ADAMTS13 is a highly specific multidomain plasma metalloprotease that regulates the multimeric size and function of von Willebrand factor (VWF) through cleavage at a single site in the VWF A2 domain. The precise role that the ADAMTS13 disintegrin-like domain plays in its function remains uncertain. Truncated ADAMTS13 variants suggested the importance of the disintegrin-like domain for both enzyme activity and specificity. Targeted mutagenesis of nonconserved regions (among ADAMTS family members) in the disintegrin-like domain identified 3 of 8 ADAMTS13 mutants (R349A, L350G, V352G) with reduced proteolytic activity. Kinetic analyses revealed a 5- to 20-fold reduction in catalytic efficiency of VWF115 (VWF residues 1554-1668) proteolysis by these mutants. These residues form a predicted exposed exosite on the surface of the disintegrin-like domain that lies approximately 26 Å from the active site. Kinetic analysis of VWF115 carrying the D1614A mutation suggested that Arg349 in the ADAMTS13 disintegrin-like domain interacts directly with Asp1614 in VWF A2. We hypothesize that this interaction assists in positioning the scissile bond within the active site of ADAMTS13 and therefore plays a major role in determining cleavage parameters (Km and kcat), as opposed to binding affinity (Kd) of ADAMTS13 for VWF, the latter being primarily determined by the spacer domain.
Introduction
Von Willebrand factor (VWF) is a large multidomain plasma glycoprotein. It circulates as covalently associated multimers that range from 2 to 40 VWF units.1 VWF has 2 hemostatic roles: (1) to mediate platelet tethering at sites of vascular injury, and (2) it acts as a carrier protein for factor VIII.1,2 Normally, plasma VWF circulates in a globular form that does not readily interact with circulating platelets. However, after vessel damage, VWF binds subendothelial collagen via its A3 domain.3,4 Once bound, the shear forces exerted by the flowing blood induce VWF unfolding that reveals further binding sites.5 The exposed VWF A1 domain can now bind to the GPIb-IX-V receptor complex on the surface of circulating platelets.6 This results in platelet tethering, and ultimately the formation of a primary platelet plug. Large VWF multimers are more hemostatically active than smaller forms. This is because they contain more platelet and collagen binding sites, and also because they unravel more readily in response to shear. VWF multimers are synthesized intracellularly by dimerization in the endoplasmic reticulum and then by multimerization in the Golgi apparatus.7 After their secretion from endothelial cells, VWF multimers can be converted to smaller less adhesive forms by the plasma metalloprotease, ADAMTS13.8,,,,–13
ADAMTS13 is expressed predominantly in the liver.14 It has also been shown to be expressed in hepatic stellate cells,15 in platelets,16 by cultured endothelial cells,17,18 and by glomerular podocytes.19 It is secreted into the blood as an active enzyme20 and circulates at a plasma concentration of approximately 5 nM.14,21,22 ADAMTS13 cleaves VWF at a single site in its A2 domain between Tyr1605 and Met1606.23,24 Physiologically, VWF is proteolysed only once it has first been unraveled in response to rheologic shear forces as in its globular form the scissile bond in VWF normally lies buried within the A2 domain.25 The lower molecular weight VWF multimers that arise after proteolysis exhibit reduced adhesive potential.8 In this way, ADAMTS13 modulates VWF platelet-tethering function.
ADAMTS13 is a large multidomain plasma protein. From its N-terminus, it consists of a metalloprotease domain, disintegrin-like domain, thrombospondin type 1 repeat (TSP1), cysteine-rich domain and spacer domain—a domain organization that is common to all ADAMTS family members. Thereafter, there are 7 additional TSP1 repeats, and uniquely among ADAMTS family members, these are followed by 2 C-terminal CUB domains.9,26
ADAMTS13 is a highly specific enzyme. It cleaves only one known substrate (VWF), and does so at just a single site (Tyr1605-Met1606). It seems that this specificity is likely conferred through extensive interactions between ADAMTS13 and VWF.27,28 The metalloprotease domain contains a Zn2+ ion in the active site that is essential for hydrolysis of the target peptide bond.9 Three histidine residues coordinate this catalytic Zn2+ ion, and the sequence (HEXXHXXGXXHD) is highly conserved among all family members.9 The metalloprotease domain also contains a high-affinity Ca2+ binding site adjacent to the active-site cleft, which is critical for ADAMTS13 activity.29 Although the metalloprotease domain is essential for cleavage of VWF, the interaction between this domain and VWF appears to be very weak.30
Several reports have demonstrated the importance of the ADAMTS13 spacer domain in the binding and proteolysis of VWF.10,30,,,–34 Interactions between the spacer and the C-terminal part of the VWF A2 domain appear to mediate much of the binding affinity (Kd ∼ 10-20 nM) between these 2 molecules.33,34 More recently, Gao et al provided evidence for the involvement of all of the N-terminal ADAMTS13 domains (metalloprotease domain through spacer) in substrate proteolysis.27 However, the precise role that each of these domains plays has not been determined. The crystal structures of the metalloprotease domain and disintegrin-like domain of ADAMTS1, 4, and 5 have recently been determined,35,–37 which has enabled homology modeling of the ADAMTS13 structure. This has suggested a role for disintegrin-like domains in substrate recognition and proteolysis. In light of this, we aimed to characterize the functional role of this domain in ADAMTS13.
Methods
Expression and purification of recombinant wild-type ADAMTS13 and truncated ADAMTS13
Recombinant human wild-type ADAMTS13 with a C-terminal Myc/His tag was expressed in HEK293 cells and purified as previously described.38,–40 Expression vectors containing the metalloprotease (MP; residues Met1-Ser292) or the metalloprotease domain and disintegrin-like domain (MP-Dis; residues Met1-Gly385) with a C-terminal His tag were generated using standard inverse polymerase chain reaction (PCR) techniques. Vectors were verified by sequencing. ADAMTS13 truncations were expressed by large-scale (2L) transient transfection of HEK293T cells using linear polyethylenimine (Polysciences, Warrington, PA). Expression and secretion of MP and MP-Dis were confirmed by Western blotting using an anti-His mAb (Invitrogen, Frederick, MD). Conditioned media were collected 4 days after transfection, cleared, and then concentrated 50-fold by tangential flow filtration. ADAMTS13 MP and MP-Dis were partially purified using 5 mL Ni2+ chelating HiTrap columns coupled to an AKTA FPLC (GE Healthcare, Little Chalfont, United Kingdom). Thereafter, they were purified to homogeneity by gel filtration using a 120-mL HiLoad 16/60 Superdex 75-pg column (GE Healthcare). MP and MP-Dis were dialyzed into 20 mM Tris (pH 7.8), 150 mM NaCl. Recombinant protein purity was confirmed by sodium dodecylsulfide–polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining (GE Healthcare). The concentration of fully purified proteins was determined using a Qubit total protein assay (Invitrogen).
Expression of recombinant ADAMTS13 disintegrin-like domain variants
Generation of disintegrin-like domain mutations (D330A, Q333A, D340A, D343A, R349A, L350G, L351G, V352G) in the full-length ADAMTS13 expression vector was performed using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA), according to the manufacturer's instructions. All vectors were verified by sequencing. For functional analysis, wild-type ADAMTS13 and its mutants were expressed transiently in HEK293T cells using linear polyethylenimine (Polysciences), as previously described.29,41 Expression and secretion of ADAMTS13 were confirmed by Western blotting. After 3 to 4 days, conditioned medium was harvested, cleared, and concentrated using 50-kDa MWCO spin columns (Millipore, Billerica, MA). Thereafter, samples were dialyzed in 20 mM Tris (pH 7.8), 150 mM NaCl. ADAMTS13 concentration in these samples was determined using a specific in-house ADAMTS13 enzyme-linked immunosorbent assay (ELISA) using antibodies purified from rabbits immunized with full-length recombinant ADAMTS13. A polyclonal anti-ADAMTS13 antibody (affinity depleted of anti-ADAMTS13 TSP1(2-4) antibodies) was used as the capture antibody, and an affinity purified biotinylated rabbit polyclonal anti-ADAMTS13 TSP1(2-4) antibody was used for detection, as previously described.21 To further verify all ADAMTS13 concentrations, we confirmed our results by equal loading of each WT and ADAMTS13 variant based on our ELISA results and subsequent Western blotting using an anti-Myc monoclonal antibody (Invitrogen). All samples gave equal intensity bands consistent with the equal amounted load predicted by the ELISA.
Molecular modeling of ADAMTS13 MP-Dis
Expression and purification of VWF115 and VWF115(D1614A)
For use as a specific ADAMTS13 substrate, the VWF A2 domain fragment, VWF115 (spanning VWF residues 1554-1668), and also VWF115(D1614A) were expressed in bacteria, purified, and quantified as previously described.39
ADAMTS13 activity assays
For qualitative analysis of the activity of full-length and truncated ADAMTS13, 5 nM or 50 nM purified recombinant ADAMTS13, MP-Dis, or MP in 20 mM Tris-HCl (pH 7.8), 150 mM NaCl, and 5 mM CaCl2 were preincubated at 37°C for 1 hour without substrate. VWF115 (6 μM) was added to start the reaction. At different time points (0-17 hours), 60-μL subsamples were removed and reactions stopped with 5 μL 0.5 M EDTA. Thereafter, proteolysis of VWF115 was visualized by SDS-PAGE and Coomassie staining. For preliminary comparison of the activity of full-length wild-type ADAMTS13 with the individual disintegrin-like domain mutants, similar experiments were set, except 0.5 nM ADAMTS13 was used, and reactions were followed from 0 to 2 hours.
For quantitative analysis of VWF115 or VWF115(D1614A) proteolysis, reactions were set up using 10 nM ADAMTS13 and 1 μM VWF115. Higher enzyme concentrations were used in these reactions than for the SDS-PAGE assay to enable faster proteolysis of the mutants and shorter time to reaction completion. To quantify proteolysis, samples were analyzed by high-performance liquid chromatography (HPLC), as previously described.39,–41 From these results, the catalytic efficiencies (kcat/Km) were derived, as previously described.39,40
Individual determination of the kinetic constants, kcat and Km, was performed essentially as previously described.39 Briefly, activity assays were set up using a range of substrate concentrations (0-20 μM). Thereafter, early time points (0-30 minutes) were taken so that the proteolysis reactions were in the linear part of the time-course curve (ie, less than 15% substrate proteolysis). Wild-type ADAMTS13 (1 nM) or ADAMTS13(R349A) (8 nM) was used to enable between 5% and 15% substrate proteolysis within 10 to 30 minutes. The rate of proteolysis (nMs−1) per nanomolar enzyme was determined and plotted as a function of substrate concentration. From these Michaelis-Menten plots, individual Km and kcat values were determined.39
For the analysis of the proteolysis of purified multimeric plasma-derived VWF, VWF was incubated with 1.5 M guanidine-HCl at 37°C for 30 minutes. The denatured VWF was then diluted 10-fold to a final concentration of 40 nM into 150 mM NaCl, 20 mM Tris-HCl, 0.5% BSA, 5 mM CaCl2 buffer, in which 1 to 5 nM ADAMTS13 had been preincubated for 60 minutes. The reaction was incubated at 37°C for 1 to 2 hours before quenching with loading buffer (10 mM Tris, 2% SDS, 8 M Urea, 1 mM EDTA, pH 8.0) and analyzed by 1% agarose gel electrophoresis and Western blotting for VWF, as previously described.44,45
Results
Importance of the ADAMTS13 disintegrin-like domain in substrate proteolysis
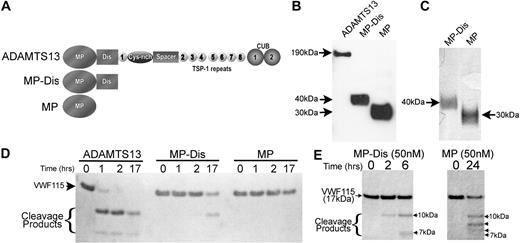
To first assess the importance of the disintegrin-like domain in VWF proteolysis, we expressed full-length ADAMTS13 and the truncated variants, MP-Dis and MP (Figure 1A). Analysis of conditioned medium from transfected cells revealed that full-length ADAMTS13 migrated as a band of approximately 190 kDa, as expected. Both MP-Dis and MP were efficiently secreted and migrated as single bands of 40 kDa and 30 kDa, respectively (Figure 1B). Ni2+-chelating chromatography and gel filtration were used to purify MP-Dis and MP to homogeneity, as ascertained by silver staining (Figure 1C). This allowed accurate determination of the concentration of these truncations using a total protein assay. The activities of purified ADAMTS13, MP-Dis, and MP were assessed using VWF115 as a substrate. Purified full-length ADAMTS13 (5 nM) proteolysed VWF115, generating 2 cleavage products of 10 kDa and 7 kDa (Figure 1D). MP-Dis proteolysed VWF115 slowly, but specific proteolysis was seen to occur after long incubations (17 hours). However, no specific proteolytic activity could be detected using MP under these conditions. As predicted, the rate of VWF115 cleavage was appreciably enhanced when 50 nM MP-Dis was used (Figure 1E). This also enabled visualization of both the 10-kDa and 7-kDa cleavage products. At this concentration, MP exhibited some proteolytic activity (although markedly less than MP-Dis), but the cleavage products generated suggested that the enzyme had lost specificity, consistent with a previous report.30 This was further confirmed by HPLC analysis of the VWF115 cleavage products by HPLC, which revealed additional peaks resulting from proteolysis by MP (data not shown). Together, these results demonstrated that the disintegrin-like domain plays a functional role in the proteolysis of the VWF A2 domain, not only in enhancing the rate of proteolysis, but probably also by imparting specificity for the target scissile bond.
Importance of the ADAMTS13 disintegrin-like domain in VWF115 proteolysis. (A) Schematic representation of the domain organization of ADAMTS13, and the truncated variants MP-Dis and MP. (B) ADAMTS13, MP-Dis, and MP detection by Western blotting in conditioned media after transfection using an anti-His mAb. (C) SDS-PAGE and silver staining of MP-Dis and MP after purification. (D) ADAMTS13, MP-Dis, or MP (5 nM each) were incubated with 6 μM VWF115 at 37°C. Reactions were stopped with EDTA and sample was analyzed by SDS-PAGE. (E) Reactions as in panel D except 50 nM MP-Dis or MP was used.
Importance of the ADAMTS13 disintegrin-like domain in VWF115 proteolysis. (A) Schematic representation of the domain organization of ADAMTS13, and the truncated variants MP-Dis and MP. (B) ADAMTS13, MP-Dis, and MP detection by Western blotting in conditioned media after transfection using an anti-His mAb. (C) SDS-PAGE and silver staining of MP-Dis and MP after purification. (D) ADAMTS13, MP-Dis, or MP (5 nM each) were incubated with 6 μM VWF115 at 37°C. Reactions were stopped with EDTA and sample was analyzed by SDS-PAGE. (E) Reactions as in panel D except 50 nM MP-Dis or MP was used.
Design and expression of ADAMTS13 disintegrin-like domain mutants
To further analyze the functional role of the ADAMTS13 disintegrin-like domain, we modeled MP-Dis based on the recent crystal structures of ADAMTS1, 4, and 5.35,–37 This model identified a linker region (amino acids shown in Figure 2A,B) that extends from the back of the metalloprotease domain and that positions the disintegrin-like domain at one end of the active-site cleft. Given its proximity to the active site, this suggested that the disintegrin-like domain might be ideally positioned to influence directly cleavage of the substrate.
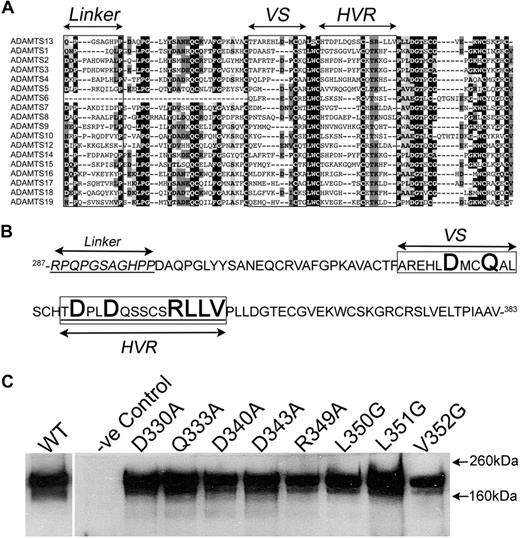
Mutagenesis and expression of ADAMTS13 disintegrin-like domain mutants. (A) Amino acid sequence alignment of the disintegrin-like domain of all ADAMTS family members. (B) Amino acid sequence of the ADAMTS13 disintegrin-like domain. In panels A and B, the linker region that wraps behind the metalloprotease domain is shown. The variable segment (VS) and the hypervariable region (HVR) are boxed. In panel B, the amino acids in bold are those that were mutated and analyzed in this study. (C) Wild-type and mutant ADAMTS13 were expressed in HEK293T cells. After 3 days, expression and secretion were analyzed by Western blot analysis of conditioned media using an anti-ADAMTS13 pAb.
Mutagenesis and expression of ADAMTS13 disintegrin-like domain mutants. (A) Amino acid sequence alignment of the disintegrin-like domain of all ADAMTS family members. (B) Amino acid sequence of the ADAMTS13 disintegrin-like domain. In panels A and B, the linker region that wraps behind the metalloprotease domain is shown. The variable segment (VS) and the hypervariable region (HVR) are boxed. In panel B, the amino acids in bold are those that were mutated and analyzed in this study. (C) Wild-type and mutant ADAMTS13 were expressed in HEK293T cells. After 3 days, expression and secretion were analyzed by Western blot analysis of conditioned media using an anti-ADAMTS13 pAb.
From the sequence alignments of the disintegrin-like domain with homologous domains of other proteins (Figure 2A), 2 surface-exposed loops have been identified in this domain that are poorly conserved, termed the variable segment (VS) and the hypervariable region (HVR).46,–48 We hypothesized that these loops may therefore represent regions that confer specific functions or mediate specific interactions that are particular to each protein. From the ADAMTS1, 4, and 5 crystal structures, and also in the ADAMTS13 model,46 many of the residues in the VS and HVR are located in close proximity to and are on the same face as the active-site cleft. These were therefore targeted for mutagenesis (Figure 2B). In the first instance, those residues within the nonconserved VS or HVR were identified as potentially important amino acids. Thereafter, we selected those amino acids within VS and HVR that were predicted to be both solvent exposed and on the same face as the active-site cleft. Some of the other residues present in the nonconserved regions were modeled on the other sides of the disintegrin-like domain, so were not targeted. We did not target residues that were likely to be important for the folding/structure of the disintegrin-like domain (ie, Cys and Pro). A total of 8 single point mutations were generated (Figure 2B), D330A, Q333A, D340A, D343A, R349A, L350G, L351G, and V352G, involving residues with the potential involvement in either electrostatic or hydrophobic interactions.
All ADAMTS13 mutants were transiently expressed in HEK293T cells. Western blot analysis of conditioned media demonstrated that all mutants were expressed and secreted very similarly to wild-type ADAMTS13 (Figure 2C), suggesting that the mutations did not by themselves induce any gross structural changes that influenced enzyme secretion.
Functional analysis of ADAMTS13 disintegrin-like domain mutants
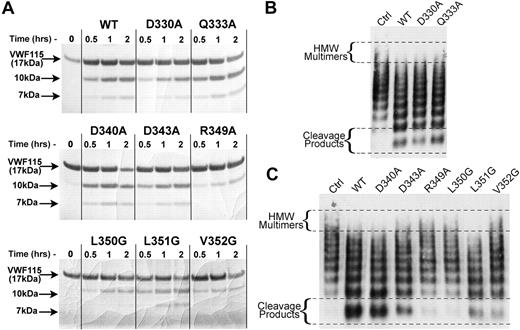
In the first instance, all ADAMTS13 disintegrin-like domain mutants were screened to qualitatively assess their activity, and so to identify those with altered proteolytic function. Reactions were set up using 0.5 nM ADAMTS13, and VWF115 as a specific substrate. From 0 to 2 hours, reactions were stopped with EDTA and analyzed by SDS-PAGE to visualize the 10-kDa and 7-kDa VWF115 cleavage products (Figure 3A). By comparison with wild-type ADAMTS13, all mutants exhibited normal or near normal activity with the exception of R349A, L350G, and V352G (Figure 3A). Each of these mutants proteolysed VWF115 appreciably slower than wild-type ADAMTS13, as evidenced by the reduced appearance of the 10-kDa and 7-kDa bands.
Identification of ADAMTS13 disintegrin-like domain residues important for proteolysis of VWF. (A) Activity assays containing 0.5 nM wild-type or mutant ADAMTS13 and 6 μM VWF115 were incubated at 37°C. At the times indicated, reactions were stopped with EDTA and analyzed by SDS-PAGE. Proteolysis was assessed by the generation of the 10-kDa and 7-kDa VWF115 cleavage products. (B) Multimeric VWF activity assays were set up using 1 nM wild-type ADAMTS13 or VS1 mutants (D330A and Q333A) under denaturing conditions. Reactions were stopped after 2 hours with EDTA followed by VWF multimer analysis. (C) Multimeric VWF activity assays were set up using 3.5 nM wild-type ADAMTS13 or HVR mutants under denaturing conditions. Reactions were stopped after 1 hour with EDTA followed by VWF multimer analysis.
Identification of ADAMTS13 disintegrin-like domain residues important for proteolysis of VWF. (A) Activity assays containing 0.5 nM wild-type or mutant ADAMTS13 and 6 μM VWF115 were incubated at 37°C. At the times indicated, reactions were stopped with EDTA and analyzed by SDS-PAGE. Proteolysis was assessed by the generation of the 10-kDa and 7-kDa VWF115 cleavage products. (B) Multimeric VWF activity assays were set up using 1 nM wild-type ADAMTS13 or VS1 mutants (D330A and Q333A) under denaturing conditions. Reactions were stopped after 2 hours with EDTA followed by VWF multimer analysis. (C) Multimeric VWF activity assays were set up using 3.5 nM wild-type ADAMTS13 or HVR mutants under denaturing conditions. Reactions were stopped after 1 hour with EDTA followed by VWF multimer analysis.
To substantiate these results, we tested all ADAMTS13 variants using multimeric full-length VWF as a substrate under denaturing conditions (Figure 3B VS mutants; Figure 3C HVR mutants).49 In Figure 3B, both wild-type ADAMTS13 and the 2 VS mutants (D330A and Q333A) caused comparable disappearance of the higher molecular weight multimers to and appearance of lower molecular weight cleavage products. This observation was consistent with the similar activities observed when using the VWF115 screening assay (Figure 3A).
Comparison of the HVR mutants with wild-type ADAMTS13 is shown in Figure 3C. These results confirmed our findings from the VWF115 assay. By comparison with wild-type ADAMTS13, both R349A and L350G mutants exhibited dramatically reduced proteolytic function (Figure 3C). The V352G mutant also exhibited reduced activity, although not as great as seen for R349A and L350G. In this full-length VWF assay, there was a suggestion that the D343A and L351G mutants also had slightly reduced activity.
Identification of the site of interaction of the ADAMTS13 disintegrin-like domain on VWF
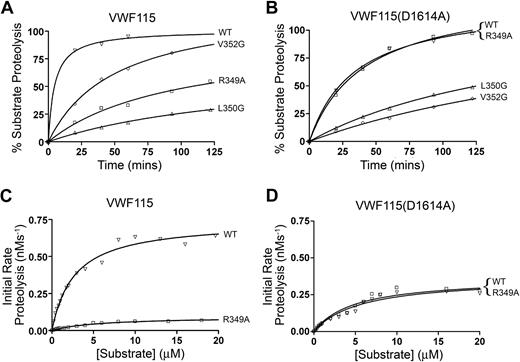
To more accurately quantify the influence upon substrate proteolysis of those disintegrin-like domain mutations that exhibited appreciably reduced proteolytic function, further time-course experiments were set up using VWF115, and cleavage was quantified by HPLC (Figure 4A). These results corroborated those seen by SDS-PAGE (Figure 3A). Comparison of these curves revealed that V352G had 4- to 5-fold reduced catalytic efficiency, whereas R349A and L350G exhibited 10- to 20-fold reduced enzymatic function (n = 2).
Kinetic analysis of ADAMTS13 disintegrin-like domain mutants. Activity assays were set up using 10 nM ADAMTS13 (or ADAMTS13 mutants, as shown) and either 1 μM VWF115 (A) or VWF115(D1614A) (B). Samples were stopped with EDTA from 0 to 2 hours and percentage of VWF115 proteolysis was determined by HPLC. The reduced proteolytic function of ADAMTS13(R349A) seen in panel A was completely lost when VWF115(D1614A) was used as a substrate (B), suggesting that the influence of R349A is dependent on D1614 in VWF115. ADAMTS13 and ADAMTS13(R349A) were further analyzed by measuring the initial rate of VWF115 (C) or VWF115(D1614A) (D) as a function of substrate concentration. Initial rates of proteolysis (< 15% cleavage) were analyzed after 20 minutes by HPLC. Using VWF115 (C), a large difference between the Vmax for ADAMTS13 and ADAMTS13(R349A) was evident, confirming the time-course results in panel A. When VWF115(D1614A) was used, this difference was completely lost (D), confirming the results in panel B. Graphs shown are single reactions, but representative of 3 separate experiments. The kinetic values derived from panels C and D are shown in Table 1 (n = 3).
Kinetic analysis of ADAMTS13 disintegrin-like domain mutants. Activity assays were set up using 10 nM ADAMTS13 (or ADAMTS13 mutants, as shown) and either 1 μM VWF115 (A) or VWF115(D1614A) (B). Samples were stopped with EDTA from 0 to 2 hours and percentage of VWF115 proteolysis was determined by HPLC. The reduced proteolytic function of ADAMTS13(R349A) seen in panel A was completely lost when VWF115(D1614A) was used as a substrate (B), suggesting that the influence of R349A is dependent on D1614 in VWF115. ADAMTS13 and ADAMTS13(R349A) were further analyzed by measuring the initial rate of VWF115 (C) or VWF115(D1614A) (D) as a function of substrate concentration. Initial rates of proteolysis (< 15% cleavage) were analyzed after 20 minutes by HPLC. Using VWF115 (C), a large difference between the Vmax for ADAMTS13 and ADAMTS13(R349A) was evident, confirming the time-course results in panel A. When VWF115(D1614A) was used, this difference was completely lost (D), confirming the results in panel B. Graphs shown are single reactions, but representative of 3 separate experiments. The kinetic values derived from panels C and D are shown in Table 1 (n = 3).
These 3 residues (Arg349, Leu350, and Val352) are predicted to form a cluster on the surface of the ADAMTS13 disintegrin-like domain immediately adjacent to the active-site cleft. Based on this observation, we estimated the distance from the catalytic Zn2+ ion to Arg349 on our model of ADAMTS13 MP-Dis to be approximately 26 Å. As the VWF A2 domain must unravel, into what we might assume to be a linear polypeptide chain, before proteolysis by ADAMTS13 can occur, we measured 26 Å from the Tyr1605-Met1606 scissile bond to estimate which part of VWF this disintegrin-like domain cluster might interact with. Interestingly, Asp1614 is located approximately 26 Å from the ADAMTS13 cleavage site in unfolded VWF. We have previously demonstrated that substitution of this amino acid in VWF115 significantly reduces its rate of proteolysis by ADAMTS13.39 For these reasons, we hypothesized that the positively charged Arg349 on ADAMTS13 might directly interact with the negatively charged Asp1614 on VWF. To test this hypothesis, we examined the proteolysis of VWF115 that carried a D1614A substitution by the 3 affected disintegrin-like domain mutants. Wild-type ADAMTS13 proteolysed VWF115(D1614A) slower than VWF115 (compare Figure 4A,B), similar to our previous report.39 However, whereas with VWF115, the R349A, L350G, and V352G disintegrin-like domain mutants all exhibited reduced proteolytic function (Figure 4A), when VWF115(D1614A) was used we no longer detected any functional difference between wild-type ADAMTS13 and ADAMTS13(R349A) (Figure 4B). As with VWF115, the L350G and V352G mutants still exhibited reduced activity compared with wild-type ADAMTS13 (Figure 4B). As the functional deficit of ADAMTS13(R349A) cannot be detected when VWF(D1614A) is used as a substrate, these results strongly suggest that the influence of Arg349 in the proteolytic function of ADAMTS13 must be mediated in a manner that is dependent on Asp1614 in VWF.
To confirm and extend these findings, we performed kinetic analysis, determining the initial rates of substrate proteolysis as a function of substrate concentration to derive the individual kinetic constants, kcat and Km, for wild-type ADAMTS13 and ADAMTS13(R349A) using both VWF115 and VWF115(D1614A) as substrates (Figure 4C,D, respectively). For proteolysis of VWF115, we derived a kcat of 0.59 (± 0.13) s−1 and Km of 1.88 (± 0.73) μM for ADAMTS13 (Table 1). For ADAMTS13(R349A), we obtained a reduced kcat of 0.09 (± 0.01) s−1 and an increased Km of 6.20 (± 0.56) μM. These results determine a catalytic efficiency (kcat/Km) of ADAMTS13(R349A) that is approximately 20-fold less than for wild-type ADAMTS13, consistent with our time-course experiments. Conversely, when we used VWF115(D1614A), both the kcat and Km derived for wild-type ADAMTS13 and ADAMTS13(R349A) were essentially indistinguishable. These data corroborated the results of the time-course experiments, and supported the hypothesis that Arg349 in ADAMTS13 interacts directly with Asp1614 in the VWF A2 domain.
Kinetic values (kcat and Km) and the catalytic efficiency (kcat/Km) derived for proteolysis of VWF115 and VWF115(D1614A) by both ADAMTS13 and ADAMTS13(R349A)
| . | VWF 115 . | VWF115(D1614A) . | ||||
|---|---|---|---|---|---|---|
| Km, μM . | kcat, s−1 . | kcat/Km, ×105 M−1s−1 . | Km, μM . | kcat, s−1 . | kcat/Km, ×105 M−1s−1 . | |
| ADAMTS13 | 1.88 ± 0.73 | 0.59 ± 0.13 | 3.24 ± 0.57 | 7.99 ± 2.38 | 0.37 ± 0.04 | 0.50 ± 0.17 |
| ADAMTS13(R349A) | 6.20 ± 0.56 | 0.09 ± 0.01 | 0.15 ± 0.01 | 8.75 ± 2.34 | 0.47 ± 0.04 | 0.57 ± 0.13 |
| . | VWF 115 . | VWF115(D1614A) . | ||||
|---|---|---|---|---|---|---|
| Km, μM . | kcat, s−1 . | kcat/Km, ×105 M−1s−1 . | Km, μM . | kcat, s−1 . | kcat/Km, ×105 M−1s−1 . | |
| ADAMTS13 | 1.88 ± 0.73 | 0.59 ± 0.13 | 3.24 ± 0.57 | 7.99 ± 2.38 | 0.37 ± 0.04 | 0.50 ± 0.17 |
| ADAMTS13(R349A) | 6.20 ± 0.56 | 0.09 ± 0.01 | 0.15 ± 0.01 | 8.75 ± 2.34 | 0.47 ± 0.04 | 0.57 ± 0.13 |
All data are mean of 3 separate experiments ± SD.
Discussion
In this study, we aimed to examine the role of the disintegrin-like domain in ADAMTS13 function. Using purified mammalian expressed MP-Dis and MP fragments, we showed that the disintegrin-like domain not only enhances ADAMTS13 activity, but also probably affects enzyme specificity (Figure 1), in accordance with a previous report.30 Although analysis of MP-mediated VWF115 proteolysis by both SDS-PAGE and HPLC revealed the expected sized cleavage products, it was clear that additional proteolytic fragments were also present. Because of the slow rate of substrate proteolysis, it was not possible to isolate enough cleavage product to characterize any alternative cleavage site(s).
MP-Dis cleaved VWF115 appreciably faster than MP, highlighting the functional role of the disintegrin-like domain. Other studies have also suggested the functional importance of the disintegrin-like domain. For example, a monoclonal antibody against the ADAMTS13 disintegrin-like domain greatly inhibits ADAMTS13 function in vitro under static50 and flow51 conditions and augments thrombus growth both in vitro and in vivo in animal models.51 Furthermore, Ai et al found that MP-Dis was more active than MTCS (ie, an ADAMTS13 truncated after the spacer domain but that lacked the disintegrin-like domain), and that MTCS was appreciably less active than MDTCS.30 In each case, the reduced activity can be attributed to the absence of the disintegrin-like domain.
For ADAMTS13 to bind and cleave VWF efficiently and specifically, it seems that multiple interactions are required. It is clear that the active-site cleft in the metalloprotease domain must interact with VWF in proximity to the Tyr1605-Met1606 scissile bond in the A2 domain. However, this interaction appears to be very weak.30,52 Binding can be enhanced with the addition of the disintegrin-like domain.30
It has been known for some time that the spacer domain plays a critical role in binding VWF and therefore also in promoting proteolysis. It was elegantly demonstrated that this domain interacts specifically with the C-terminal part of the VWF A2 domain,33,34 and this appears to mediate much of the binding affinity between ADAMTS13 and VWF. However, binding and proteolysis can be separate events, and additional interactions may be required once binding has occurred. This is highlighted by the finding that whereas the binding affinity (Kd) of ADAMTS13 for VWF115 is approximately 20 nM,39 the Km for VWF115 proteolysis is approximately 1.9 μM (ie, ∼ 100-fold higher; Table 1). This suggests that whereas tight binding between VWF and ADAMTS13 is mediated by interactions largely between the spacer domain and the C-terminal aspect of the VWF A2 domain, additional lower affinity interactions are important for optimal proteolysis, and are important functional determinants of the Km. In support of this, Gao et al recently suggested that VWF A2 domain residues 1641 to 1659 may contain a site of interaction with the ADAMTS13 cysteine-rich domain, and that VWF residues 1597 to 1623 may contain the MP-Dis sites of interaction.27 Deletion of the residues 1609 to 1623 in this region completely abolished substrate proteolysis.
The recent crystal structures of the MP-Dis from ADAMTS1, 4, and 5 have suggested that the ADAMTS family disintegrin-like and cysteine-rich domains share structural homology.35,–37 These structural data have allowed us to model the ADAMTS13 MP-Dis (Figure 5). From the structures of other disintegrin-like and cysteine-rich domains, as well as the amino acid alignments of these regions, it has been noted that there are 2 regions that exhibit low sequence conservation.46 These regions have been termed VS and HVR and are predicted to be largely surfaced exposed. We hypothesized that the amino acids within VS and HVR might contribute to enzyme-specific function. Targeted mutagenesis of residues within these regions enabled functional characterization of the disintegrin-like domain. Of the 8 disintegrin-like domain mutants that we generated, we identified 3 (R349A, L350G, and V352G) that exhibited appreciably reduced proteolytic function using VWF115 as a substrate. Importantly, this finding was confirmed using multimeric VWF as a substrate demonstrating that the effects of these substitutions are pertinent to the physiologic substrate of ADAMTS13.
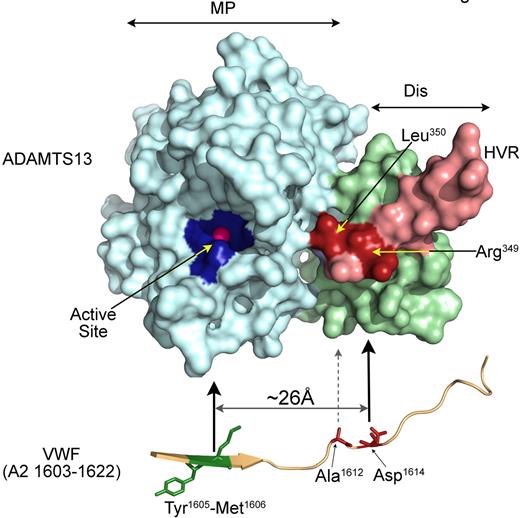
Model depicting the proposed role of the ADAMTS13 disintegrin-like domain. Homology model of the ADAMTS13 MP-Dis. The metalloprotease domain is shown in light blue showing the 3 active site His and catalytic residues Glu (dark blue) that coordinate a catalytic Zn2+ ion (pink). The disintegrin-like domain is depicted in light green, light pink, and red. The hypervariable region (HVR) is highlighted in light pink with Arg349 and Leu350 highlighted in red. These amino acids lie adjacent to the active-site cleft. Arg349 is located approximately 26 Å from the active site Zn2+. The catalytic efficiency of ADAMTS13 is reduced 10- to 20-fold when either of these residues is mutated. We propose a model in which Asp1614 in the VWF A2 domain (located ∼ 26 Å from the Tyr1605-Met1606 scissile bond) interacts with Arg349 in ADAMTS13. This in turn helps position the scissile bond into the active-site cleft.
Model depicting the proposed role of the ADAMTS13 disintegrin-like domain. Homology model of the ADAMTS13 MP-Dis. The metalloprotease domain is shown in light blue showing the 3 active site His and catalytic residues Glu (dark blue) that coordinate a catalytic Zn2+ ion (pink). The disintegrin-like domain is depicted in light green, light pink, and red. The hypervariable region (HVR) is highlighted in light pink with Arg349 and Leu350 highlighted in red. These amino acids lie adjacent to the active-site cleft. Arg349 is located approximately 26 Å from the active site Zn2+. The catalytic efficiency of ADAMTS13 is reduced 10- to 20-fold when either of these residues is mutated. We propose a model in which Asp1614 in the VWF A2 domain (located ∼ 26 Å from the Tyr1605-Met1606 scissile bond) interacts with Arg349 in ADAMTS13. This in turn helps position the scissile bond into the active-site cleft.
This cluster of amino acids is predicted to be located adjacent to the end of the metalloprotease domain active-site cleft (Figure 5), which makes these residues ideally positioned to interact directly with the VWF A2 domain. The positively charged Arg349 lies a predicted distance of approximately 26 Å from the active site Zn2+ (Figure 5). Given that the Tyr1605-Met1606 bond must be positioned over this ion for cleavage to occur, we hypothesized that the site of interaction of Arg349 must be approximately the same distance from this bond. Intriguingly, the negatively charged Asp1614 in VWF is in this position if the substrate is fully unraveled. We previously reported that substitution of this amino acid in VWF115 results in a markedly reduced rate of substrate proteolysis.39 We therefore rationalized that important ionic interactions between Arg349 and Asp1614 would explain these results. Of course, alternative explanations for reduced activity of ADAMTS13(R349A) are possible, but these would involve indirect changes induced by the substitution.
Our hypothesis was strengthened by our kinetic analyses that demonstrated that the functional difference between wild-type ADAMTS13 and the R349A mutant is completely lost when the D1614A substitution is introduced into VWF115 (Figure 4). Conversely, the other residues in the disintegrin-like domain cluster still exhibited impaired activity when VWF115(D1614A) was used, suggesting that the effect of these substitutions are independent of this residue. This would fit with the hypothesis that Leu350 and Val352 are more likely involved in hydrophobic (rather than ionic) interactions. Inspection of the amino acids flanking Asp1614 reveals Ala1612 to be ideally positioned to form such a hydrophobic interaction with Leu350 (Figure 5).
Examination of the kinetic data reveals that the ADAMTS13(R349A) has a 20-fold reduced kcat/Km compared with wild-type ADAMTS13 using VWF115 (Figure 4C; Table 1). When VWF115(D1614A) is used a substrate (Figure 4D; Table 1), both the kcat and Km values for ADAMTS13(R349A) and wild-type ADAMTS13 were essentially the same. Interestingly, there was an increase in the kcat for R349A using VWF115(D1614A), which was the major determinant of a similar (3.8-fold) increase in kcat/Km of ADAMTS13(R349A) as derived from the time-course analysis (Figure 4B). These results suggest that when the negatively charged Asp1614 of VWF is confronted with an alanine in the complementary binding site of ADAMTS13 (as in R349A), the binding is disrupted (as reflected in the increase of the Km). In addition, it may be that some form of incompatibility between the enzyme and the substrate arises that prevents a proper positioning of the scissile bond toward the active site of the ADAMTS13 metalloprotease domain, and therefore an optimal turnover of the substrate. As judged from the kcat values, this incompatibility between enzyme and substrate is lessened when only VWF115 is mutated, or both ADAMTS13(R349A) and VWF(D1614A) are used. Although ADAMTS13(R349A) had an approximately 3.5-fold increase in Km, we do not think that this would measurably alter the overall binding with VWF, as this is mediated primarily via the spacer domain interactions.
Based on our results, we now suggest the following model for the binding and cleavage of VWF by ADAMTS13. We propose that the tight binding between ADAMTS13 and VWF is provided by cysteine-rich and spacer domain interactions and residues 1641 to 1658 and 1659 to 1668 of VWF A2 domain, respectively. This approximates the 2 molecules, but is not sufficient for proteolysis to occur. Thereafter, weaker binding between the disintegrin-like domain involving Arg349, Leu350 in ADAMTS13 with Asp1614, and Ala1612 in VWF assist in positioning the Tyr1605-Met1606 bond into the active-site cleft, and this interaction influences the Km and kcat for substrate proteolysis. This disintegrin-like domain interaction is therefore critical to efficient substrate cleavage, as when this is disrupted proteolytic function is compromised despite the normal interactions of the cysteine-rich/spacer domains, and the fully functional metalloprotease domain.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr T. McKinnon for providing plasma purified VWF.
This work was supported by British Heart Foundation (London, United Kingdom) grants RG/06/007 and FS/06/002 (J.T.B.C. and D.A.L.) and by an unrestricted grant from Amgen funding a studentship. We are grateful for support from the National Institute for Health Research Biomedical Research Centre (London, United Kingdom) funding scheme.
Authorship
Contribution: R.d.G. designed the research, performed experiments, analyzed results, and wrote the paper; A.B. and N.R. performed experiments; D.A.L designed the research, analyzed results, and wrote the paper; and J.T.B.C. designed the research, performed experiments, analyzed results, wrote the paper, and made the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James T. B. Crawley, Department of Haematology, Imperial College London, 5th Fl, Commonwealth Bldg, Hammersmith Hospital Campus, Du Cane Rd, London W12 0NN, United Kingdom; e-mail j.crawley@imperial.ac.uk.