Abstract
CS1 is highly expressed on tumor cells from the majority of multiple myeloma (MM) patients regardless of cytogenetic abnormalities or response to current treatments. Furthermore, CS1 is detected in MM patient sera and correlates with active disease. However, its contribution to MM pathophysiology is undefined. We here show that CS1 knockdown using lentiviral short-interfering RNA decreased phosphorylation of ERK1/2, AKT, and STAT3, suggesting that CS1 induces central growth and survival signaling pathways in MM cells. Serum deprivation markedly blocked survival at earlier time points in CS1 knockdown compared with control MM cells, associated with earlier activation of caspases, poly(ADP-ribose) polymerase, and proapoptotic proteins BNIP3 and BIK. CS1 knockdown further delayed development of MM tumor and prolonged survival in mice. Conversely, CS1 overexpression promoted myeloma cell growth and survival by significantly increasing myeloma adhesion to bone marrow stromal cells (BMSCs) and enhancing myeloma colony formation in semisolid culture. Moreover, CS1 increased c-maf–targeted cyclin D2-dependent proliferation, -integrin β7/αE-mediated myeloma adhesion to BMSCs, and -vascular endothelial growth factor-induced bone marrow angiogenesis in vivo. These studies provide direct evidence of the role of CS1 in myeloma pathogenesis, define molecular mechanisms regulating its effects, and further support novel therapies targeting CS1 in MM.
Introduction
CS1 is a cell surface glycoprotein that was recently identified as a novel target for multiple myeloma (MM) treatment because of its expression on tumor cells from the majority of MM patients.1,2 It is characterized by 2 extracellular immunoglobulin (Ig)-like domains and an intracellular signaling domain with immune receptor tyrosine-based switch motifs.3-7 CS1 mRNA and protein are specifically expressed at high levels in normal and malignant plasma cells, but not normal organs, solid tumors, or CD34+ stem cells. Only a small subset of resting lymphocytes, including natural killer (NK) cells and a subset of CD8+ T cells, express detectable but low levels of CS1.1,8 Unlike other potential antibody targets for MM treatment, such as CD138 (syndecan-1), CD38, and CD40, which are also expressed in other normal tissues,9-13 this restricted expression pattern makes CS1 an attractive target for therapeutic antibodies. The humanized anti-CS1 monoclonal antibody (mAb) elotuzumab (formerly known as HuLuc63) mediates significant antibody-dependent cellular cytotoxicity against allogeneic and autologous CS1-expressing MM cells and inhibits tumor cell growth in several xenograft models of human MM.2 Elotuzumab is currently under evaluation in phase 1 clinical trials for the treatment of relapsed MM
Currently, the function of CS1 in MM cells is unknown. In NK cells, CS1 acts as a self-ligand and mediates homophilic interaction.14 Immunofluorescence studies showed that CS1 is colocalized with CD138 in the subcellular uropod membranes of MM cell lines and patient MM cells, suggesting that CS1 might be involved in MM cell adhesion.2 Because the interaction of MM cells with bone marrow stroma supports tumor cell growth, survival, and chemoresistance by inducing key factors, such as interleukin-6, B cell–activating factor of the TNF family, and vascular endothelial growth factor (VEGF),15,16 CS1 might promote MM cell growth in the bone marrow microenvironment. CS1 gene is localized in the long arm of chromosome 1 (1q23.1-q24.1), and CS1 gene and protein amplification has been identified in MM cell lines (ie, OPM2, H929, and KMS20).17 Because gains of chromosome 1q are frequent chromosomal alterations in malignant CD138+ patient MM cells and frequently associated with disease progression,18 CS1 overexpression might contribute to the pathophysiology of MM. Most recently, we detected CS1 protein in MM patient sera, but not in sera from persons with monoclonal gammopathy of undetermined significance or in healthy donors; moreover, circulating CS1 levels correlated with disease activity. These studies further suggest a potential role for CS1 in MM pathogenesis.
In the present study, we characterized the activity of CS1 in MM pathophysiology both by inhibiting CS1 using lentiviral CS1shRNA in CS1-expressing MM cells and by overexpressing CS1 in CS1-low-expressing MM cells. We used microarray profiling to identify genes up-regulated in CS1-overexpressing cells and down-regulated in CS1-null MM cells. We found that CS1 expression promotes MM cell adhesion to bone marrow stromal cells (BMSCs), clonogenic growth, and tumorigenicity in vivo via coregulation of c-maf transactivation. These results establish a pathophysiologic role of CS1 in MM and strongly support novel therapies using anti-CS1 mAb elotuzumab in MM.
Methods
Cell culture and BMSCs
CS1-expressing OPM2 and MM1S (kindly obtained by sources previously described)2,19 as well as U266 cells (ATCC, Manassas, VA) weakly expressing CS12 were grown in RPMI 1640 (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS; HyClone Laboratories, Logan, UT), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). BMSCs were obtained from the CD138-negative fraction separated from CD138-positive patient multiple myeloma cells as described.19 When a confluent layer of adherent cells was obtained, cells were trypsinized and cultured in RPMI 1640/10% fetal calf serum.
Lentiviral CS1 shRNA transduction
Lentiviral CS1 shRNA was generated as described previously.2,20 The sense oligonucleotide sequence CS1 siRNAs was as follows: clone 1, target sequence 5′-GCAGCCAATGAGTCCCATAAT-3′; clone 2, target sequence 5′-CCCTCACACTAATAGAACAAT-3′;clone 3, target sequence 5′-GTCGGGAAACTCCTAACATAT-3′; and clone 4, target sequence 5′-GCTCAGCAAACTGAAGAAGAA-3′. Lentiviral CS1 shRNA and control shRNA were produced in 293t packaging cells and then transduced into MM cell lines, followed by selection in puromycin (2 μg/mL, Invitrogen) to obtain CS1null and control MM cell lines.
Cell viability assays
CS1null OPM2 cells and control OPM2 cells were incubated with 0.1% FBS/RPMI 1640 medium in triplicate in 96-well plates for 3 days. Apoptosis was assayed by individual caspase activity assay (Promega, Madison, WI). MM1S and U266 transfectants were plated at 5000, 7500, and 10 000 cells per well in 10% FBS/RPMI 1640 in triplicate. Cell viability was assessed by the yellow tetrazolium (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) (MTT) assay (ATCC). Absorbance of treated cells was divided by that of control MM cells to calculate survival fraction. Cell proliferation was assayed by [3H]thymidine incorporation.19
Immunoblotting and flow cytometric analysis
Total cell lysates were subjected to 10% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes, as previously reported.19 All Abs were obtained from Cell Signaling Technology (Danvers, MA), except mouse anti-CS1 mAb Luc90 (provided by PDL BioPharma, Redwood City, CA) and anti-c-maf Ab (Santa Cruz Biotechnology, Santa Cruz, CA). To determine whether CS1 knockdown altered phosphorylation of kinases, lysates from cntOPM2 and CS1null OPM2 cells were subjected to Kinetworks phospho-site KPSS-1.3 analysis using Kinexus Bioinformatics (Vancouver, BC).21
CS1 expression was further monitored by flow cytometric analysis using anti–CS1-phycoerythrin mAb (R&D Systems, Minneapolis, MN) and phycoerythrin-conjugated mouse isotype control IgG2a mAb.2
Transfection
MM cells were transfected with a CS1-expression plasmid pflagCS1 (kindly provided by Dr Daniel Afar, PDL BioPharma) or control vector plasmid using the Cell Line Nucleofector Kit V Solution (Amaxa Biosystems, Walkersville, MD). Two days after transfection, U266 cells overexpressing CS1 were isolated with magnetic microbeads followed by selection with blasticidin (5 μg/mL, Invitrogen) to generate stable U266CS1 transfectants. Immunoblotting using an antiflag Ab confirmed exogenous CS1 expression in U266CS1 cells. Mock control U266 transfectants were also selected by blasticidin. Cell growth images were first examined with a Leica DM LB research microscope using Leica IM50 Image Manager (Leica Microsystems, Deerfield, IL) and processed using Adobe Photoshop Software, version 7.0 (Adobe Systems, San Jose, CA).
Cell adhesion assays
MM cells (5 × 106/mL) were labeled with calcein-AM (Invitrogen), washed, and resuspended in culture medium.22 Cells were added to BMSC-coated 96-well plates at 37°C for 30 minutes; unbound cells were then washed out, and adherence was assessed using a fluorescence plate reader (Wallac VICTOR2; PerkinElmer Life & Analytical Sciences, Waltham, MA).
Methylcellulose colony formation assays
A total of 500 and 1000 cells per well in quadruplicate were plated in 1 mL of human methylcellulose-based medium (R&D Systems) in 24-well culture plates. If 48-well culture plates were used, 100, 200, and 400 cells per well were plated. Colonies consisting of more than 40 cells were scored at 10 days and stained. Images were acquired as described in “Transfection” and processed using Adobe Photoshop Software, version 7.0 (Adobe).
Gene expression profiling
Total RNA was isolated from OPM2, cntOPM2, 4 CS1null OPM2, U266, and U266CS1 MM cell lines in duplicate using QIAGEN RNeasy kit (QIAGEN, Valencia, CA).15,19 Affymetrix U133A Plus2 arrays (Affymetrix, Santa Clara, CA) were hybridized with biotinylated in vitro transcription products (10 μg/chip) within the DFCI Microarray Core Facility. DNA chips were analyzed with a Gene Array Scanner (Affymetrix), and CEL files were obtained using Affymetrix Microarray Suite 5.0 software. All analyses were performed using Bioconductor packages23 and R program. Packages “simplyaffy” and “affyPLM” were used for quality assessment of the CEL files. The Robust Multi-Array normalization method24 was used to normalize and obtain the summarized expression values. The complete dataset has gone through several filtering steps to remove noninformative or nonexpressed genes, and 10 896 genes were used in formal statistical analysis. The Rank Product method (“RankProd” package)25 was used to identify differentially expressed genes in (1) U266CS1 versus U266 cells, and (2) CS1null OPM2 versus cntOPM2 cells. Genes were identified using false discovery rate (fdr) of 0.05 and P = .01. The microarray data have been deposited into GEO under accession number GSE14680 (NIH Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo).
VEGF ELISA
U266CS1 or U266 cells were added to BMSC-coated plates (BMSCs/MM cells at 3:1), and supernatants harvested from 2-day cultures were tested for VEGF secretion by enzyme-linked immunosorbent assay (ELISA; R&D Systems). The minimum detectable level of VEGF was 10.0 pg/mL.
Human plasmacytoma xenograft model
All animal studies were approved by the Dana-Farber Cancer Institute Animal Care and Use Committee. The xenograft tumor model was performed as previously described.19 CB17 severe combined immunodeficiency (SCID) mice (Charles River Breeding Laboratories, Portage, MI) were subcutaneously inoculated with OPM2 (4.0 × 106) or U266 (106) cells in 100 μL RPMI 1640 medium. Tumor size was measured every third day in 2 dimensions using calipers, and tumor volume was calculated using the following formula:
V = 0.5 × a × b2
where a and b are the long and short diameter of the tumor, respectively. Animals were killed when their tumors reached 2 cm3 in volume or when paralysis or a major compromise in their quality of life occurred. At the time of the animals' death, tumors were excised. Survival was evaluated from the first day of treatment until death.
siRNA transduction
The c-maf siRNA oligonucleotide GAAGACUACUACUGGAUGA (Dharmacon RNA Technologies, Lafayette, CO) was transduced into U266CS1 cells by electroporation (Amaxa Biosystems) using nucleofaction. The negative control siRNA of Dharmacon RNA Technologies (ON-TARGETplus siCONTROL NonTargeting siRNA) was used as a control. Live cells were separated by Ficoll centrifugation 48 hours after transfection and plated in 96-well tissue plates to assay for proliferation.
Statistical analysis
Statistical significance of differences observed in CS1null or CS1-overexpressing MM cells and mice injected with CS1null or CS1-overexpressing MM cells compared with respective control groups was determined using Student t test. Student t test is also used for other experiments in the current study. The minimal level of significance was P less than .05. Tumor formation changes and survival of mice were determined using the GraphPad PRISM (GraphPad Software, San Diego, CA).
Results
CS1 depletion on cell membrane inhibits phosphorylation of ERK, AKT, and STAT3
To investigate the function of CS1 in MM cells, its expression was knocked down with lentiviral CS1 shRNAs. Four lentiviral CS1 shRNAs were generated targeting different regions in the CS1 mRNA. Specific CS1 down-regulation was confirmed by inhibition of CS1 mRNA and protein expression, whereas CS1 was expressed in parental OPM2 and OPM2 cells infected with control lentiviral vector (cntOPM2; Figure 1A).
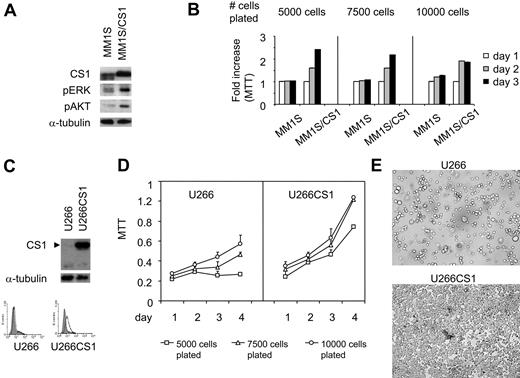
CS1 knockdown affects phosphorylation of ERK1/2, STAT3, and AKT. (A) CS1 knockdown after lentiviral CS1 shRNA infection. RNA was prepared from OPM2 cells infected with individual lentiviral CS1shRNA (targeting 4 regions on CS1 cDNA) or control shRNA, followed by gene expression analysis. Arbitrary unit of CS1 mRNA is shown. Immunoblotting and flow cytometric analysis further confirmed CS1 membrane depletion in CS1null OPM2 cells. Solid histogram represents CS1 expression; open histogram, isotype control. (B) Phosphorylated ERK1/2, STAT3, and AKT in C1null OPM2 versus cntOPM2 cells were examined using specific phosphorylated Abs. α-Tubulin was used as a loading control. (C) Cell lysates were examined for the phosphorylation status of more than 37 phospho-sites in 29 signaling proteins with a panel of 37 highly validated phospho-site-specific Abs (Kinetworks KPSS-1.3 Phosphosite Screen, Kinexus Biosystems). Representative multiple immunoblots and the migration of target phosphoproteins are shown. (D) The intensity of the ECL signals (counts per minute) was quantified from the multiple immunoblots for the total cell lysates of CS1null OPM2 and cntOPM2. The fold changes of phosphorylation in the CS1null OPM2 (■) relative to cntOPM2 (□) are presented. The averaged results from 2 phospho-site analyses are shown.
CS1 knockdown affects phosphorylation of ERK1/2, STAT3, and AKT. (A) CS1 knockdown after lentiviral CS1 shRNA infection. RNA was prepared from OPM2 cells infected with individual lentiviral CS1shRNA (targeting 4 regions on CS1 cDNA) or control shRNA, followed by gene expression analysis. Arbitrary unit of CS1 mRNA is shown. Immunoblotting and flow cytometric analysis further confirmed CS1 membrane depletion in CS1null OPM2 cells. Solid histogram represents CS1 expression; open histogram, isotype control. (B) Phosphorylated ERK1/2, STAT3, and AKT in C1null OPM2 versus cntOPM2 cells were examined using specific phosphorylated Abs. α-Tubulin was used as a loading control. (C) Cell lysates were examined for the phosphorylation status of more than 37 phospho-sites in 29 signaling proteins with a panel of 37 highly validated phospho-site-specific Abs (Kinetworks KPSS-1.3 Phosphosite Screen, Kinexus Biosystems). Representative multiple immunoblots and the migration of target phosphoproteins are shown. (D) The intensity of the ECL signals (counts per minute) was quantified from the multiple immunoblots for the total cell lysates of CS1null OPM2 and cntOPM2. The fold changes of phosphorylation in the CS1null OPM2 (■) relative to cntOPM2 (□) are presented. The averaged results from 2 phospho-site analyses are shown.
CS1 activates ERK1/2 and PI3K/AKT in CS1-expressing NK cells.26 We therefore determined whether CS1 knockdown affects phosphorylation of signaling cascades in OPM2 MM cells. Whole cell lysates from OPM2 cells infected with lentiviral CS1 shRNA or control shRNA were subjected to immunoblotting using specific phosphorylated Abs. As seen in Figure 1B, phosphorylation of ERK1/2, STAT3, and AKT was significantly down-regulated in OPM2 cells transduced with lentiviral CS1 shRNA (CS1null OPM2), but not control shRNA (cntOPM2). These lysates were further analyzed for the phosphorylated forms of various signaling proteins using a panel of 37 different phospho-site Abs in the Kinetworks KPSS-1.3 Screen. Consistent down-regulation of phosphorylated ERK1/2, STAT3, and AKT was confirmed in CS1null OPM2 versus cntOPM2 cell lysates (Figure 1C,D). In addition, changes in phosphorylation of various kinases were seen in CS1null OPM2 versus cntOPM2 lysates. Thus, CS1 down-regulation decreased phosphorylation of ERK1/2, STAT3, and AKT, as well as altered phosphorylation of multiple kinases, suggesting that CS1 can induce signaling cascades in MM cells.
Stable CS1 knockdown promotes apoptosis after serum deprivation
In an attempt to generate stable CS1null transfectants, we transduced MM1S, OPM2, and H929 MM cells with lentiviral CS1 or control shRNA and then selected with puromycin for 2 months to assess CS1 expression. We were able to generate stable CS1null transfectant OPM2 cells, which were cultured for 4 months under drug selection. Although growth and survival of CS1null OPM2 versus control transfectants did not significantly differ in 3-day MTT or [3H]thymidine incorporation assays, viability was reduced in CS1null OPM2 compared with cntOPM2 cells between 10 and 14 days after plating (data not shown). Using caspase activity, poly(ADP-ribose) polymerase (PARP) cleavage, and MTT assays, we next determined whether serum deprivation induced earlier cell death in CS1null OPM2 than cntOPM2 cells. As seen in Figure 2A, earlier activation of caspases 8 and 9 followed by caspase 3 was observed in pools of CS1null OPM2 than cntOPM2 cells. After 2 days of serum deprivation, PARP cleavage was induced in CS1null OPM2, but not cntOPM2, cells (Figure 2B). These results correlated with a significant reduction in cell survival of CS1null OPM2 cells; cell viability was inhibited by 3.8-fold at day 6 after serum withdrawal in CS1null versus cntOPM2 cells (Figure 2C). Moreover, induction of proapoptotic molecules BINP3 and BIK mRNA and protein was observed in CS1null OPM2 versus cntOPM2 cells (> 4-fold alteration, P < .001), evidenced by expression profiling and by immunoblotting (Figure 2D). Therefore, serum deprivation induced earlier apoptosis in CS1null OPM2 cells associated with earlier activation and cleavage of caspases, as well as elevated proapoptotic protein expression.
CS1 down-regulation induces earlier apoptosis after serum deprivation. (A) Pools of CS1null OPM2 and cntOPM2 cells in quadruplicate were incubated with serum-free medium for the indicated periods followed by individual caspase activity assays. ■ represents CS1null OPM2 cells; □, cntOPM2 cells. (B) Cell lysates were prepared from indicated cells after indicated time periods and subjected to immunoblotting using anti-PARP Ab. (C) Cell viability was assayed by MTT at indicated time periods. ■ represents CS1null OPM2 cells; □, cntOPM2 cells. (D) mRNA transcripts (top panel) and protein levels (bottom panel) of proapoptotic molecules BNIP3 and BIK differed in CS1null OPM2 versus control OPM2 cells.
CS1 down-regulation induces earlier apoptosis after serum deprivation. (A) Pools of CS1null OPM2 and cntOPM2 cells in quadruplicate were incubated with serum-free medium for the indicated periods followed by individual caspase activity assays. ■ represents CS1null OPM2 cells; □, cntOPM2 cells. (B) Cell lysates were prepared from indicated cells after indicated time periods and subjected to immunoblotting using anti-PARP Ab. (C) Cell viability was assayed by MTT at indicated time periods. ■ represents CS1null OPM2 cells; □, cntOPM2 cells. (D) mRNA transcripts (top panel) and protein levels (bottom panel) of proapoptotic molecules BNIP3 and BIK differed in CS1null OPM2 versus control OPM2 cells.
CS1 knockdown severely blocks tumor growth in vivo
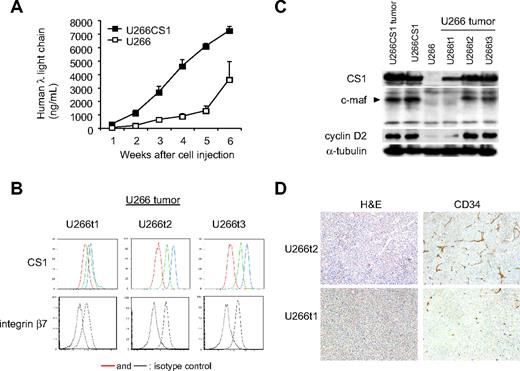
We next asked whether CS1 knockdown blocked MM cell growth in vivo. CB17 SCID mice were injected subcutaneously with CS1null OPM2 versus cntOPM2 cells to compare the development of tumor and detection of human immunoglobulin λ light chain in sera. All 8 mice injected with cntOPM2 cells rapidly developed tumors, requiring death within 3 weeks. In contrast, mice injected with CS1null OPM2 cells developed tumors at a significantly later time point (Figure 3A,B, P < .01). Tumor onset in mice receiving CS1null OPM2 cells was 28 days compared with 8 days for mice injected with cntOPM2 cells (P < .01). At the end of the 80-day observation, 1 of 8 mice receiving CS1null OPM2 cells still did not develop tumor. Importantly, tumors that developed in mice receiving CS1null OPM2 cells expressed CS1, unlike the cells originally injected in these animals (Figure 3C). CS1 expression in these tumors was similar to tumors removed from mice receiving cntOPM2 cells. These results indicate that CS1 expression is required for tumor formation in vivo.
CS1 knockdown blocks in vivo tumor formation. (A) Mice (n = 8 per group) received pools of CS1null OPM2 cells or control OPM2 cells. Tumor development (top panel) and human λ light chain (ng/mL) in blood (bottom panel) were compared (P < .01). ○ represents CS1null OPM2 cells; ●, cntOPM2 cells. (B) Survival was evaluated after cell inoculation until death (mice were killed when tumors reached 2 cm3 in volume). P < .01. (C) Flow cytometric analysis of injected cells and tumors in mice injected with CS1null OPM2 cells. Solid histogram represents CS1 expression; open histogram, isotype control.
CS1 knockdown blocks in vivo tumor formation. (A) Mice (n = 8 per group) received pools of CS1null OPM2 cells or control OPM2 cells. Tumor development (top panel) and human λ light chain (ng/mL) in blood (bottom panel) were compared (P < .01). ○ represents CS1null OPM2 cells; ●, cntOPM2 cells. (B) Survival was evaluated after cell inoculation until death (mice were killed when tumors reached 2 cm3 in volume). P < .01. (C) Flow cytometric analysis of injected cells and tumors in mice injected with CS1null OPM2 cells. Solid histogram represents CS1 expression; open histogram, isotype control.
CS1 overexpression promotes growth and survival in MM cells
To validate our CS1 siRNA studies and to exclude possible off-target and other nonspecific effects in RNA interference studies, we independently overexpressed CS1 in MM1S and U266 MM cells using a pflagCS1 expression vector. Enhanced CS1 expression in MM1S cells was observed after transfection with pflagCS1, but not control, expression vectors (Figure 4A). Immunoblotting using antiflag Ab further confirmed the specificity of overexpressed CS1 by plasmid pflagCS1 (data not shown). In addition, immunoblotting demonstrated increased phosphorylation of ERK1/2 and AKT in MM1S/CS1 transfectants versus MM1S cells transfected with control vector (Figure 4A). In parallel, these transfectants were plated at an increasing number of cells per well in 96-well culture plates, followed by MTT assay. CS1 increased MM1S cell growth and survival because viability was significantly higher in MM1S/CS1 than control MM1S cells (Figure 4B; P < .01). This effect of CS1 was most prominent when cells were plated at lowest density, where control cells did not proliferate. More than 90% of MM cell lines tested express CS1, and U266 is one of a few MM cell lines that express low levels of CS1.2 Similar results were observed when CS1–low-expressing U266 cells were transfected in a similar fashion. Stable U266CS1 and control transfectants were obtained after selection with blasticidin for 2 weeks. CS1 overexpression was validated in CS1 transfectants by immunoblotting and immunofluorescence (Figure 4C). Importantly, time- and dose-dependent CS1-induced proliferation and survival effects were observed (Figure 4D). In addition to increased total cell numbers, U266CS1 cells grew adherent to standard tissue flasks, whereas control U266 cells grew in suspension (Figure 4E). Of note, U266 as well as control U266 transfectants grow mostly as nonadherent cells, with only a small fraction of adherent cells (< 5%). In contrast, all U266CS1 cells exhibited growth as adherent cells.
MM cell growth and survival were up-regulated by CS1 overexpression. (A) MM1S cells were transfected with pflagCS1 (MM1S/CS1) or control vector (MM1S). An augmented CS1 protein level in MM1S/CS1 was confirmed by immunoblotting using specific Abs. Cell viability was measured by MTT in panel B (MM1S/CS1 vs MM1S) and panel D (U266CS1 vs U266). Error bars indicate the SE from 3 independent experiments. (C) CS1 expression after transfection with pflagCS1 or control vector in U266 cells. Open histogram represents CS1; solid histogram, isotype control. (E) Cells were plated at the same density at day 0, and cell morphology was demonstrated after 3 days of culture. Images were taken using a Leica DFC300FX with a 20×/0.35 NA objective and Leica IM50 Image Manager (original magnification, ×200).
MM cell growth and survival were up-regulated by CS1 overexpression. (A) MM1S cells were transfected with pflagCS1 (MM1S/CS1) or control vector (MM1S). An augmented CS1 protein level in MM1S/CS1 was confirmed by immunoblotting using specific Abs. Cell viability was measured by MTT in panel B (MM1S/CS1 vs MM1S) and panel D (U266CS1 vs U266). Error bars indicate the SE from 3 independent experiments. (C) CS1 expression after transfection with pflagCS1 or control vector in U266 cells. Open histogram represents CS1; solid histogram, isotype control. (E) Cells were plated at the same density at day 0, and cell morphology was demonstrated after 3 days of culture. Images were taken using a Leica DFC300FX with a 20×/0.35 NA objective and Leica IM50 Image Manager (original magnification, ×200).
CS1 promotes MM cell adhesion and clonogenic growth
We next determined whether CS1 increased MM cell adhesion to BMSCs. In a 30-minute adhesion assay, MM1S/CS1 and U266CS1 transfectants demonstrated significantly increased adherence to BMSCs relative to control cells (P < .01; Figure 5A), consistent with our previous studies demonstrating reduced adhesion of CS1-knockdown MM1S cells transfected by lentiviral CS1 shRNA.2 Although CS1 protein level was low in U266 control cells, CS1 expression was detected by immunoblotting after long exposure (> 5 minutes) time of films (Figure 5B). In addition, CS1 mRNA was expressed in control U266 cells (Figure 6A). We further separated U266 cells in suspension from adherent U266 cells and examined CS1 expression on both fractions. Adherent U266 cells expressed CS1, whereas floating U266 cells did not (Figure 5B). These results strongly indicate a correlation between CS1 expression and adherent MM cell growth.
CS1 enhances MM cell adhesion to bone marrow stroma. (A) Binding of CS1-overexpressing or control MM cells to BMSCs was determined by 30-minute adhesion assay in a fluorescence plate reader. Experiments were done in triplicates, and a representative experiment was shown. a.u. indicates arbitrary units. (B) CS1 was detectable in control U266 cells after long exposure (7 minutes) compared with short exposure (40 seconds) in immunoblotting analysis. CS1 expression in U266 cells in suspension versus adherent U266 cells was analyzed by flow cytometry and immunoblotting. (C) Cell morphology and colony formation were examined in methylcellulose-based culture at 6 days after plating U266CS1 versus control U266 transfectants. Images were taken using a Leica DFC300FX with a 40×/0.6 NA objective and Leica IM50 Image Manager (original magnification, ×400). (D) Colonies were stained and visualized with a Leica DM LB research microscope and quantitated in panel E. P < .01.
CS1 enhances MM cell adhesion to bone marrow stroma. (A) Binding of CS1-overexpressing or control MM cells to BMSCs was determined by 30-minute adhesion assay in a fluorescence plate reader. Experiments were done in triplicates, and a representative experiment was shown. a.u. indicates arbitrary units. (B) CS1 was detectable in control U266 cells after long exposure (7 minutes) compared with short exposure (40 seconds) in immunoblotting analysis. CS1 expression in U266 cells in suspension versus adherent U266 cells was analyzed by flow cytometry and immunoblotting. (C) Cell morphology and colony formation were examined in methylcellulose-based culture at 6 days after plating U266CS1 versus control U266 transfectants. Images were taken using a Leica DFC300FX with a 40×/0.6 NA objective and Leica IM50 Image Manager (original magnification, ×400). (D) Colonies were stained and visualized with a Leica DM LB research microscope and quantitated in panel E. P < .01.
CS1 up-regulates c-maf transactivation in MM cells. (A) Transcriptional and protein changes of CS1, c-maf, and cyclin D2 in CS1-overexpressing U266CS1 and CS1null OPM2 MM cells. The representative probe sets for each gene were used for mRNA expression analysis (P < .003), with confirmation of protein levels by immunoblotting using specific Abs. (B) Transcriptional changes of other c-maf-transactivation targets in U266 versus U266CS1 or OPM2 versus CS1null OPM2 cells (P < .003). Fold changes of listed target mRNA in these cells are shown in Table 1. (C) Integrin β7 expression is enhanced in U266CS1 versus U266 cells, as analyzed by flow cytometry. (D) Top panel: VEGF secretion by U266CS1 or U266 cells, alone or in the presence or absence of BMSCs, as well as in BMSCs alone. Experiments were done in triplicate; error bars represent SE. Fold increase in VEGF induced by MM cell adhesion to BMSCs is shown in the bottom panel. P < .03.
CS1 up-regulates c-maf transactivation in MM cells. (A) Transcriptional and protein changes of CS1, c-maf, and cyclin D2 in CS1-overexpressing U266CS1 and CS1null OPM2 MM cells. The representative probe sets for each gene were used for mRNA expression analysis (P < .003), with confirmation of protein levels by immunoblotting using specific Abs. (B) Transcriptional changes of other c-maf-transactivation targets in U266 versus U266CS1 or OPM2 versus CS1null OPM2 cells (P < .003). Fold changes of listed target mRNA in these cells are shown in Table 1. (C) Integrin β7 expression is enhanced in U266CS1 versus U266 cells, as analyzed by flow cytometry. (D) Top panel: VEGF secretion by U266CS1 or U266 cells, alone or in the presence or absence of BMSCs, as well as in BMSCs alone. Experiments were done in triplicate; error bars represent SE. Fold increase in VEGF induced by MM cell adhesion to BMSCs is shown in the bottom panel. P < .03.
To extend this observation and assay for clonogenic capacity, U266CS1 and control U266 cells were cultured in methylcellulose at equivalent cell numbers. U266CS1 cells formed significant colonies 1 week after cell plating, whereas control U266 cells did not (Figure 5C). U266CS1 exhibited adherent morphology with protrusions and homotypic adhesion, as seen in Figure 4E. Two weeks later, U266CS1 cells had markedly increased ability to form colonies compared with controls in a dose-dependent manner (Figure 5D,E). Therefore, CS1 significantly promoted U266 growth in methylcellulose, evidenced by increased colony number and size.
Identification of CS1 expression associated with c-maf transactivation
To further define the molecular mechanisms of CS1-induced adhesion and growth in MM, we used microarrays to profile gene expression in 4-individual CS1null OPM2 cells, control OPM2 cells including OPM2 and cntOPM2 cells, as well as U266CS1 and control U266 cells, with 2 independent replicates of each cell. Data analysis aimed to identify genes commonly altered in CS1null OPM2 versus control OPM2 cells, as well as in U266CS1 versus control U266 cells. At fdr of 0.05 and P of .01, we identified 1135 (659 up-regulated, 476 down-regulated) and 676 (331 up-regulated, 345 down-regulated) differentially expressed genes in CS1null OPM2 compared with control OPM2 cells and in U266CS1 compared with control U266 cells, respectively. Notably, c-maf and its target genes were identified with large-scale expression difference under at least one comparison (listed in the previous sentence). As expected, CS1 mRNA was significantly increased in U266CS1 versus control U266 cells but decreased in CS1null OPM2 cells versus control OPM2 cells (Table 1; Figure 6A). c-maf and cyclin D2 were up-regulated in U266CS1 versus control U266 cells; conversely, CS1null OPM2 cells had reduced c-maf and cyclin D2 transcription compared with control OPM2 cells. Immunoblotting further confirmed up-regulation of c-maf and cyclin D2 protein in U266CS1 versus control U266 cells (Figure 6A). Similar patterns of increased and reduced expression of other c-maf-transactivated genes integrin β7, CCR1, and integrin αE were demonstrated in U266CS1 versus U266 cells and CS1null OPM2 versus control OPM2 cells (Figure 6B). Flow cytometric analysis further confirmed up-regulation of integrin β7 in U266CS1 versus U266 cells (Figure 6C). Because VEGF is induced by c-maf–increased MM cell adhesion to bone marrow stroma,13,27,28 we examined whether VEGF induced by MM cell adhesion to BMSCs was further increased by adherence of U266CS1 compared with U266 cells. Significantly, more VEGF was induced when U266CS1 cells adhered to BMSCs than U266 control cells (Figure 6D, P < .03). These results indicate that CS1 modulates MM cell interaction with BMSCs, associated with c-maf–transactivated growth and survival.
Fold changes in mRNA expression in U266CS1 versus U266 and CS1null OPM2 versus OPM2 cells
| Gene . | Gene ID . | Probe set . | Fold change . | FDR . | P . | |
|---|---|---|---|---|---|---|
| U266CS1/ U266 . | CS1null OPM2/ OPM2 . | |||||
| CS1 | 57823 | 219159_s_at | 6.20 | 0.026 | 0.039 | 1.00E−04 |
| c-maf | 4094 | 206363_at | 3.16 | 0.078 | 0.004 | < 1.00E−04 |
| 4094 | 209348_s_at | 2.69 | 0.033 | 0.045 | < 1.00E−04 | |
| cyclin D2 | 894 | 200953_s_at | 1.89 | 0.004 | 0.001 | < 1.00E−04 |
| CCR1 | 1230 | 205098_at | 1.01 | 0.009 | 0.045 | < 1.00E−04 |
| Integrin β7 | 3695 | 205718_at | 1.74 | 0.005 | 0.015 | < 1.00E−04 |
| Gene . | Gene ID . | Probe set . | Fold change . | FDR . | P . | |
|---|---|---|---|---|---|---|
| U266CS1/ U266 . | CS1null OPM2/ OPM2 . | |||||
| CS1 | 57823 | 219159_s_at | 6.20 | 0.026 | 0.039 | 1.00E−04 |
| c-maf | 4094 | 206363_at | 3.16 | 0.078 | 0.004 | < 1.00E−04 |
| 4094 | 209348_s_at | 2.69 | 0.033 | 0.045 | < 1.00E−04 | |
| cyclin D2 | 894 | 200953_s_at | 1.89 | 0.004 | 0.001 | < 1.00E−04 |
| CCR1 | 1230 | 205098_at | 1.01 | 0.009 | 0.045 | < 1.00E−04 |
| Integrin β7 | 3695 | 205718_at | 1.74 | 0.005 | 0.015 | < 1.00E−04 |
FDR indicates false discovery rate.
CS1 coregulates c-maf transactivation in vivo
To determine whether CS1 coregulates c-maf transactivation in vivo and confirm its role in MM growth in vivo, U266CS1 and control U266 cells were injected subcutaneously in the flanks of CB17 SCID mice. All 8 mice injected with U266CS1 developed tumors within 21 to 31 days, whereas mice injected with CS1–low-expressing control U266 cells developed fewer tumors at a later time point (Figure 7A). Importantly, 3 tumors developing in mice injected with control U266 cells had significant CS1 and integrin β7 expression at the time of death (Figure 7B), unlike the cells originally injected in these animals (Figure 6C). CS1 and integrin β7 levels were similar to those in U266CS1 tumors. Furthermore, integrin β7 was enhanced in tumors with increased CS1 expression. C-maf and cyclin D2 were also up-regulated in tumors with higher CS1 expression (Figure 7C). These tumors further exhibited growth as adherent cells in culture (data not shown). Tumors derived from U266CS1 cells and those originally injected into these animals had significant CS1 and c-maf as well as cyclin D2 expression (Figure 7C). Moreover, increased angiogenesis, as assessed by immunohistochemistry using anti-CD34 mAb, was seen in tumors expressing elevated CS1 (U266t2) compared with those with moderate CS1 expression (U266t1; Figure 7D). These results indicate that enforced CS1 expression promotes the in vivo growth and survival of CS1-expressing MM, associated with c-maf transactivation.
CS1 up-regulates tumor formation via c-maf in vivo. (A) Human λ light chain levels in blood were assayed by ELISA to assess tumor development in mice injected with indicated cells. (B) Cell surface expression of CS1 and integrin β7 on tumors in mice injected with U266 cells (U266t1-3) was analyzed by flow cytometric analysis. Red and dotted lines represent isotype control; green line, CS1 levels on excised tumor; blue line, CS1 level after 4-day culture of excised tumors. (C) Immunoblotting analysis of tumors in mice injected with indicated cells using specific Abs. (D) Hematoxylin and eosin and CD34 staining by immunohistochemistry of tumors expressing high (U266t2) and low (U266t1) levels of CS1. Images were taken using a Leica DFC300FX with a 10×/0.22 NA objective and a Leica IM50 Image Manager (original magnification, ×100).
CS1 up-regulates tumor formation via c-maf in vivo. (A) Human λ light chain levels in blood were assayed by ELISA to assess tumor development in mice injected with indicated cells. (B) Cell surface expression of CS1 and integrin β7 on tumors in mice injected with U266 cells (U266t1-3) was analyzed by flow cytometric analysis. Red and dotted lines represent isotype control; green line, CS1 levels on excised tumor; blue line, CS1 level after 4-day culture of excised tumors. (C) Immunoblotting analysis of tumors in mice injected with indicated cells using specific Abs. (D) Hematoxylin and eosin and CD34 staining by immunohistochemistry of tumors expressing high (U266t2) and low (U266t1) levels of CS1. Images were taken using a Leica DFC300FX with a 10×/0.22 NA objective and a Leica IM50 Image Manager (original magnification, ×100).
Knockdown of c-maf inhibits U266CS1 cell proliferation and adhesion to BMSCs
c-maf expression was knocked down by c-maf siRNA in U266CS1 cells. Specific down-regulation of c-maf, cyclin D2, and integrin β7 was validated by immunoblotting and flow cytometric analysis, whereas CS1 expression was unchanged (Figure 8A). c-maf siRNA transduction further reduces the proliferation and adhesion to BMSCs of c-maf-overexpressing U266CS1 cells (Figure 8B). These results further confirmed that CS1-induced MM cell growth is dependent on c-maf transactivation.
Inhibition of c-maf inhibits U266CS1 cell proliferation. (A) c-maf, cyclin D2, and cell surface integrin β7 expression was analyzed by immunoblotting and flow cytometry in U266CS1 cells 72 hours after transduction with a c-maf siRNA oligonucleotide. (B) U266CS1 cells transduced with c-maf siRNA (□) or control siRNA (■) were assayed by the incorporation of [H3]thymidine. A representative experiment performed in quadruplicate is shown. *P < .03. (C) Binding of U266CS1 transduced with c-maf siRNA or control siRNA to BMSCs was determined by an adhesion assay in a fluorescence plate reader. A representative experiment performed in triplicates was shown. a.u. indicates arbitrary units. *P < .03.
Inhibition of c-maf inhibits U266CS1 cell proliferation. (A) c-maf, cyclin D2, and cell surface integrin β7 expression was analyzed by immunoblotting and flow cytometry in U266CS1 cells 72 hours after transduction with a c-maf siRNA oligonucleotide. (B) U266CS1 cells transduced with c-maf siRNA (□) or control siRNA (■) were assayed by the incorporation of [H3]thymidine. A representative experiment performed in quadruplicate is shown. *P < .03. (C) Binding of U266CS1 transduced with c-maf siRNA or control siRNA to BMSCs was determined by an adhesion assay in a fluorescence plate reader. A representative experiment performed in triplicates was shown. a.u. indicates arbitrary units. *P < .03.
Discussion
A potential role of CS1 in MM pathophysiology was suggested by its wide expression on MM cell lines and patient cells regardless of cytogenetic abnormalities (> 95% tested samples) and its potential involvement in cell adhesion. However, to date, there was no direct evidence to implicate CS1 in the pathogenesis of MM. Our present study demonstrates that cell surface CS1 induces central growth and survival signaling pathways in MM cells. The data suggest that CS1 enhances MM adhesion to BMSCs as well as homotypic adhesion between tumor cells. Importantly, continued CS1 expression appears to be required for clonogenic growth of MM cells in semisolid medium and for MM tumor formation in vivo, associated with c-maf modulation of MM-BMSC interactions.
CS1 is a cell membrane protein containing cytoplasmic tyrosine-based motifs and immune receptor tyrosine-based switch motifs. Our studies showed that changes in CS1 expression alter the phosphorylation status of multiple kinases, including ERK1/2, STAT3, and AKT signaling cascades in MM cells, suggesting that it can induce signaling in MM cells. To date, however, there is only one report of CS1-induced signaling in NK cells.26 Specifically, stimulation of CS1 led to recruitment of Ewing's sarcoma's/FLI1-transcripts 2 (EAT-2) but not signaling lymphocytic activation molecule-associated protein, thereby promoting CS1 phosphorylation through a Src-kinase and downstream signaling to elicit NK cell–mediated cytotoxicity.26 Whether MM cells use similar mechanisms to mediate the phosphorylation of CS1 and its downstream signaling is still unclear. Nevertheless, our results from CS1-knockdown and CS1-overexpressing MM cell models suggest that membrane CS1 could mediate its downstream effects by activation of these major MM signaling pathways, including ERK1/2, STAT3, and AKT.
It is well known that cell growth under standard culture conditions is affected by cell density. Moreover, cell-cell contact is a critical stimulus for cell proliferation as well as inducing growth and survival factors. Here we demonstrate that CS1 overexpression dramatically changed the morphology of U266 cells from growth in suspension to adherent growth on both standard culture flasks and BMSCs. Increased homotypic adhesion associated with CS1 overexpression triggered growth and survival signaling, that is, pERK1/2, pSTAT3, and pAKT, which in turn enhanced MM proliferation and tumor formation. All U266CS1 cells as well as CS1-expressing tumor cells cultured from mice injected with CS1-low-expressing U266 cells demonstrate enhanced growth as adherent cells and increased adhesion to BMSCs. Moreover, these 2 phenotypes correlated with CS1 and c-maf expression, not only in the current U266 study model but across many MM lines. For example, MM1S and MM1R cells express high levels of CS1, significantly adhere to BMSCs, and grow with an adherent phenotype. In addition, all tumors derived from mice injected with U266CS1 appeared to exhibit higher levels of CS1 than at the time of injection (data not shown). All escape U266 tumors express CS1 at various and significantly higher levels than U266 cells for injection into mice. These results strongly support the view that CS1 regulates MM cell interaction with BMSCs in a CS1-dependent manner in vitro and is also required for MM cell growth in vivo.
We showed that CS1 associated with c-maf transactivation promotes MM adhesion, clonogenic growth, and tumor formation. This is the first study to define the molecular mechanism whereby CS1 contributes to MM pathophysiology by augmenting c-maf function. c-maf defines a new class of oncogenes, not only increasing cell proliferation but further enhancing interactions between stromal cells and tumors cells by up-regulating adhesion molecules integrin β7/αΕ, CCR1, and VEGF secretion.27,29,30 c-maf was overexpressed in approximately 50% of MM patients; thus, it is one of the most frequent oncogenic events in MM. Although c-maf translocation involving the IgH locus provides a molecular mechanism to account for its overexpression, the majority of MM cells overexpressing c-maf (∼ 50%) are without c-maf translocation (> 40%).31 In addition to MM, c-maf transforms T cells in mice and human, and significantly, extremely elevated c-Maf levels were found in angioimmunoblastic T-cell lymphoma.32,33 Here we show a strong association of CS1 expression with c-maf pathway activation in vitro and in vivo. CS1 expression is associated with c-maf, integrin β7, and cyclin D2 activation, as well as enhanced angiogenesis, in MM xenograft tumors. Enhanced angiogenesis in CS1-overexpressing tumors further confirmed an increase in MM cell adhesion-triggered VEGF from BMSCs by CS1-augmented adhesion in vivo. Importantly, prominent CS1 protein expression was observed in escape tumors formed in either CS1 knockdown or CS1 overexpression in vivo study models, further suggesting its pathogenic role in MM. Moreover, inhibition of c-maf by c-maf siRNA in U266CS1 cells specifically down-regulated c-maf, integrin β7, and cyclin D2, but not CS1, consistent with integrin β7 and cyclin D2 as c-maf-transactivated genes.27 Concurrently, reduction of c-maf in U266CS1 cells decreased the proliferation and adhesion to BMSCs, confirming that CS1-enhanced MM cell growth and adhesion are dependent on c-maf transactivation in MM cells. These results imply that CS1 is involved in up-regulating c-maf in MM patients without c-maf translocation.
Increased c-maf, integrin β7, and cyclin D2 in U266 escape tumors with increased CS1 expression strongly support that CS1 up-regulates c-maf pathway in MM. Ongoing studies are investigating how CS1 regulates c-maf expression. Because serum CS1 levels correlate with disease activity,2 it is plausible that CS1 amplifies MM growth and survival via c-maf. Our results therefore strongly support targeting CS1 as a novel therapeutic approach to improve patient outcome in MM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Sophia Adamia and Dr Alexei Protopopov (Dana-Farber Cancer Institute) for microscopy and helpful discussions and Dr William Hahn (Dana-Farber Cancer Institute) for providing lentiviral CS1 shRNA constructs.
This work was supported by a Multiple Myeloma Research Foundation Research Award (Y.-T.T.), National Institutes of Health grants (RO-1 50947, PO1-78378, and SPORE P50CA100707), and the Lebow Fund to Cure Myeloma (K.C.A.).
National Institutes of Health
Authorship
Contribution: Y.-T.T. designed and analyzed data and wrote the manuscript; E.S., W.S., K.K., X.-F.L., and P.B. performed and verified flow data analysis; M.F., X.-F.L., and P.B. designed and performed animal studies; X.-F.L., W.S., M.J.R., P.B., S.N., K.K., K.P., and T.H. performed and analyzed in vitro experiments; F.H. and G.T. performed gene expression profiling and statistical analysis; D.E.H.A. provided pflagCS1 plasmid and antiCS1 mAbs for immunoblottings; N.C.M., R.D.C. and G.T. provided reagents and input to studies; and D.E.H.A., R.D.C., G.T., and K.C.A. critically evaluated and edited the manuscript. All coauthors subsequently collaborated on completing the article.
Conflict-of-interest disclosure: D.E.H.A. is an employee of Facet Biotech, whose product was used in this research. The remaining authors declare no competing financial interests.
Correspondence: Yu-Tzu Tai, Department of Medical Oncology, Dana-Farber Cancer Institute, M551, 44 Binney St, Boston, MA 02115; e-mail: yu-tzu_tai@dfci.harvard.edu; or Kenneth C. Anderson, Department of Medical Oncology, Dana-Farber Cancer Institute, M557, 44 Binney St, Boston, MA 02115; e-mail: kenneth_anderson@dfci.harvard.edu.






![Figure 8. Inhibition of c-maf inhibits U266CS1 cell proliferation. (A) c-maf, cyclin D2, and cell surface integrin β7 expression was analyzed by immunoblotting and flow cytometry in U266CS1 cells 72 hours after transduction with a c-maf siRNA oligonucleotide. (B) U266CS1 cells transduced with c-maf siRNA (□) or control siRNA (■) were assayed by the incorporation of [H3]thymidine. A representative experiment performed in quadruplicate is shown. *P < .03. (C) Binding of U266CS1 transduced with c-maf siRNA or control siRNA to BMSCs was determined by an adhesion assay in a fluorescence plate reader. A representative experiment performed in triplicates was shown. a.u. indicates arbitrary units. *P < .03.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/18/10.1182_blood-2008-10-183772/6/m_zh80170934210008.jpeg?Expires=1769089471&Signature=yfMVb1E2JhGwuSqnm~SxJmraEFqMsyaCF-OEUELUB0-pnzSW4V4Z8Ki11faWTUpZtndDGeHWXFWqyYAIcCoy1~UJA88CtK0AKs0cGop-AB7Gq5oaA6nT3ljmtUDjTz8RIjvMNOvmDBczciSl7uBeHhJx1Oazcb~OBNx6wXhtwnJlCpJ-KCH5R3a3Z8oP65uuWvPMLuWVHRYr9VnEBUaW-PfI-wWacZHsW~IJ71Cl6LkJ-708O5GMpmzEZrUafYTkXBnXBnE-0dhV98ic728DMCVBG~VqIy7MYA8D~RbcNoG0fJk4RWASD6KfjunPFV8Ybr2nM7i3cuCx5kM3IiOLZg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal