An additional experiment was undertaken in which 3 mice each received a transplant of 3% of a CD34+ cell–enriched light density fraction of cells from a third CB (CB-6). STELA was again conducted on subpopulations isolated 6 weeks after transplantation, and compared with populations isolated from the pretransplantation sample. The changes in telomere length between the regenerated CD34+ cells and the terminally differentiated cells (CD15/66b+ and CD3+) appeared similar in all 3 mice (Figure 6; Table 1). Similar to previous experiments, telomere decreases were most pronounced in the most primitive cells after transplantation, and the telomere loss in fully differentiated cells can be mostly attributed to this initial decline. All 3 mice also showed very similar decreases in telomere length in the regenerated CD34+ populations (2.60 kb, 2.68 kb, and 2.80 kb). However, comparing across the 3 different CBs (CB-4, CB-5, and CB-6) revealed different deceases in telomere length in the regenerated CD34+ populations (3.97 kb, 2.79 kb, and 2.69 kb [mean], respectively; Figure 6). Future experiments will determine whether telomere lengths of regenerated CD34+ cells from mice that received a transplant of the same CB are similar due to the function of that particular CB or to the dose of cells administered to each mouse.
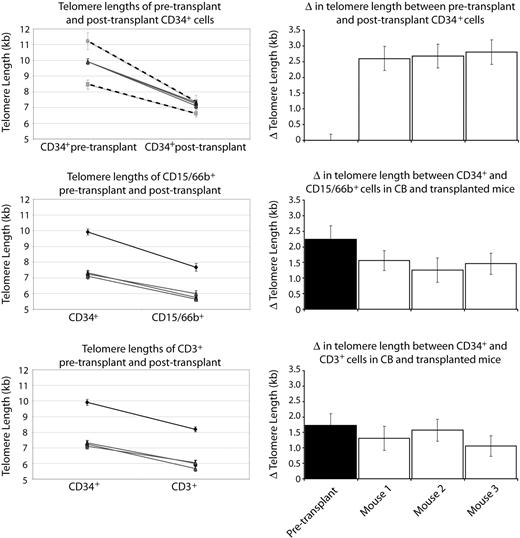
Results from a single CB transplanted into 3 mice. Top graph shows the comparison between 3 mice that each received a transplant of 3% of whole CB-6 (gray lines), and the 2 separate CB transplantation experiments (CB-4 and CB-5; dashed lines). Although the independent transplantation experiments yield differing degrees of telomere loss after transplantation, the mice that received a transplant within the same experiment with the same CB show indistinguishable telomere loss. Subsequent graphs compare the telomere length of primitive CD34+ cells with terminally differentiated CD15/66b+ or CD3+ cells before and after transplantation (black and gray lines, respectively), and show similar changes in telomere length in the 3 mice that received a transplant of the same CB. Right panel shows the degree of telomere loss between pretransplantation and posttransplantation CD34+ cells (top bar chart), and the degree of telomere loss between CD34+ and differentiated cells in untransplanted CB, and the 3 mice that underwent transplantation (subsequent bar charts).
Results from a single CB transplanted into 3 mice. Top graph shows the comparison between 3 mice that each received a transplant of 3% of whole CB-6 (gray lines), and the 2 separate CB transplantation experiments (CB-4 and CB-5; dashed lines). Although the independent transplantation experiments yield differing degrees of telomere loss after transplantation, the mice that received a transplant within the same experiment with the same CB show indistinguishable telomere loss. Subsequent graphs compare the telomere length of primitive CD34+ cells with terminally differentiated CD15/66b+ or CD3+ cells before and after transplantation (black and gray lines, respectively), and show similar changes in telomere length in the 3 mice that received a transplant of the same CB. Right panel shows the degree of telomere loss between pretransplantation and posttransplantation CD34+ cells (top bar chart), and the degree of telomere loss between CD34+ and differentiated cells in untransplanted CB, and the 3 mice that underwent transplantation (subsequent bar charts).
Discussion
Telomere length measurements offer a powerful strategy to obtain information about the replicative history of cells. Here we have exploited this approach to make comparisons of the number of cell divisions that occur during the process of hematopoietic cell differentiation under 3 different conditions: during the expansion of the hematopoietic system at birth, after G-CSF activation, and during the regeneration of the system after transplantation. To achieve the precision required for analysis of the small numbers of primitive cells available, we used the STELA methodology. We show that STELA generates comparative data to Q-FISH, although it is possible that telomeres more than 18 kb are undetected due to PCR limitations. However, most adult MPB have telomeres less than 10 kb in length, even in the CD34+CD38− population,38 whereas CBs have an average telomere length of 12 kb in CD34+CD38− cells,16 with only 1% of CB samples showing lymphocytes and granulocytes with average lengths more than 13 kb.21 The XpYp telomere has been used previously to assess telomere lengths,29,30,33 and is average in length with respect to other telomere ends within a cell.39
As predicted, STELA measurements performed on subsets of cells purified from CB and MPB samples showed that telomere length erodes as cells differentiate in a hierarchic manner. In addition to providing new support for the current model of hematopoietic cell differentiation in healthy humans both in the neonate and in adults, this study suggests a novel approach for the classification of new lineage markers using comparative telomere length measurements to rank cell types within a hierarchy. Moreover, in the absence of active telomerase, the technique presented here can be applied to any suggested cellular hierarchy.
The same data also revealed a significant increase in the number of differentiated cells with telomeres that had atypically shortened in the adult MPB samples, presumably due to DNA damage or aberrant replication rather than typical incomplete end-replication. These abruptly shortened telomeres are important as they could be short enough to be capable of triggering replicative senescence or genomic instability. Data presented here show that in MPB, ultrashort telomeres are seen at a low frequency in primitive cells and become more prevalent in fully differentiated cells. Given the importance of stem cells within the hematopoietic hierarchy, it appears that mechanisms are in place to limit atypical erosion in these cells.
It is possible that because primitive cells have undergone the least number of cell divisions, they may be less susceptible to rapid telomere shortening from unresolved replication errors or stalls because there have been fewer DNA replication events. In more differentiated cells, however, rapid turnover may result not only in more replication-mediated critical telomere shortening, but also an accumulation of such events from earlier divisions. Alternatively, there are several mechanisms that could be influencing the rate of DNA damage in different cell populations. First, because critically short telomeres are preferentially elongated by telomerase,40,41 telomerase in more primitive cells could be better adapted to telomere repair. Second, it is possible that the HSC niche protects them from damaging signals. It has been reported previously that reactive oxygen species (ROSs) can trigger senescence42 and that the resulting DNA lesions are repaired at telomeres less efficiently.43 Although the average telomere length is not significantly affected in normal cells subjected to high oxidative stress,10 it has been speculated that it may lead to a small subset of ultrashort telomeres, which in turn triggers cell senescence.20 In addition to the external environment, it has been shown that mitochondrial superoxide production in fibroblasts also increases with replicative age, a process proposed to be a major determinant of telomere-dependent senescence.44
It is interesting that the CB samples showed no evidence of a corresponding increase in atypically short telomeres in the more differentiated cells. It is possible that the number of telomeres analyzed in CB samples was insufficient to detect significant frequencies of outliers or that the greater variation in telomere length characteristic of the CB populations obscured the detection of outliers. Alternatively, abruptly shortened telomeres may become more prevalent later in life due to a greater exposure to damage from replication and/or the environment in which the cells are produced or ultrashort telomeres more rapidly induce an apoptotic response in CB cells leading to their elimination. It is also possible that telomerase levels in CB cells are higher than in MPB and are therefore better able to repair abruptly truncated telomeres.
It has been previously noted that the telomeres in hematopoietic cells display rapid erosion after transplantation, a process that has been postulated to be a cause of late graft failure in bone marrow recipients via stem cell exhaustion.24 By comparing the rate of telomere loss from CD34+CD38+ cells to terminally differentiated cells, it is possible to predict the number of cell divisions required during differentiation. This allows us to address and important question: is the telomere loss observed after transplantation due to the expansion of progenitor cells before differentiation, or are all cell types in the hematopoietic hierarchy subject to increased turnover? In the case of the former, the number of cell divisions from expanded CD34+CD38+ cells to terminally differentiated cells should be similar to the rate before transplantation. In the case of the latter, the number of divisions is susceptible to considerable alteration at a later stage of differentiation under differing conditions of blood cell demand.
Here we have carried out transplantations in mice to examine directly the extent of telomere loss as transplanted cells differentiate. Our data show that telomere loss occurs in the most primitive detectable cells 6 weeks after transplantation, indicating that the limited telomerase levels in hematopoietic stem cells is unable to offset the number of cell divisions required for these cells to repopulate the bone marrow. Indeed, assuming a 100-bp loss of telomeric DNA per cell division, transplanted cells from CB-4 and CB-5 have undergone 32 and 21 more divisions from Lin−CD34+CD38−Rho− to CD34+CD38+, respectively, than the control CB cells. This assumption likely results in an underestimate of cell turnover as the impact of telomerase activity during repopulation has not been assessed. In contrast, similar numbers of cell divisions were calculated between CD34+CD38− cells and terminally differentiated cells in transplant and CB samples. Thus, telomere length changes after transplantation can be attributed to losses almost exclusively in the primitive compartment, at least in the xenogeneic host model studied here. Using telomere length as a marker for replicative history, these data strongly suggest that primitive cells undergo considerable expansion directly after transplantation as previously shown by vector marking of clones45 and stem cell turnover measurements46 in similar models, and this expanded population then produces differentiated cells.
Interestingly, CD19/20+ cells showed only a modest telomere loss after transplantation with respect to pretransplantation samples, and the average length in these cells was higher than in the CD34+CD38− cells. This indicates that CD19/20+ cells either differentiate early, before the expansion of CD34+CD38− cells, or they can reactivate telomerase. It has been shown previously that a subset of B cells has long telomeres, and that memory B cells have, on average, 2-kb longer telomeres than naive B cells.47 Telomerase has been implicated in the maturation of B cells within germinal centers, but this is the first evidence that telomerase can be reactivated in pre-B cells developing in a xenograft model.
Taken together, our data suggest that the principal mechanism for telomere shortening in the cells produced in recipients of a hematopoietic transplant is the rapid expansion of very primitive cells rather than an expansion of lineage-restricted cells or their more differentiated progeny. Thus, with the notable exception of a subset of CD19/20+ cells, both myeloid and lymphoid lineages show transplantation-related losses of telomere repeats that, if severe, could result in telomere-mediated late graft failure.
In addition to examining telomere dynamics in a defined hierarchy, telomeres themselves are important in the hematopoietic system. It has been shown previously that mutations in either the telomerase RNA component (hTERC) or the reverse-transcriptase component (hTERT) are implicated in several hematologic disorders. In addition to mutations in the dyskerin gene on the X chromosome, mutations in hTERT and hTERC can cause dyskeratosis congenita, an inherited multisystem disorder characterized by early bone marrow failure.48 Several telomerase mutations have also been identified in patients suffering from aplastic anemia49 and familial idiopathic pulmonary fibrosis.50 Telomere length is likely to be an important factor in progression of these disorders, presumably because HSCs with shortened telomeres have a lower replicative capacity, eventually leading to exhaustion of the stem cell pool. However, telomerase levels in HSCs are low, and insufficient to maintain telomere length, so presumably these disorders reflect short telomeres (inherited and from a lack of telomerase during development) rather than telomerase expression in adult HSCs.21 It is possible that mutations in telomerase components may also affect the ability of a cell to repair critically short telomeres.
Finally, telomere attrition could also have serious implications for ex vivo HSC expansion strategies. If telomeres are significantly eroded during the expansion process, recipient donors could also face the possibility of telomere-associated late graft failure. Thus it may be useful to use large numbers of cells for transplantation and consider methods for introducing telomerase during this process to avoid such complications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Flow Cytometry Facility of the Terry Fox Laboratory, the Animal Resource Center of the BC Cancer Agency, and Mike Schertzer for technical assistance. We are most grateful to Geraldine Aubert and Ester Falconer for advice and comments on the paper.
This work was funded by grants to P.M.L. from the National Institutes of Health (AI29524), the Canadian Institute of Health Research (MOP38075), and the National Cancer Institute of Canada (with funds from the Terry Fox Run), and grants to C.J.E. from the Canadian Institute of Health Research and the Stem Cell Network.
National Institutes of Health
Authorship
Contribution: M.H. carried out STELA and FACS sorting, designed analysis software, analyzed the results, and designed all figures; K.L. carried out antibody staining and FACS isolation of all cells analyzed, performed the xenotransplantations, and retrieved subsets of human cells for telomere analysis; E.A.C. carried out Q-FISH experiments; C.J.E. provided CB and MPB samples and oversaw the studies in mice; P.M.L. oversaw the telomere studies; and all authors contributed to the overall research plan and the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter M. Lansdorp, Terry Fox Laboratory, BC Cancer Agency, 675 West 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: plansdor@bccrc.ca.