Abstract
The mechanisms regulating key fate decisions such as self-renewal and differentiation in hematopoietic stem and progenitor cells (HSPC) remain poorly understood. We report here a screening strategy developed to assess modulators of human hematopoiesis using a lentiviral short hairpin RNA (shRNA) library transduced into cord blood-derived stem/progenitor cells. To screen for modifiers of self-renewal/differentiation, we used the limited persistence of HSPCs under ex vivo culture conditions as a baseline for functional selection of shRNAs conferring enhanced maintenance or expansion of the stem/progenitor potential. This approach enables complex, pooled screens in large numbers of cells. Functional selection identified novel specific gene targets (exostoses 1) or shRNA constructs capable of altering human hematopoietic progenitor differentiation or stem cell expansion, respectively, thereby demonstrating the potential of this forward screening approach in primary human stem cell populations.
Introduction
Identifying the genes and pathways that regulate self-renewal and differentiation in somatic stem cells is a central goal in stem cell and cancer biology. Although the mechanisms that regulate stem cell fate have remained relatively poorly defined, intense efforts have been devoted to development of conditions that would enable amplification of transplantable stem cells, particularly in the field of hematopoiesis. Approaches using cytokines or molecules defined by their association with leukemia have provided considerable, but limited, insight.1
To date, most experimental approaches to study stem cells in mammalian systems have been limited to the assessment of individual candidate genes, generally in murine systems. The recent creation of large short hairpin RNA (shRNA) viral vector libraries, however, has now made it possible to perform unbiased forward genetic screens in mammalian cells.2-6 Most RNAi screens published to date are based on cell line systems or the use of cells with genetic alterations predisposing to a certain phenotype.2-6 Here we have explored the potential of pooled lentiviral shRNA libraries in screens for modifications that directly regulate the fate of primary human hematopoietic stem and progenitor cells (HSPC). HSPCs have several features that, compared with most other primary human cell types, make them particularly attractive for such screens. They are readily accessible from several sources such as umbilical cord blood (CB), bone marrow, or mobilized peripheral blood, and they can easily be purified and transduced with lentiviral vectors while retaining their fundamental properties.7 Moreover, HSPCs can be assayed in vivo and in vitro for a variety of functional outcomes. Importantly, however, when cultured without support from stroma cells, HSPCs can only maintain an undifferentiated state for a limited time. We decided to use this limited maintenance ex vivo as a basis for a screen aimed at finding modifiers of self-renewal/differentiation. We hypothesized that by targeting large numbers of cells with a pooled lentiviral shRNA library, it would be possible to functionally select for perturbations leading to prolonged maintenance or expansion of undifferentiated HSPCs.
Methods
Vector and virus production
The shRNA library used in this study was a pooled version of the Broad Institute RNAi Consortium (TRC) lentiviral library with approximately 7000 shRNA vectors targeting around 1300 human genes. The pLKO.1 self-inactivating lentiviral vector drives shRNA expression from a human U6 promoter and carries the puromycin-resistance gene under control of a phosphoglycerate kinase promoter.4 Lentiviruses were produced by transient transfection of the vector plamids in human 293T cells along with packaging plasmid (pCMVΔ8.9), and envelope plasmid (vsv-g pHCMV-G) provided by the RNAi Consortium. All shRNA sequences that were individually assessed in the study are listed in Table 1.
List of all shRNAs used in this study
| Name . | Target sequence (sense strand) . |
|---|---|
| shGFP | GGTGCGCTCCTGGACGTAGCC |
| shEXT1 | GAAGAACACAGCGGTAGGAAT |
| shEXT1–1 | AGAGCCAGATTGTGCCAACTA |
| shEXT1–2 | CCTGCTTCGATTTCACCCTTT |
| shEXT1–3 | CCCAACTTTGATGTTTCTATT |
| shEXT1–4 | GCACTTAGACAGCAGACACAA |
| shEXT1–5 | CCTTCGTTCCTTGGGATCAAT |
| shPLCZ1 | CGTGAATGTCTACTGTTTAAA |
| shSTK38 | CAGCAAGGGCCATGTGAAACT |
| shSTK38–1 | GCTCGGAAGGAAACAGAGTTT |
| shSTK38–2 | GCAGAAACAGTATTAGCCATA |
| shSTK38–3 | CCCACCAATATGTCATAGTAA |
| shSTK38–4 | CCCACCAAGTATTTCCTTTAT |
| shSTK38–5 | CAGAGACATCAAACCAGACAA |
| shSTK38–6 | CATGTGAAACTTTCTGACTTT |
| shEGLN3 | CTACGTCAAGGAGAGGTCTAA |
| shMLH1 | TATTCCATCCGGAAGCAGTAC |
| shABCC12 | GCGATGTTACTAGCAGCAGAA |
| Name . | Target sequence (sense strand) . |
|---|---|
| shGFP | GGTGCGCTCCTGGACGTAGCC |
| shEXT1 | GAAGAACACAGCGGTAGGAAT |
| shEXT1–1 | AGAGCCAGATTGTGCCAACTA |
| shEXT1–2 | CCTGCTTCGATTTCACCCTTT |
| shEXT1–3 | CCCAACTTTGATGTTTCTATT |
| shEXT1–4 | GCACTTAGACAGCAGACACAA |
| shEXT1–5 | CCTTCGTTCCTTGGGATCAAT |
| shPLCZ1 | CGTGAATGTCTACTGTTTAAA |
| shSTK38 | CAGCAAGGGCCATGTGAAACT |
| shSTK38–1 | GCTCGGAAGGAAACAGAGTTT |
| shSTK38–2 | GCAGAAACAGTATTAGCCATA |
| shSTK38–3 | CCCACCAATATGTCATAGTAA |
| shSTK38–4 | CCCACCAAGTATTTCCTTTAT |
| shSTK38–5 | CAGAGACATCAAACCAGACAA |
| shSTK38–6 | CATGTGAAACTTTCTGACTTT |
| shEGLN3 | CTACGTCAAGGAGAGGTCTAA |
| shMLH1 | TATTCCATCCGGAAGCAGTAC |
| shABCC12 | GCGATGTTACTAGCAGCAGAA |
shRNA indicates short hairpin RNA.
Isolation, culture and transduction of cells
Mononuclear cells were isolated from umbilical cord blood on Ficoll gradient and CD34+ cells were isolated from the mononuclear cells by double MACS columns (Miltenyi Biotec, Bergisch Gladbach, Germany) as previously described.8 Cells were cultured in SFEM StemSpan medium (StemCell Technologies, Vancouver, BC), with recombinant human thrombopoietin, stem cell factor, and Flt-3 ligand (all from R&D Systems, Minneapolis, MN) at 100 ng/mL each. Cells (200 000 cells per well) were cultured for 1 day and then transduced overnight in 24-well plates coated with RetroNectin (Takara Bio, Otsu, Japan) at a multiplicity of infection (MOI) of 2, rendering 20% to 30% transduction efficiency of the CD34+ cells.7 After transduction, cells were replated in fresh StemSpan medium and maintained with biweekly half-media changes and wells were split as needed. Cord blood samples were obtained by a protocol approved by the Institutional Review Board of Massachusetts General Hospital/Harvard Medical School.
CFC and LTC-IC assays
Colony-forming cell (CFC) assays were established by plating cells in methylcellulose medium H4445 (StemCell Technologies) with or without 2.5 μg/mL puromycin. Cells were plated according to day 0 cell counts. Two weeks after plating, cultures were evaluated for the presence of colony-forming units granulocyte-macrophage (CFU-GM), CFU-erythroid (CFU-E), and burst-forming units erythroid (BFU-E). For long-term culture initiating cell (LTC-IC) assays, AFT024 stromal cells (Dr K. Moore, Mount Sinai School of Medicine, New York, NY) were grown to confluence in 96-well plates and irradiated at 1500 rad 24 hours before establishing LTC-IC cultures at limit dilution as previously described.8 LTC-IC cultures were maintained for 5 weeks, and the medium was then replaced with clonogenic methylcellulose medium. After 2 weeks, wells were evaluated for the presence of hematopoietic colonies.
Transplantation assays
Cells were intravenously injected into 8- to 12-week-old sublethally irradiated (350 rad) NOD/SCID mice. Contribution of human cells from bone marrow (BM) was measured by FACS after 7 weeks. For secondary transplantations, a half femur equivalent of BM cells from primary transplanted mice was injected into new mice. All mice studies were performed at Lund University and were approved by the local ethical committee (approval no. M211-06).
FACS analysis
Cells were stained with anti–human CD34, CD38, CD33, CD45, CD19, and CD15, all APC, PE, or FITC (Becton Dickinson, Franklin Lakes, NJ) and analyzed using a FACSCalibur (Becton Dickinson).
PCR, cloning, and sequencing
To sequence proviral shRNA inserts, methylcellulose colonies from each screening pool were harvested for DNA extraction. Polymerase chain reaction (PCR) amplification of proviral shRNAs was performed using the primer pair 5′-GGATGAATACTGCCATTTGTCTCG-3′ (left) and 5′-AGGCCGAAGGAATAGAAGA-3′ (right). PCR products were cloned using TOPO TA kit (Invitrogen, Carlsbad, CA) and sequenced (10-12 clones per pool). Quantitative reverse transcription (RT)–PCR was performed using Taqman kits from Applied Biosystems (Foster City, CA).
Results
RNAi screen in human CB CD34+ cells
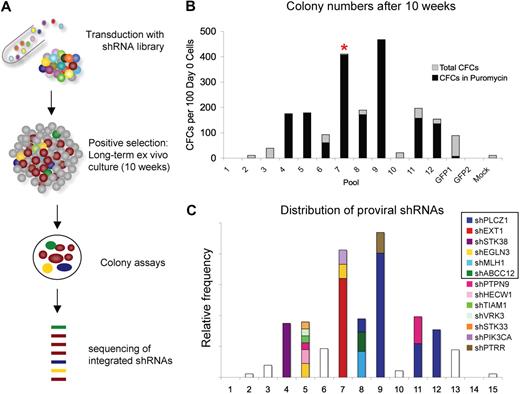
On the basis of the limited ability of HSPCs to sustain primitive potential under ex vivo culture conditions, we designed a strategy for pooled screens where perturbations conferring enhanced self-renewal/proliferation potential could be detected by positive selection (Figure 1A). A pooled version of a lentiviral shRNA library from the Broad RNAi Consortium (TRC) targeting 1300 human genes (mainly kinases, phosphatases, and proteases), with on average 5 shRNA vectors per gene, was used in this study.4 The library vector confers puromycin resistance. Twelve pools of umbilical cord blood derived CD34+ cells (200 000 cells per pool) were each transduced with an aliquot of the complete pooled shRNA library and subsequently passaged in long-term liquid cultures (10 weeks) followed by CFC assays to positively select for clones that had acquired enhanced proliferation ability combined with a sustained primitive potential. It should be noted that under the conditions used persistence of cells with this phenotype is extremely rare, providing an attractive baseline for the screening method. The transduction efficiency was kept relatively low (20%-30%) to avoid doubly transduced cells. After 10 weeks in culture, 7 of the 12 library-transduced pools exhibited a 2- to 5-fold increase in the number of CFCs compared with control-transduced pools (Figure 1B). All of these 7 pools now contained close to 100% puromycin resistant cells showing that the long-term culture had mediated a selection for library-transduced cells (Figure 1B). Of note, 1 pool (no. 7) exhibited high levels of burst-forming-unit/colony-forming unit erythroid (BFU/CFU-E) colonies in contrast to the predominantly myeloid-type colonies seen in the other pools. The colonies from each screening pool were harvested in bulk for DNA extraction and subsequent PCR amplification of the proviral inserts. The PCR products were cloned in bacteria, and 10 to 12 clones per pool were sequenced to determine the distribution of shRNAs. Figure 1C shows the distribution of shRNAs between the different pools. The most prominent hits were shEXT1 (exostoses 1) which dominated pool 7, shPLCZ1 (phospholipase C zeta 1) which dominated pools 9 and 12 and was present in pools 8 and 11, and shSTK38 (serine threonine kinase 38) which dominated pool 4. For functional validation experiments with individual shRNAs, we selected these 3 hits along with 4 additional shRNAs that either showed a frequency of more than 30% in a given pool or could be detected in more than one screening pool (shPTPN9, shEGLN3, shMLH1, and shABCC12). The shPTPN9 vector did not, however, produce sufficient virus titers for the validation assays.
RNAi screen in human CB CD34 cells. (A) Overall design of the screening strategy. Large numbers of primary CB CD34+ cells are infected with the lentiviral short hairpin RNA (shRNA) library and subsequently passaged in long-term cultures (10 weeks) followed by colony-forming cell (CFC) assays to positively select for clones that have acquired enhanced self-renewal/proliferation ability. Potential hits are identified by sequence analysis of proviral inserts from the selected cells. (B) CFC levels after 10 weeks culture. Twelve pools of library-transduced cells were independently assayed. Control pools were transduced with shRNA against GFP in 2 pools, and 1 pool was left untransduced (mock). Pool 7 showed high levels of BFU-E growth indicated by *. (C) Distribution of proviral shRNAs among the screening pools that showed increased CFC levels. The graph shows relative abundance of shRNAs in each screening pool as an overlay on the colony numbers shown in panel B.
RNAi screen in human CB CD34 cells. (A) Overall design of the screening strategy. Large numbers of primary CB CD34+ cells are infected with the lentiviral short hairpin RNA (shRNA) library and subsequently passaged in long-term cultures (10 weeks) followed by colony-forming cell (CFC) assays to positively select for clones that have acquired enhanced self-renewal/proliferation ability. Potential hits are identified by sequence analysis of proviral inserts from the selected cells. (B) CFC levels after 10 weeks culture. Twelve pools of library-transduced cells were independently assayed. Control pools were transduced with shRNA against GFP in 2 pools, and 1 pool was left untransduced (mock). Pool 7 showed high levels of BFU-E growth indicated by *. (C) Distribution of proviral shRNAs among the screening pools that showed increased CFC levels. The graph shows relative abundance of shRNAs in each screening pool as an overlay on the colony numbers shown in panel B.
Validation of candidate shRNAs
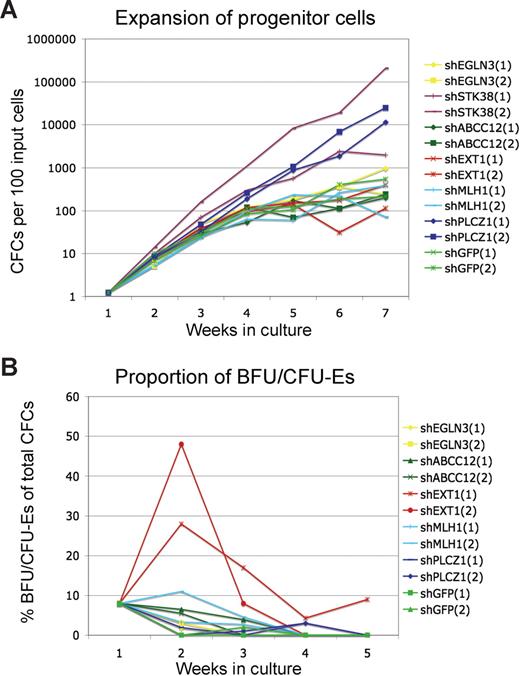
Figure 2 summarizes the outcome of the validation assays. Cells transduced with either shPLCZ1 or shSTK38 exhibited a more than 1000-fold increased expansion of progenitors during culture (Figure 2C). Despite an exponential growth rate for several weeks, these progenitor cells had not become immortalized and subsequently stopped growing after 12 to 15 weeks. Cells transduced with shEXT1 did not show the same overall expansion but produced dramatically higher frequencies of BFU/CFU-Es (Figure 2B), similar to the observation in the primary screen. Interestingly, the marked effect of shEXT1 on BFU/CFU-E formation was only seen after 1 to 2 weeks, whereas in the initial screen, this was observed after 10 weeks of culture. Although we occasionally saw increased BFU/CFU-E frequency at later time points, the only consistent effect was at 1 to 2 weeks. It is possible that the effect at later time points required an optimal integration site of the vector or an initial target cell with certain rare qualities. Nevertheless, the 3 most prominent hits from the screen represented strong and reproducible perturbations specifically mediated by shRNAs.
Validation of candidate shRNAs. Cells were transduced with individual shRNA vectors and cultured for several weeks. Total colony-forming cell numbers (A) as well as the frequency of CFU-E/BFU-E colonies (B) were measured weekly. Results from 2 experiments are shown.
Validation of candidate shRNAs. Cells were transduced with individual shRNA vectors and cultured for several weeks. Total colony-forming cell numbers (A) as well as the frequency of CFU-E/BFU-E colonies (B) were measured weekly. Results from 2 experiments are shown.
Knockdown of EXT1 leads to increased BFU/CFU-E production
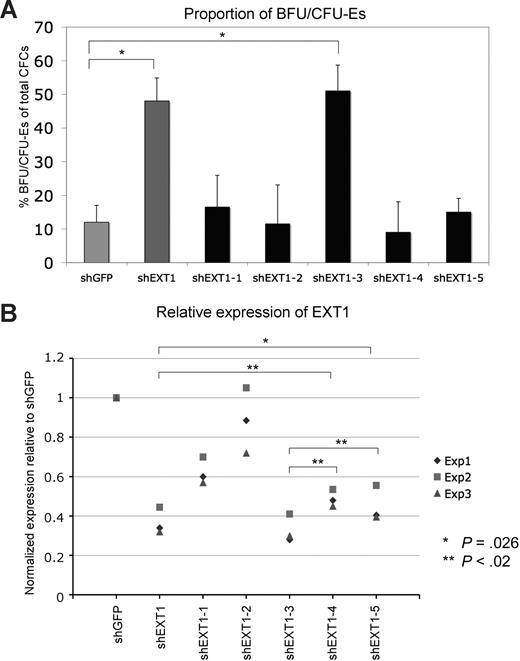
We next asked whether the higher levels of BFU/CFU-E colonies triggered by shEXT1 could be reproduced with additional non-overlapping shRNAs targeting EXT1. The library contained 5 additional shRNAs against EXT1 that we tested functionally in the BFU/CFU-E assays as well as for knockdown of the EXT1 transcript. It should be noted that the aim when assessing knockdown is to test relative efficiency of the different shRNAs and that the level of transcript suppression must be interpreted with caution when testing heterogenous populations of primary cells. The level of knockdown in each cell will vary depending on differentiation stage, vector copy number, and integration site. The knockdown values from the transduced populations can mainly be used as a guide for relative potency of shRNAs rather than as an indicator of absolute levels.
We found one additional shRNA that conferred the erythroid phenotype and also knocked down the EXT1 transcript with similar efficiency as the original construct (Figure 3A,B). The other 4 constructs did not render a phenotype and showed significantly less efficient knockdown (Figure 3B).
Target validation for EXT1. (A) shRNA vectors targeting different regions of the EXT1 transcript were tested for their ability to induce an effect on erythroid progenitor activity as measured by frequency of erythroid progenitors 1 week after transduction Results are given as mean values (± SEM); *P < .05 (Student t test). (B) The same constructs were tested for their ability to knock down the EXT1 transcript as measured by quantitative (q)–PCR. Results from 3 independent experiments are shown. To assess differences in relative knockdown efficiency, a paired t test was used to test significance when comparing shEXT1 and shEXT1-3 with any of the other shRNAs. P values are indicated for the comparisons with shEXT1-4 and shEXT1-5. For comparisons with the other shRNAs P values were below .02.
Target validation for EXT1. (A) shRNA vectors targeting different regions of the EXT1 transcript were tested for their ability to induce an effect on erythroid progenitor activity as measured by frequency of erythroid progenitors 1 week after transduction Results are given as mean values (± SEM); *P < .05 (Student t test). (B) The same constructs were tested for their ability to knock down the EXT1 transcript as measured by quantitative (q)–PCR. Results from 3 independent experiments are shown. To assess differences in relative knockdown efficiency, a paired t test was used to test significance when comparing shEXT1 and shEXT1-3 with any of the other shRNAs. P values are indicated for the comparisons with shEXT1-4 and shEXT1-5. For comparisons with the other shRNAs P values were below .02.
This strongly suggests that the effect on BFU/CFU-E production is specifically mediated by the loss of EXT1. EXT proteins are required for the biosynthesis of heparan sulfate proteoglycans, and studies in Drosophila have shown that they thereby regulate the presentation of certain secreted growth factors such as Wnt and TGF-beta at the cell surface.9,10 We are currently investigating this potential link between EXT1 and growth factor signaling in hematopoietic cells.
shSTK38 expands candidate human hematopoietic stem cells
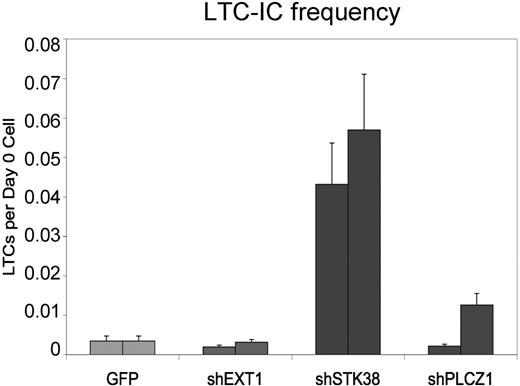
To further characterize the dramatic expansion effects from shPLCZ1 and shSTK38 and assess whether self-renewal of stem cells was affected, we performed long-term culture initiating cell (LTC-IC) assays that can detect cells in a quantitative manner that are primitive and may reflect repopulating HSPCs.11 Cells transduced with shSTK38, but not shPLCZ1 or shEXT1, displayed significantly higher numbers of LTC-ICs, as measured by limiting dilution assays after 1 week in culture, indicative of an immediate effect on HSPC self-renewal (Figure 4). We, therefore, focused our investigations on shSTK38 and first attempted to validate the target identity using 6 additional shRNA vectors against STK38. While several constructs knocked down the STK38 transcript to similar levels or even better than the original vector (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), they did not trigger a similar expansion effect, suggesting that the phenotype is mediated, at least partly, through an “off-target effect” on another gene. We were, however, still intrigued by the effect from shSTK38 because it appeared to be modifying HSPC self-renewal and decided to further study its effects. To more stringently assess the potential for stem cell expansion, shSTK38 transduced CD34+ cells were transplanted into NOD/SCID mice either immediately after transduction or after 11 and 26 days of ex vivo culture. The level of human engraftment 7 weeks after transplantation for 50 000 input cells transplanted 1 day after transduction did not differ between control and test cells (Figure 5A). When transplanting the progeny of only 1000 input cells after 26 days of culture, however, we observed higher engraftment of shSTK38 transduced cells compared with the 50-fold higher input dose transplanted at day 1 (Figure 5A). These data indicate a dramatic (> 50-fold) expansion of NOD/SCID repopulating activity over 26 days of culture. The cells expanded by shSTK38 were fully functional NOD/SCID repopulating cells as defined by both B-lymphoid and myeloid engraftment (Figure 5B) and also showed low but clearly detectable engraftment in secondary recipients (Figure 5E) consistent with stem cell self-renewal. Thus, we could show by assays stringently assessing for stem cell rather than progenitor cell function that shSTK38 induces substantial ex vivo expansion of human hematopoietic stem cells.
Markedly increased numbers of LTC-ICs induced by shSTK38. The frequency of LTC-ICs 1 week after transduction is shown for 2 independent experiments. The error bars give the 95% confidence interval from the limiting-dilution LTC-IC calculations.
Markedly increased numbers of LTC-ICs induced by shSTK38. The frequency of LTC-ICs 1 week after transduction is shown for 2 independent experiments. The error bars give the 95% confidence interval from the limiting-dilution LTC-IC calculations.
Expansion of NOD/SCID repopulating cells by shSTK38. (A) Transduced CD34+ cells were transplanted into NOD/SCID mice, either 1 day after transduction or after 11 and 26 days of ex vivo culture. The transplanted cell doses were expansion equivalents of the indicated numbers of input day 0 CD34+ cells before transduction. The level of human engraftment in bone marrow 7 weeks after transplantation is shown. (B) Day 26 shSTK38 expanded cells show lymphoid and myeloid engraftment in NOD/SCID mice. FACS plots from a representative mouse. (C) Day 26 shSTK38 expanded cells engraft in secondary recipient NOD/SCID mice. Half a femur equivalent of bone marrow from primary recipient mice was transplanted to a total of 10 secondary recipients. Engraftment levels were analyzed in bone marrow after 7 weeks. FACS plots showing engrafted human cells in 1 of 2 recipients with a clear contribution of human cells.
Expansion of NOD/SCID repopulating cells by shSTK38. (A) Transduced CD34+ cells were transplanted into NOD/SCID mice, either 1 day after transduction or after 11 and 26 days of ex vivo culture. The transplanted cell doses were expansion equivalents of the indicated numbers of input day 0 CD34+ cells before transduction. The level of human engraftment in bone marrow 7 weeks after transplantation is shown. (B) Day 26 shSTK38 expanded cells show lymphoid and myeloid engraftment in NOD/SCID mice. FACS plots from a representative mouse. (C) Day 26 shSTK38 expanded cells engraft in secondary recipient NOD/SCID mice. Half a femur equivalent of bone marrow from primary recipient mice was transplanted to a total of 10 secondary recipients. Engraftment levels were analyzed in bone marrow after 7 weeks. FACS plots showing engrafted human cells in 1 of 2 recipients with a clear contribution of human cells.
Discussion
Modifying HSPC function to achieve specific differentiation events or to expand the number of engrafting repopulating cells is of considerable importance for the field of hematopoiesis. Unbiased candidate modifiers screened against a functional outcome of interest has been extremely powerful in other systems, and we tested the feasibility of such an approach in primary human stem/progenitor cells. We identified what might be considered 3 types of outcomes associated with 3 specific shRNAs that are informative about such screens. First, shRNA targeting EXT1 appears to induce an EXT1 specific effect and is an example of an unpredicted candidate gene target that alters at least ex vivo hematopoietic activity. Second, shPLCZ1 modified a population of progenitor cells in vitro that did not score in subsequent more stem cell specific assays. Third, shSTK38 revealed a consistent and potent effector of human hematopoietic stem cells resulting in dramatic expansion of the cells. Yet, its specific molecular targets remain ill-defined and do not appear to be the sequence against which the shRNA was engineered. This outcome is of greatest interest by virtue of its functional effect but represents what is the equivalent of a chemical modifier without known gene target. It will be of considerable utility as a reagent for identifying the gene products regulating profound human hematopoietic stem cell expansion. It may also be of interest as a sequence used in a non-integrating adenoviral vector or a synthetic siRNA molecule to alter primitive human hematopoietic cells.12,13
Two general aspects about the screening strategy are important to consider. First, while this first screen most likely had coverage of the entire library (7000 shRNAs) in progenitor cells (CFCs), it did not saturate the library with respect to the targeting of the most primitive populations such as LTC-ICs and SCID repopulating cells (SRCs). Based on our experience from assaying these populations from fresh CB CD34+ cells and given a 30% transduction efficiency of the shRNA library, we estimate that this screen had a 5-fold coverage of the library in CFCs, 50% coverage in LTC-ICs, and 10% coverage in SRCs. Thus, additional screens with higher cell numbers will be necessary for a deep assessment of the library in the most primitive populations. Second, considering the use of integrating viral vectors and the selective pressure in our screening assays, functional outcomes could be associated with insertional mutagenesis events, for example, from activation of proto-onocogenes.14 We, therefore, transduced the cells at a relatively low MOI to avoid multiple integrations of the library vectors. It is still a possible that the candidate shRNAs not showing a clear phenotype in the validation experiments initially scored due to insertional mutagenesis. The most prominent outcomes from the primary screen, however, were not associated with such mechanisms but were specifically mediated by shRNA. It should also be noted that all phenotypes reported here for individual shRNAs were consistently reproduced in multiple experiments, arguing against any involvement of lentiviral integration effects at that level. Taken together, we have applied an unbiased forward RNAi screen in primary human HSPCs and conclude that the approach holds great promise for the further unraveling of regulatory mechanisms in human stem cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (Bethesda, MD) grants to D.T.S. and grants from the Swedish Cancer Foundation (Stockholm, Sweden) and the Swedish Childhood Cancer Foundation (Stockholm, Sweden) to J.L.
National Institutes of Health
Authorship
Contribution: N.A. and J.L. performed the majority of laboratory experiments; C.K. performed target validations for shSTK38; M.A. produced viruses and performed qPCR; G.H., N.H., D.T.S, and J.L. designed and planned the study; N.H. provided the lentiviral shRNA library and shRNA vectors for validation; and N.A., D.T.S., and J.L. prepared the manuscript.
Conflict-of-interest disclosure: D.T.S. is a stockholder and consultant for Fate Therapeutics and a consultant for Genzyme and VasGen. The remaining authors declare no competing financial interests.
Correspondence: David T. Scadden, Center for Regenerative Medicine, 185 Cambridge St, CPZN-4265A, Boston, MA 02114; e-mail: scadden.david@mgh.harvard.edu; or Jonas Larsson, Molecular Medicine and Gene Therapy, BMC A12, 221 84 Lund, Sweden; e-mail: jonas.larsson@med.lu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal