Abstract
Retinoids triggers differentiation of acute promyelocytic leukemia (APL) blasts by transcriptional regulation of myeloid regulatory genes. Using a microarray approach, we have identified a novel retinoid-responsive gene (CXXC5) encoding a nuclear factor, retinoid-inducible nuclear factor (RINF), that contains a CXXC-type zinc-finger motif. RINF expression correlates with retinoid-induced differentiation of leukemic cells and with cytokine-induced myelopoiesis of normal CD34+ progenitors. Furthermore, short hairpin RNA (shRNA) interference suggests for this gene a regulatory function in both normal and tumoral myelopoiesis. Interestingly, RINF localizes to 5q31.3, a small region often deleted in myeloid leukemia (acute myeloid leukemia [AML]/myelodysplasia [MDS]) and suspected to harbor one or several tumor suppressor gene.
Introduction
Differentiation of hematopoietic stem cells to terminally mature granulocytes is a multistage process requiring coordinate expression of genes orchestrated by lineage-restricted transcription factors.1 So far, a relatively small number of transcription factors have been demonstrated to be essential for myelopoiesis.1,2 Deregulation or mutations of these factors can switch the cell fate from differentiation to proliferation and contribute to acute myeloid leukemia (AML), a group of malignant hemopathies characterized by a maturation arrest and an accumulation of immature blasts in the bone marrow, blood, and other tissues.
The AML-M3 subtype (according to the French-American-British [FAB] classification) also known as acute promyelocytic leukemia (APL), corresponds to clonal expansion of leukemic blasts blocked at the promyelocytic stage of granulocytic differentiation. This pathology whose genetic hallmark is the t(15;17) translocation, represents the first cancer treated by a transcription-based therapy reestablishing terminal differentiation. Indeed, in this pathology, pharmacologic doses of all-trans retinoic acid (ATRA) trigger terminal maturation of leukemic blasts.3,4 At the molecular level, ATRA is known to act as a ligand for retinoic acid receptors (RARα, PML-RARα) in APL cells and regulates transcriptional activation of downstream target genes. Despite extensive studies using an experimental model, the NB4 cell line,5 only a few target genes encoding transcription factors have so far been shown to be really essential for retinoid-induced differentiation of promyelocytic cells.6-8
In the present study, using a microarray approach, we searched for novel genes early induced by retinoids in NB4 cells and encoding transcription factors potentially involved in the reestablishment of terminal differentiation by ATRA. Nine gene candidates were identified. One of these 9 genes induced by retinoid treatment encodes a nuclear factor that we named retinoid-inducible nuclear factor (RINF). RINF invalidation by RNA interference (RNAi) abrogates the differentiating action of ATRA in NB4 cells by enforcing a retinoid-resistance phenotype and delays cytokine-induced granulocytic differentiation of normal CD34+ myeloid progenitors, thus suggesting a key and general regulatory role for RINF in myeloid differentiation.
Methods
Cell culture, treatments, and RNA preparation
Human breast carcinoma cells (MCF7) and myeloid cells (NB4, NB4-LR1, NB4-LR2, K562, LAMA-84, and HL60) were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Biochrom AG, Berlin, Germany), 2 mM L-glutamine, 50 U/mL penicillin G, and 50 μg/mL streptomycin (Invitrogen) and were incubated at 37°C in the dark, in a 5% CO2-humidified atmosphere. For in vitro expansion of human bone marrow primary CD34+ cells (StemCell Technologies, Vancouver, BC), we supplemented the above medium with 20 ng/mL interleukin 3 (IL3), 20 ng/mL granulocyte colony-stimulating factor (G-CSF), and 50 ng/mL stem cell factor (SCF) purchased from Peprotech (Rocky Hill, NJ). Maturation was evaluated by morphology with May-Grünwald-Giemsa (MGG) staining and by nitroblue tetrazolium (NBT) reduction assay as previously described.9 Cell density was determined using a Coulter counter (Beckman Coulter, Fullerton, CA). Cell proliferation was represented as population doublings (PD) calculated by the formula: PD = Log (N/No)/Log2, where N is the number of cells counted and No the number of cells seeded at day 0. Cells treated or not with ATRA (Sigma-Aldrich, St Louis, MO) were collected together and directly stored at −80°C for RNA preparation with Trizol (Invitrogen) or the RNeasy mini kit (QIAGEN, Valencia, CA). Yield and quality of the extracted RNA was evaluated by the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Microarray hybridization, data analysis, and gene classification
All microarray experiments were performed using Human Genome Survey Microarray V1.0 chips (Applied Biosystems, Foster City, CA) that contain 31 700 oligonucleotide probes (60-mer) representing 27 868 individual human genes. A total of 6 microarrays were used for the analysis that was performed using J-Express Pro V2.7 software (MolMine, Bergen, Norway).10 Minimum information about a microarray experiment (MIAME)–compliant documentation of the microarray experiments have been deposited in Array Express at the European Bioinformatics Institute (http://www.ebi.ac.uk/arrayexpress) under the accession number E-BASE-7. A detailed description of the protocol can be found in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Quantitative reverse transcription polymerase chain reaction
First-strand cDNA synthesis (RT) were carried out starting with total RNA (0.1 to 1 μg) in a 20-μL volume using oligo-dT primers with Transcriptor Reverse Transcriptase (Roche, Indianapolis, IN) in accordance with the manufacturer's instructions. Quantitative polymerase chain reactions (PCRs) were performed using the SYBRgreen detection kit on a Light Cycler 480 machine (Roche). All specific primers and conditions are available on request. For each gene, relative mRNA expressions were normalized to RPLP2 gene expression.11
Chromatin immunoprecipitation
Briefly, NB4 cells were cross-linked with 1% formaldehyde, their nuclei isolated, resuspended in chromatin immunoprecipitation (ChIP) lysis buffer (0.1% sodium dodecyl sulfate [SDS], 0.1% Na-deoxycholate, 1% Triton X-100, 1 mM ethylenediaminetetraacetic acid [EDTA], 140 mM NaCl, 10 mM Tris, pH 8.0, and protease inhibitor cocktail) and sonicated to obtain DNA fragments of 500-1000 bp in length. Anti-RARα (Abcam-H1920) or anti-PML antibodies (sc-966; Santa Cruz Biotechnology, Santa Cruz, CA) were added to the fragmented chromatin mixture for IP that was performed by adding salmon sperm DNA/protein A agarose beads (Upstate, Millipore, Billerica, MA). Immunoprecipitated DNA fragments, input, and no Ab fractions were then recovered, uncross-linked (by addition of salt and heating), and purified by the phenol-chloroform method before being analyzed by semiquantitative PCR using pairs of primers that encompass the retinoic acid responsive elements located in promoter regions of RINF or RARβ2. A more detailed description of the protocol (including primers sequences) is available online (Document S1).
Western blot analysis
Western blot analysis was carried out as previously described.12 Customized rabbit polyclonal peptide-specific antibody against RINF was produced by Biogenes (Berlin, Germany). The immunogen peptide corresponds to amino acids 45-58 of the RINF protein. Antibody specificity was confirmed by competitive inhibition of the Western blot analysis signal by addition of the immunogene peptide to the primary antibody solution. Briefly, blots were incubated with primary antibody against RINF (polyclonal antibody), poly(ADP-ribose) polymerase (PARP; monoclonal mouse immunoglobulin G [IgG], no. AM30; Calbiochem, San Diego, CA), or actin (polyclonal rabbit IgG, no. A2066; Sigma-Aldrich), and then with an appropriate peroxydase-conjugated secondary antibody. Detection of proteins was performed using a chemiluminescent detection system (Amersham Pharmacia Biotech, Uppsala, Sweden). Blot with human tissue extracts was purchased from Millipore (TB300) that provided us the actin loading control. The protocol describing the preparation of cytoplasmic and nuclear fractions is available online (Document S1).
Immunofluorescence
Plasmid encoding Flag-tagged RINF was constructed from NB4 cells cDNA. PCR products were inserted into the pFlag-CMV-4 expression vector (Sigma-Aldrich). MCF7 cells were grown on coverslips and transfected with Flag-tagged RINF constructs according to the Fugene HD manufacturer's protocol (Roche). Two days posttransfection, cells were washed once in phosphate-buffered saline (PBS) and fixed for 15 minutes in 2% paraformaldehyde. Fixed cells were washed 3 times in PBS, permeabilized in 0.1% Triton X-100 for 10 minutes, and then incubated in blocking buffer (PBS, 2% BSA) for 30 minutes. Cells were then incubated overnight with the primary antibody (rabbit polyclonal anti-Flag M2; Sigma-Aldrich) diluted 1:3000 in blocking buffer (PBS, 2% BSA), washed twice in PBS, and incubated with the secondary antibody (Alexa-488–conjugated anti-rabbit from Molecular Probes, Invitrogen) diluted 1:1000 (PBS, 2% BSA), for 1 hour at room temperature. Subsequently, cells were washed 3 times in PBS and were mounted in Vectashield mounting medium with 4′,6-diaminidine-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) to counterstain nuclei. Immunofluorescent images were acquired by confocal microscopy on a Zeiss LSM510 META confocal laser microscope with a Plan Apochromat 63×/1.4 NA oil-immersion objective using the LSM510 software v4.0 (Carl Zeiss, Thornwood, NY).
Plasmids for lentiviral infections
Lentiviral plasmids (pLKO.1/short hairpin RNA [shRNA]/RINF) targeting RINF expression were purchased from Sigma-Aldrich (MISSION shRNA Bacterial Glycerol Stock), and control vectors (pLKO.1/TRC and pLKO.1/shRNA/scramble controls) were kindly provided by David Root13 and David M. Sabatini14 (both from Massachusetts Institute of Technology, Cambridge, MA) through a material transfer agreement (Addgene plasmids 10879 and 1864). Briefly, production of lentiviral particles were performed by transient cotransfection (Fugene HD) of HEK 293T cells with the second generation packaging system (eg, packaging plasmid psPAX2 and envelope plasmid pMD2.G) developed by D. Trono's laboratory (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; Addgene plasmids 12260 and 12259). Viral supernatants were harvested and filtered 2 days posttransfection and then applied to growing cells for spin infection (2400 rpm for 1 hour at room temperature), which was carried out in presence of 5 μg/mL proteamine sulfate. Two days after infection, NB4 cells were selected for at least 2 days with puromycine (Sigma-Aldrich) at 1 μg/mL.
URLs
Exon-intron structure and genomic organization of CXXC5 gene was performed using fast DB15 (http://www.fast-db.com). Theoretical molecular mass of RINF protein was calculated at Expert Protein Analysis System (ExPASy) proteomic server website (http://www.expasy.ch/tools/pi_tool.html). In silico analysis of the putative nuclear localization signal (NLS) motif was performed using PredictNLS (http://cubic.bioc.columbia.edu/predictNLS/). Microarray data from patients can be accessed from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) and Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
Results
Identification of CXXC5, a gene early induced by retinoic acid in the NB4 cell line
To identify new target genes of retinoic acid in APL, we performed microarray experiments using NB4 cells treated during short periods of time (90 minutes and 3 hours) with 1 μM ATRA. Microarray expression analysis allowed us to identify 35 genes consistently up-regulated at 90 minutes of treatment, with at least a 2-fold induction compared with untreated control, and that remained up-regulated after 3 hours (Table S1). Upon careful analysis and classification of these 35 genes, 9 genes encoding well-known or putative transcription factors (Table 1) were identified. The other 26 genes, listed in Table S2, did not meet the criteria for being putative transcription factors according to gene ontology and/or sequence analysis. Seven of the 9 candidate genes encode well-characterized transcription factors, known to be expressed in the myeloid tissue during granulocytic6,16-20 or monocytic21,22 differentiation. Importantly, 6 of these genes have previously been identified as early induced by retinoic acid in NB4 cells, 5 being direct targets (Table 1). These observations confirmed and validated the accuracy of our microarray screen.
List of (putative) transcription factors identified as early induced by retinoic acid in NB4 cells
| Gene symbol . | Full name . | Accession no. . | Direct target of retinoic acid* . | Expressed in myeloid tissue‡ . | Reference(s) . |
|---|---|---|---|---|---|
| CEBPB | CAAT/enhancer binding protein, beta | NM_005194 | Yes† | Yes | Duprez et al6 |
| CHOP | C/EBP-homologous protein | NM_004083 | Yes | Yes | Gery et al16 |
| ID1 | Inhibitor of DNA-binding 1 | NM_002165 | Yes | Yes | Nigten et al17 ; Jankovic et al18 |
| ID2 | Inhibitor of DNA-binding 2 | NM_002166 | Yes | Yes | Nigten et al17 |
| IRF1 | Interferon regulatory factor 1 | NM_002198 | Yes | Yes | Matikainen et al19 |
| BHLHB2 | Basic helix-loop-helix domain containing, class B, 2 | NM_003670 | Yes | ?§ | Boudjelal et al20 |
| ZC3H12A | Zinc-finger CCCH-type containing 12A | NM_025079 | ? | Yes | Zhou et al21 ; Liang et al22 |
| ZNF767 | Zinc-finger containing protein 767 | NM_024910 | ? | ? | |
| CXXC5 (RINF) | CXXC zinc-finger 5 (retinoid-inducible nuclear factor) | NM_016463 | ? | ? |
| Gene symbol . | Full name . | Accession no. . | Direct target of retinoic acid* . | Expressed in myeloid tissue‡ . | Reference(s) . |
|---|---|---|---|---|---|
| CEBPB | CAAT/enhancer binding protein, beta | NM_005194 | Yes† | Yes | Duprez et al6 |
| CHOP | C/EBP-homologous protein | NM_004083 | Yes | Yes | Gery et al16 |
| ID1 | Inhibitor of DNA-binding 1 | NM_002165 | Yes | Yes | Nigten et al17 ; Jankovic et al18 |
| ID2 | Inhibitor of DNA-binding 2 | NM_002166 | Yes | Yes | Nigten et al17 |
| IRF1 | Interferon regulatory factor 1 | NM_002198 | Yes | Yes | Matikainen et al19 |
| BHLHB2 | Basic helix-loop-helix domain containing, class B, 2 | NM_003670 | Yes | ?§ | Boudjelal et al20 |
| ZC3H12A | Zinc-finger CCCH-type containing 12A | NM_025079 | ? | Yes | Zhou et al21 ; Liang et al22 |
| ZNF767 | Zinc-finger containing protein 767 | NM_024910 | ? | ? | |
| CXXC5 (RINF) | CXXC zinc-finger 5 (retinoid-inducible nuclear factor) | NM_016463 | ? | ? |
As suggested in the literature by at least cycloheximide experiment (not necessarily for the presence of a putative retinoid-responsive element in the gene promoter).
CEBPB would be a direct target of retinoic acid in PML-RARα expressing cells (ie, in APL cells but apparently not in other cells).
As suggested at least by Northern blot, RT-PCR, and/or Western blot experiments (data that are only based on microarray experiments were not considered here).
A question mark (?) indicates that there is no evidence in the literature to our knowledge.
We focused our interest on CXXC5 (Table 1), a genomic sequence only mentioned previously in an in silico study23 and so far not subjected to functional characterization.
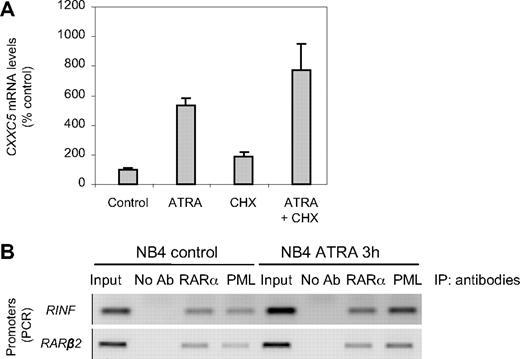
Our choice was made after 2 decisive experimental findings. First, quantitative RT-PCR analysis (Figure 1A) exquisitely confirmed ATRA induction of CXXC5 mRNA expression (here observed at 4 hours of retinoic acid treatment). Second, pretreatment of NB4 cells with the protein synthesis inhibitor cycloheximide, 30 minutes before retinoid addition and continued for 4 hours, did not block the CXXC5 mRNA up-regulation by ATRA (Figure 1A). These experiments indicate that ATRA-mediated transcription of CXXC5 does not require de novo protein synthesis and suggest that CXXC5 is a primary target of retinoic acid. This has been confirmed by ChIP experiments (Figure 1B) that demonstrated a direct binding of retinoid receptors (RARα and/or PML-RARα) to the promoter of CXXC5 gene in NB4 cells.
CXXC5 (RINF) is a direct target of retinoic acid. (A) RINF mRNA expression levels were measured by quantitative RT-PCR after 4 hours of 1 μM ATRA treatment. Inhibition of translation with cycloheximide (CHX) used at 10 μg/mL did not block ATRA-induced increase in RINF mRNA level, demonstrating that this process does not require de novo protein synthesis, and strongly suggests that RINF is a primary target of retinoic acid. The histogram represents means from 2 independent cell-culture treatments (± SEM). (B) In vivo binding of RARα and PML-RARα to the RINF promoter in NB4 cells. Cross-linked chromatin was prepared from NB4 cells treated or not with 1 μM ATRA for 3 hours and immunoprecipitated with anti-RARα or anti-PML antibodies. The precipitates were subjected to PCR analysis using primer pairs spanning the human RINF or RARβ2 gene promoters. The primers were designed to encompass the putative retinoid-responsive element found in the RINF promoter (ggagttcatgaggtgagc) or the well-established RARE (ggttcaccgaaagttca) in RARβ2 promoter (here used as a positive control); bold letters in the sequences represent the direct repeats. Input, PCRs performed on total chromatin from NB4 cells before IP; No Ab (no antibody control), PCRs performed on sample obtained after IP with an irrelevant antibody or without any antibody.
CXXC5 (RINF) is a direct target of retinoic acid. (A) RINF mRNA expression levels were measured by quantitative RT-PCR after 4 hours of 1 μM ATRA treatment. Inhibition of translation with cycloheximide (CHX) used at 10 μg/mL did not block ATRA-induced increase in RINF mRNA level, demonstrating that this process does not require de novo protein synthesis, and strongly suggests that RINF is a primary target of retinoic acid. The histogram represents means from 2 independent cell-culture treatments (± SEM). (B) In vivo binding of RARα and PML-RARα to the RINF promoter in NB4 cells. Cross-linked chromatin was prepared from NB4 cells treated or not with 1 μM ATRA for 3 hours and immunoprecipitated with anti-RARα or anti-PML antibodies. The precipitates were subjected to PCR analysis using primer pairs spanning the human RINF or RARβ2 gene promoters. The primers were designed to encompass the putative retinoid-responsive element found in the RINF promoter (ggagttcatgaggtgagc) or the well-established RARE (ggttcaccgaaagttca) in RARβ2 promoter (here used as a positive control); bold letters in the sequences represent the direct repeats. Input, PCRs performed on total chromatin from NB4 cells before IP; No Ab (no antibody control), PCRs performed on sample obtained after IP with an irrelevant antibody or without any antibody.
In silico characterization of CXXC5 gene
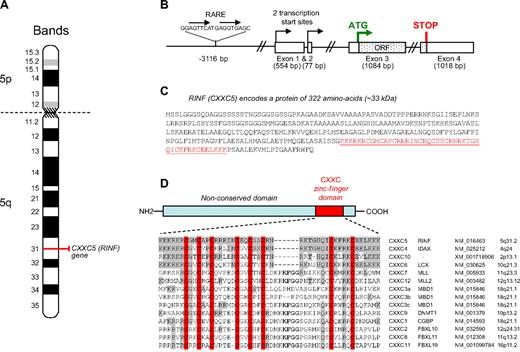
The main genomic features of CXXC5 (ENSG00000171604) available from databases are briefly summarized in Figure 2. The CXXC5 gene is located on the long arm of chromosome 5, at 5q31.3 (Figure 2A), a region frequently deleted in AML and myelodysplasia (MDS; see “Discussion”). The gene spans 35.5 kb and is organized in 4 exons (Figure 2B). The upstream promoter region of this gene has not yet been functionally analyzed, but relevant for our study, it contains a potential retinoid-responsive element at −3116 bp from the transcription start site. Despite the existence of 2 potential alternative transcription start sites, one in exon 1 and the other in exon 2, the 2 mRNA sequences reveals the same open reading frame with the start codon in exon 3. Conceptual translation predicts a protein of 322 amino acids (Figure 2C) and a theoretical molecular weight of 32.98 kDa. Protein sequence analysis revealed a unique conserved domain, a typical CXXC zinc-finger motif (Cx2Cx2Cx4-5Cx2Cx2C9-15Cx4C) between amino acids 257 and 302, in proximity to the C-terminal end. This motif known to contain 8 conserved cysteine residues coordinating 2 zinc ions24 is found in 11 other proteins (often chromatin-associated proteins) and is assumed to recognize unmethylated CpG.24-26 Figure 2D presents an alignment of all known members of the protein family bearing this motif.
CXXC5 gene structure and protein product (RINF). (A) The CXXC5 (RINF) gene localizes to chromosome 5q31.3. The gene starts 139 008 130 bp from pter and ends 139 043 651 bp from pter. It is oriented in plus strand direction and is 35.522 kb long. (B) Genomic organization of CXXC5 (RINF) gene. The gene is organized in 4 exons and 95% of the open reading frame is located in exon 3. A putative retinoic acid–responsive element (RARE) of direct repeat 2 (DR2) type is situated at −3116 bp upstream of the exon 1 transcription start site. (C) Predicted amino acid sequence of RINF according to one-letter amino acid representation. The open reading frame predicts a protein sequence of 322 amino acid residues and a theoretical molecular weight of 32.98 kDa (http://www.expasy.ch/tools/pi_tool.html). The zinc-finger domain is underlined in red. (D) Except the CXXC zinc finger domain, no conserved structural domain was found. An alignment of the CXXC motif of RINF protein with its human paralogs is presented. Amino acids that are invariant among CXXC motifs are in gray, and the conserved cysteine residues are in red. The consensus sequence for CXXC-type zinc-finger can be defined by Cx2Cx2Cx4-5Cx2Cx2C9-15Cx4C.
CXXC5 gene structure and protein product (RINF). (A) The CXXC5 (RINF) gene localizes to chromosome 5q31.3. The gene starts 139 008 130 bp from pter and ends 139 043 651 bp from pter. It is oriented in plus strand direction and is 35.522 kb long. (B) Genomic organization of CXXC5 (RINF) gene. The gene is organized in 4 exons and 95% of the open reading frame is located in exon 3. A putative retinoic acid–responsive element (RARE) of direct repeat 2 (DR2) type is situated at −3116 bp upstream of the exon 1 transcription start site. (C) Predicted amino acid sequence of RINF according to one-letter amino acid representation. The open reading frame predicts a protein sequence of 322 amino acid residues and a theoretical molecular weight of 32.98 kDa (http://www.expasy.ch/tools/pi_tool.html). The zinc-finger domain is underlined in red. (D) Except the CXXC zinc finger domain, no conserved structural domain was found. An alignment of the CXXC motif of RINF protein with its human paralogs is presented. Amino acids that are invariant among CXXC motifs are in gray, and the conserved cysteine residues are in red. The consensus sequence for CXXC-type zinc-finger can be defined by Cx2Cx2Cx4-5Cx2Cx2C9-15Cx4C.
Gene expression profile of CXXC5 in promyelocytic cells treated with retinoids
In agreement with our microarray data, time-course analysis of CXXC5 mRNA levels (by quantitative RT-PCR) demonstrated a 2-fold induction as early as 90 minutes of ATRA treatment (Figure 3A). The transcript level of CXXC5 reached its highest level at 12 hours of treatment (∼ 6-fold the expression level observed in untreated control). Then, its level remained stably up-regulated for at least 48 hours (Figure 3A). To corroborate CXXC5 expression and induction at the protein level, we customized a polyclonal antibody directed against amino acids 45-58 of the predicted CXXC5 polypeptide chain (see “Methods”). Basal expression level of CXXC5 protein was very low in untreated NB4 cell extracts (Figure 3B), but a strong band, specific to CXXC5 protein (verified by competition with the immunogenic peptide, not shown), appeared at the expected molecular weight of 33 kDa within 4 hours after onset of ATRA treatment. The kinetic of induction of the CXXC5 protein is in agreement with the induction observed at the mRNA level. Interestingly, compared with NB4 cells, only a very weak induction of CXXC5 protein was noticed after ATRA treatment of the maturation-resistant subclones NB4-LR1 and NB4-LR2 (Figure S1). These observations were compatible with a potential participation of CXXC5 during terminal maturation of APL cells. Moreover, data extracted from publicly available microarray datasets (Figure S2), confirmed the early induction of CXXC5 mRNA by ATRA in primary cells from APL patients,27 validating the data obtained with the NB4 cell line.
RINF expression and subcellular localization in NB4 cells and other myeloid cell lines and tissues. (A) Relative expression of RINF mRNA levels (measured by quantitative RT-PCR) during 1 μM ATRA treatment of NB4 cells. (B) Expression of RINF protein in total extracts from NB4 cells treated or not treated with 1 μM ATRA. RINF was detected with our customized polyclonal rabbit antibody (see “Methods”) that detects a specific band at 33 kDa. Actin was used as a loading control. (C) Expression of RINF protein in nuclear and cytosolic fractions of NB4 cells treated or not treated with 1 μM ATRA. RINF was detected with the polyclonal antibody. In this experiment, PARP and actin were used as controls to evaluate quality, purity, and loading of the nuclear and cytosolic extracts. (D) FLAG-tagged RINF (green; Alexa Fluor 488–conjugated anti-FLAG monoclonal antibody) localizes in the nucleus (stained in blue with DAPI) of MCF7 cells analyzed by confocal and differential interference contrast (DIC) microscopy. Scale bar, 10 μm. (E) RINF mRNA expression in various myeloid cell lines measured by quantitative RT-PCR with or without ATRA at 4 hours of treatment (see “Methods”). Expression of RINF protein in various myeloid cell lines (F) treated or not treated with 1 μM ATRA during 4 hours and in various human tissues (G). The same amounts of protein (determined with bicinchoninic acid [BCA] assay test) were loaded for each of the myeloid cell line (20 μg) and human tissue extracts (65 μg; see “Methods”). Actin was used as a loading control.
RINF expression and subcellular localization in NB4 cells and other myeloid cell lines and tissues. (A) Relative expression of RINF mRNA levels (measured by quantitative RT-PCR) during 1 μM ATRA treatment of NB4 cells. (B) Expression of RINF protein in total extracts from NB4 cells treated or not treated with 1 μM ATRA. RINF was detected with our customized polyclonal rabbit antibody (see “Methods”) that detects a specific band at 33 kDa. Actin was used as a loading control. (C) Expression of RINF protein in nuclear and cytosolic fractions of NB4 cells treated or not treated with 1 μM ATRA. RINF was detected with the polyclonal antibody. In this experiment, PARP and actin were used as controls to evaluate quality, purity, and loading of the nuclear and cytosolic extracts. (D) FLAG-tagged RINF (green; Alexa Fluor 488–conjugated anti-FLAG monoclonal antibody) localizes in the nucleus (stained in blue with DAPI) of MCF7 cells analyzed by confocal and differential interference contrast (DIC) microscopy. Scale bar, 10 μm. (E) RINF mRNA expression in various myeloid cell lines measured by quantitative RT-PCR with or without ATRA at 4 hours of treatment (see “Methods”). Expression of RINF protein in various myeloid cell lines (F) treated or not treated with 1 μM ATRA during 4 hours and in various human tissues (G). The same amounts of protein (determined with bicinchoninic acid [BCA] assay test) were loaded for each of the myeloid cell line (20 μg) and human tissue extracts (65 μg; see “Methods”). Actin was used as a loading control.
Subcellular localization of retinoid-inducible nuclear factor, RINF (CXXC5)
Western blot analysis of nuclear and cytoplasmic fractions of ATRA-treated NB4 cells clearly detected expression of CXXC5 in the nuclear extracts (Figure 3C). This increased expression level lasted at least for 48 hours. Because our polyclonal antibody barely detected the relevant epitope (amino acids 45-58) of the folded native protein, we generated a plasmid encoding a FLAG-tagged CXXC5 for in situ immunochemistry experiments. In situ confocal analysis of MCF7 cells transiently transfected with this construct showed a strong nuclear staining (Figure 3D) and confirmed the nuclear localization pattern of CXXC5 protein. Of note, from cell to cell and according to the level of overexpression, the fluorescence pattern was partly associated to a fine chromatin matrix, nucleoplasm, and/or discrete punctuated structures in the nucleus. However, we did not detect any fluorescent signal at the nuclear membrane or in the nucleoli.
Based on these experimental evidences, we propose to rename CXXC5 as RINF, for retinoid-inducible nuclear factor. In keeping with this proposed name, a detailed analysis of the RINF (CXXC5) sequence revealed a putative NLS between amino acid residues 257 and 262 (KKKRKR), located to the N-terminal basis of the zinc-finger domain. This NLS motif is highly conserved with the typical monopartite sequence originally described28 and is also found in 14 other proteins that have been experimentally demonstrated to localize in the nucleus according to the PredictNLS browser.29
Expression patterns of RINF in different myeloid cell lines and various human tissues
At this stage, it became important to evaluate basal expression levels of the RINF gene in other myeloid cell lines (Figure 3E,F) and in various human tissues (Figure 3G) as well as its potential regulation by retinoids. The more immature cell lines tested, K562 and LAMA-84 cells, showed the highest RINF mRNA levels (16- and 12-fold the basal level of NB4 cells, respectively) and also the highest RINF protein expression. Neither RINF mRNA nor protein levels seemed up-regulated upon ATRA treatment in these cell lines (Figure 3E-F). In comparison, the HL60 (M2-subtype) and NB4 (M3-subtype) cell lines both harbor low basal levels of RINF mRNA and protein. Importantly, treatment of the latter cell lines with ATRA indicated that HL60 cells (known to embark on terminal differentiation upon such a treatment30 ) behave similarly to NB4 cells with respect to RINF induction (here observed at 4 hours), both at the mRNA and protein level. Importantly, this induction in HL60, a cell line lacking the expression of the chimeric receptor PML-RARα, demonstrates that PML-RARα is not required for RINF induction by ATRA. In agreement with this observation, we noticed that ATRA also induces RINF mRNA expression in 2 other cell lines with a basal expression level similar to NB4 cells (data not shown), the A549 lung carcinoma (a 3-fold increase), and the HeLa cervix cancer cells (a 2-fold increase). Finally, to substantiate the fact that RINF expression was not restricted to myeloid tissue, we performed a Western blot analysis on various protein extracts from different organs (Figure 3G). RINF protein showed different expression levels in the human tissues tested; the highest expression level was in placenta and the lowest in brain. Altogether, these data indicate that the regulated expression and the putative function of RINF is not dependent on PML-RARα expression and encompass a field of interest much broader than the APL pathology.
RINF expression is required for differentiation of promyelocytic leukemia cells
To investigate the potential involvement of RINF in terminal differentiation of NB4 cells after ATRA treatment, we performed an RNAi approach using shRNAs delivered by lentiviral vectors (Figure 4A). After infection and selection of NB4 cells stably expressing each of the 5 different shRNA constructs directed against the RINF mRNA, their efficiency (to target basal RINF mRNA expression level) was compared with control vectors, one expressing scrambled shRNA and one being the empty vector. The 2 best knockdown efficiencies were observed with shRNA/RINF-3 (61%) and shRNA/RINF-4 (85%), with which no proliferation changes or morphologic alterations were noticed in NB4 cells in the absence of retinoid. With such a silencing efficiency (Figure 4A), we could reasonably expect to efficiently block RINF mRNA induction by ATRA in an important proportion of the cells and, in this manner, evaluate the consequence on the cell phenotype after ATRA treatment.
shRNA-mediated silencing of RINF imparts resistance to ATRA-induced terminal differentiation of NB4 cells. (A) Lentiviral shRNA vectors and their mRNA target sequences used to knock down RINF expression. After infection and selection of NB4 cells (see “Methods”) with the lentiviral vector constructs, their efficiencies to target basal RINF expression were monitored by quantitative RT-PCR measuring basal RINF mRNA expression levels (indicated in percent of mock control, the pLKO.1/empty vector). The most efficient knockdowns were obtained with shRNA/RINF-3 and shRNA/RINF-4 constructs (61% and 85%, respectively, in the absence of ATRA) indicated in red. (B) Cell growth (population doublings) of NB4 cells, stably expressing the shRNA constructs, in the presence of 1 μM ATRA. The presented kinetic experiment (here during 12 days of ATRA treatment) is representative of 3 separate experiments (performed from different batches of infections). † indicates postmaturation cell death of control cells. (C) RINF mRNA expression was assessed by quantitative RT-PCR (percent of untreated mock control at 12 hours of culture), terminal differentiation was assessed by cell morphology at day 4 (scale bar, 25 μm), and NBT reduction assay was assessed at day 2 (scale bar, 25 μm) of NB4 cells infected with empty, shRNA/scramble, shRNA/RINF-3, and shRNA/RINF-4 vectors. Cells were treated or not treated with 1 μM ATRA for 4 days (first round of ATRA, d0-4, left panel). After 2 more weeks of culture in the absence of ATRA, shRNA/RINF-3 and shRNA/RINF-4 cells that escaped the first round of ATRA (indicated by an arrow [→]) were retreated (second round of ATRA, days 20 to 24, right panel) for 4 days with 1 μM ATRA.
shRNA-mediated silencing of RINF imparts resistance to ATRA-induced terminal differentiation of NB4 cells. (A) Lentiviral shRNA vectors and their mRNA target sequences used to knock down RINF expression. After infection and selection of NB4 cells (see “Methods”) with the lentiviral vector constructs, their efficiencies to target basal RINF expression were monitored by quantitative RT-PCR measuring basal RINF mRNA expression levels (indicated in percent of mock control, the pLKO.1/empty vector). The most efficient knockdowns were obtained with shRNA/RINF-3 and shRNA/RINF-4 constructs (61% and 85%, respectively, in the absence of ATRA) indicated in red. (B) Cell growth (population doublings) of NB4 cells, stably expressing the shRNA constructs, in the presence of 1 μM ATRA. The presented kinetic experiment (here during 12 days of ATRA treatment) is representative of 3 separate experiments (performed from different batches of infections). † indicates postmaturation cell death of control cells. (C) RINF mRNA expression was assessed by quantitative RT-PCR (percent of untreated mock control at 12 hours of culture), terminal differentiation was assessed by cell morphology at day 4 (scale bar, 25 μm), and NBT reduction assay was assessed at day 2 (scale bar, 25 μm) of NB4 cells infected with empty, shRNA/scramble, shRNA/RINF-3, and shRNA/RINF-4 vectors. Cells were treated or not treated with 1 μM ATRA for 4 days (first round of ATRA, d0-4, left panel). After 2 more weeks of culture in the absence of ATRA, shRNA/RINF-3 and shRNA/RINF-4 cells that escaped the first round of ATRA (indicated by an arrow [→]) were retreated (second round of ATRA, days 20 to 24, right panel) for 4 days with 1 μM ATRA.
Strikingly, expression of these 2 shRNA constructs gives the NB4 cell population the capacity to proliferate in the presence of pharmacologic doses of ATRA (Figure 4B). Indeed, while the cells expressing control vectors ineluctably differentiated and died within a week upon ATRA treatment, both cell cultures infected with either shRNA/RINF-4 or shRNA/RINF-3 lentiviral vectors bypassed the growth arrest (Figure 4B) and the terminal differentiation process (Figure 4C). The efficiency of our RNAi strategy to block ATRA-induced RINF mRNA expression was evaluated by quantitative RT-PCR (Figure 4C) at 12 hours of treatment, a time at which RINF mRNA expression was maximum (see also Figure 3A). Importantly, knockdown levels obtained with shRNA/RINF-3 (at ∼ 50%) and shRNA/RINF-4 (60%) compared with control vectors treated with ATRA, were enough to rescue approximately 30% to 50% NB4 cells from terminal differentiation as assessed by reduction of NBT and morphologic analysis (Figure 4C left panel).
Of note, these cells (shRNA/RINF-3 and shRNA/RINF-4) continued to proliferate in the presence of retinoids, and even after 2 weeks of culture without ATRA (from day 6 to 20), their resistance to ATRA was confirmed and remained closely associated to an efficient block in RINF mRNA expression (Figure 4C right panel). Altogether, these results demonstrate that shRNA-mediated RINF knockdown impairs ATRA-induced terminal differentiation of promyelocytic leukemia cells and strongly suggest that RINF expression is required for retinoid-induced terminal maturation of NB4 cells.
RINF contribution was then evaluated in the HL60 cell line committed into the granulocytic differentiation pathway with pharmacologic doses of ATRA (Figure S3). As observed with NB4 cells, the 2 shRNAs efficiently target RINF expression (Figure S3) and delayed the maturation process assessed by NBT and morphologic analysis, underpinning the functional involvement of RINF during granulocytic differentiation. However, in contrast to NB4-shRNA/RINF cells that continued to grow in the presence of retinoids, ATRA-treated HL60-shRNA/RINF cells inevitably declined and died within 12 days.
Finally, we wondered whether RINF overexpression would be sufficient to induce granulocytic maturation of NB4 or HL60 cells in the absence of ATRA (Figure S4). Our data indicate that ectopic RINF expression (Figure S4B) is necessary but not sufficient to trigger granulocytic differentiation of leukemic cells. Indeed, even after 10 days of RINF overexpression, we did not notice any sign of differentiation neither at the morphologic level (Figure S4C), nor at the molecular level (using anti-CD11b surface marker, data not shown).
RINF expression and function during cytokine-driven myelopoiesis of normal CD34+ progenitor cells
Our above findings clearly indicate a broad spectrum of RINF expression in hematopoietic and other human tissues. These data indicated that although inducible by pharmacologic concentrations of ATRA and required for differentiation of some myeloid leukemia cells, RINF expression may well be regulated physiologically by cytokines during normal progenitor myelopoiesis. In this case, its regulated expression could represent a more general event also occurring during normal development along the granulocytic lineage.
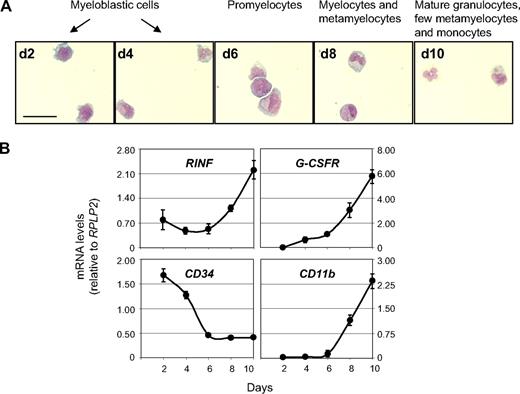
To test this hypothesis, we examined RINF expression during cytokine-induced granulocytic differentiation of CD34+ cells isolated from bone marrow of a healthy adult donor. Differentiation was assessed by morphologic changes (Figure 5A) and analysis of CD34, CD11b, and G-CSFR expression comparatively to RINF (Figure 5B). The time-course expression profile of RINF mRNA levels was determined by quantitative RT-PCR analysis. After an initial decrease (from day 2 to day 4), the RINF mRNA level reached its minimal expression between day 4 and 6, a time at which most of the cells in culture were at the blastic and promyelocytic stage. Later on, between day 6 and day 8, RINF mRNA expression increased approximately 3.5-fold from its minimum level, concomitantly to terminal cell maturation into myelocytes, metamyelocytes, and ultimately into short-lived polynuclear neutrophils (Figure 5A,B). The detection of RINF mRNA in CD34+ progenitors and in normal hematopoietic cells during cytokine-driven differentiation confirms that its expression is not restricted to APL cells (and is therefore not dependent on PML-RARα expression), and most importantly that its induction is not restricted to ATRA pharmacologic signaling. These data are consistent with its potential role during normal myelopoiesis.
RINF expression during normal myelopoiesis. (A) Cytokine-induced (IL3 and G-CSF at 20 ng/mL, SCF at 50 ng/mL) granulocytic differentiation of myeloid CD34+ cells (from a healthy donor). For cell morphology (scale bar, 25 μm), cells were spread on a glass slide by cytospin, air-dried, and stained with MGG at different time points of culture (from day 2 to day 10). For each day of culture recorded, the main stage of myelopoiesis observed is indicated. At day 10, most of the cells were terminally differentiated into polynuclear neutrophil granulocytes. In our experimental conditions, a few monocytic cells were also noticed (not shown). (B) Relative mRNA expression of various genes (measured by quantitative RT-PCR) during cytokine-induced granulocytic differentiation of CD34+ progenitors (± SEM).
RINF expression during normal myelopoiesis. (A) Cytokine-induced (IL3 and G-CSF at 20 ng/mL, SCF at 50 ng/mL) granulocytic differentiation of myeloid CD34+ cells (from a healthy donor). For cell morphology (scale bar, 25 μm), cells were spread on a glass slide by cytospin, air-dried, and stained with MGG at different time points of culture (from day 2 to day 10). For each day of culture recorded, the main stage of myelopoiesis observed is indicated. At day 10, most of the cells were terminally differentiated into polynuclear neutrophil granulocytes. In our experimental conditions, a few monocytic cells were also noticed (not shown). (B) Relative mRNA expression of various genes (measured by quantitative RT-PCR) during cytokine-induced granulocytic differentiation of CD34+ progenitors (± SEM).
To evaluate the functional relevance of RINF expression during normal myelopoiesis, CD34+ progenitor cells directed toward the granulocytic lineage by cytokine treatment were infected with the lentiviral shRNA constructs to knock down RINF expression. Like noninfected cells, most of the cells infected with control vectors (shRNA/scramble and mock control) matured into polynuclear granulocytes (Figures 5A,6) before dying and progressively disintegrated in cell cultures (not shown). In the 3 cell cultures tested from different donors, granulocytic differentiation occurred with different kinetics (see Figure 6), all cultures ending with cell maturation in polynuclear neutrophils that rapidly died. Only few monocytes and plastic-adherent macrophages, known to have a much longer life, persisted in the cultures. Importantly, cell populations infected under the same conditions with shRNA/RINF-3 and shRNA/RINF-4 (Figure 6) showed significant accumulation of unmatured cells (promyelocytes and myelocytes). This difference in granulocytic maturation, observed with the 2 shRNAs compared with the control cultures, exquisitely mirrored the results obtained with the ATRA-treated leukemic cell lines NB4 and HL60 and suggests a similar role of RINF during normal myelopoiesis.
Functional involvement of RINF during normal myelopoiesis. CD34+ myeloid progenitors isolated from 3 healthy donors (A-C) were infected with each of the 4 lentiviral shRNA constructs, either targeting RINF expression (shRNA/RINF-3 and shRNA/RINF-4) or control vectors (empty and shRNA/scramble). The 12 cell populations were then treated in the same conditions with cytokines (IL3 and G-CSF at 20 ng/mL, SCF at 50 ng/mL) to drive them into granulocytes. For each cell population, myeloid differentiation was evaluated, every 2 to 3 days of culture and during several weeks, by cell morphology analysis after cytospin and MGG staining (scale bar, 25 μm). According to the donor, cell differentiation of cultured progenitors developed with different kinetics, and the figure shows morphology of the cell populations at the most informative days recorded, respectively, after 14, 30, and 18 days of culture for donors A, B, and C. Note that cell cultures infected with shRNA–RINF-3 or shRNA–RINF-4 constructs display more immature cells at the promyelocytic/myelocytic stage (indicated by arrows [↑]) than the controls (cell cultures infected with empty or scramble vectors). For donor A, the kinetic of granulocytic differentiation was fast (until day 10), and only a few adherent monocytes/macrophages persisted in control cultures at day 14.
Functional involvement of RINF during normal myelopoiesis. CD34+ myeloid progenitors isolated from 3 healthy donors (A-C) were infected with each of the 4 lentiviral shRNA constructs, either targeting RINF expression (shRNA/RINF-3 and shRNA/RINF-4) or control vectors (empty and shRNA/scramble). The 12 cell populations were then treated in the same conditions with cytokines (IL3 and G-CSF at 20 ng/mL, SCF at 50 ng/mL) to drive them into granulocytes. For each cell population, myeloid differentiation was evaluated, every 2 to 3 days of culture and during several weeks, by cell morphology analysis after cytospin and MGG staining (scale bar, 25 μm). According to the donor, cell differentiation of cultured progenitors developed with different kinetics, and the figure shows morphology of the cell populations at the most informative days recorded, respectively, after 14, 30, and 18 days of culture for donors A, B, and C. Note that cell cultures infected with shRNA–RINF-3 or shRNA–RINF-4 constructs display more immature cells at the promyelocytic/myelocytic stage (indicated by arrows [↑]) than the controls (cell cultures infected with empty or scramble vectors). For donor A, the kinetic of granulocytic differentiation was fast (until day 10), and only a few adherent monocytes/macrophages persisted in control cultures at day 14.
Discussion
The multistage process of hemopoietic cell differentiation has long served as a model study for the understanding of tumor etiology and for the design of therapeutic strategies. Disturbances in the developmental programs that rule the production of mature functional cells frequently result from genetic or epigenetic alterations (gene deletions, mutations, methylation, etc) in a limited number of key regulatory genes, whose functions of predilection are signal transduction and/or gene transcription control. In the particular case of hemopoietic malignancies, the last decade has brought major advances in the finding of these key regulatory genes,1 but uncertainty remains on their functional hierarchy.
In the present study, we have identified and characterized a novel member of the zinc-finger family, CXXC5 (RINF), and shown its implication during retinoid-induced terminal differentiation of myelocytic leukemia cells, but also during cytokine-driven normal myelopoiesis. Zinc-finger–containing proteins constitute one of the largest protein superfamilies in the mammalian genome and can be classified into evolutionary and functionally divergent protein families, with structurally different conserved domains interacting with DNA, RNA, lipids, or other proteins.31 The CXXC-type zinc finger is found in a small subset of proteins (Figure 2) involved in chromatin remodeling through their histone methyltransferase (MLL, MLL2), histone demethylase (FBXL-10, FBXL-11, FBXL-19), DNA methyltransferase (DNMT1), or CpG-binding (CGBP, MBD1) activities. The CXXC domains of MLL, CGBP, and MBD1 have been shown to bind to nonmethylated CpG,32-34 and the conserved KFGG motif (in the second linker of the zinc finger) is essential for this recognition.24 The paralogs lacking the KFGG motif (CXXC-4, CXXC-5, CXXC-6, and CXXC-10; Figure 2) are expected not to share the latter property, and one of the future challenges will be to identify the molecular target(s) recognized by this specific subset of proteins with conserved CXXC domains but lacking the KFGG motif. Interestingly, 3 members of the CXXC-type family (containing or not the KFGG motif), MLL, MLL2, and LCX, have previously been involved in leukemogenesis through chromosomal translocations or internal rearrangements.35-38 MLL is one of the most frequently translocated genes in leukemia, and in spite of more than 30 different fusion partners that have been described, the CXXC domain is retained in all known MLL fusion proteins, and the domain appears to be essential for myeloid transformation,25 underpinning the biologic importance of this zinc-finger subtype even if the mechanism of action remains to be clarified.
The expression and the induction patterns of the CXXC5 (RINF) gene reported here supported a functional involvement of RINF in both normal and tumoral myelopoiesis, at least in the latest stages of the maturation process (promyelocyte, myelocyte). Knockdown experiments, using specific RNAi targeting, further demonstrated that RINF expression was necessary for the terminal differentiation process of promyelocytic leukemia cells. However, and importantly, our study clearly supports the idea that RINF pathophysiology is not restricted to APL, but may have broader implication in other hemopoietic malignancies and normal hemopoiesis. Thus, the RINF status (such as mutations, aberrant expression/induction) may have predictive value for ATRA responsiveness, and therefore being an important tool for decision making on therapeutic regimens for AML patients.
In the clinical context, an exciting prospect concerning CXXC5 (RINF) biology will be to evaluate whether its deregulated expression could contribute to the etiology of some hematologic diseases. Indeed, interstitial deletion or complete loss of the long arm of chromosome 5 are recurrent chromosome abnormalities in malignant myeloid disorders39,40 characterized by abnormal myelopoiesis and an excess of blasts in the bone marrow, like MDS (often described as a preleukemic disorder) and AML. Despite extensive studies that have delineated the major commonly deleted region (CDR) to chromosome band 5q31,41-44 candidate tumor suppressor genes presumed to contribute to leukemogenesis remain to be identified in this region of 4 megabases (Mb) in size.43,45 It is important to note that, inside this segment, RINF localizes less than 20 kb from the distal (telomeric) marker D5S594 that delineate the smallest (< 1 Mb) CDR described at this date.42 Surprisingly, the RINF gene has so far escaped genetic and functional investigation, probably because the gene has been considered to localize outside the CDR. Even so, the close proximity of RINF to the CDR may affect RINF expression due to loss of regulatory sequences upstream of the identified gene, as well as more distant deletions within 5q31. Our findings combined with data in the literature led us to consider RINF to be a strong candidate for a tumor suppressor gene of myeloid cell transformation. Interestingly, mining publicly available microarray databases for myeloid pathologies,46 we brought to light a lower RINF mRNA expression in CD34+ cells from myelodysplastic patients with deletion 5q compared with normal healthy donors (Figure S5). This observation is compatible with a putative haploinsufficiency mechanism, and it will be important to evaluate the genuine contribution of RINF deregulation in these hemopathies, as it has been recently described for the ribosomal gene RPS14 (located at 5q33.1 in a distinct CDR) in another subgroup of MDS with refractory anemia, the 5q− syndrome.47
Because RINF expression is not restricted to myeloid tissue, this gene may also be involved in development and/or homeostasis of other tissues, as it has very recently been suggested in rat neural stem cells.48 Its direct induction by retinoids, which does not require de novo synthesis of an intermediate protein regulator, suggests that RINF might mediate, at least in part, some of the pleiotropic effects of retinoids, for instance such as their antiproliferative action in various solid tumors, even independently of any differentiation. Taken together, our findings support the hypothesis that RINF expression, pharmacologically inducible by retinoids in different tissues, may have a broad interest because of its likely implication in several developmental processes and pathologies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Nina Glomnes, Pablo Badin, Raphael Ehre, Audrey Astori, and Josette Hillion for technical assistance; Marc Niere, Line Myklebust, Stian Knappskog, and Thomas Arnesen for continuous scientific discussions; Anne-Kristin Stavrum, Kjell Pettersen, Rita Holdhus, and Harald Breilid for their technical assistance with microarray data; Christelle Doliger for the confocal microscopy; and Marianne Enger for the cell sorting.
This work was supported by the University of Bergen (Bergen, Norway), the European Commission (Brussels, Belgium), Inserm (Paris, France), the Norwegian Cancer Society (Oslo, Norway; J.R.L.), the Association pour la Recherche contre le Cancer (Paris, France), the Fondation de France (Paris, France), Cent pour Sang la Vie (Paris, France), and Capucine (Paris, France; grants to E.S.B). F.P. was supported by International Agency for Research on Cancer (Lyon, France), Marie-Curie Actions EIF (555319) and ERG (046567), University of Bergen, and Inserm for postdoctoral fellowships.
Authorship
Contribution: F.P. conceived the study, performed most of the experiments, and mainly wrote the paper; E.N. carried out Western blot analysis, prepared cell fractions, and helped edit the manuscript; together with F.P., I.J., and B.D. designed and analyzed the microarray experiments; A.A. performed the ChIP experiments; M.L. participated in discussing the data and writing the manuscript; E.S.-B. helped conceive the study, discussed the data, and participated in writing the manuscript; and J.R.L. contributed to study design, provided guidance and supervision of the data analysis, and participated in writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Frédéric Pendino, Inserm U685, Hôpital Saint-Louis, Institut Universitaire d'Hématologie, 1, avenue Claude Vellefaux, 75010, Paris, France; e-mail: frederic.pendino@inserm.fr; or Johan Lillehaug, Department of Molecular Biology, University of Bergen, Thormøhlensgt 55, 5008 Bergen, Norway; e-mail: johan.lillehaug@mbi.uib.no.

![Figure 3. RINF expression and subcellular localization in NB4 cells and other myeloid cell lines and tissues. (A) Relative expression of RINF mRNA levels (measured by quantitative RT-PCR) during 1 μM ATRA treatment of NB4 cells. (B) Expression of RINF protein in total extracts from NB4 cells treated or not treated with 1 μM ATRA. RINF was detected with our customized polyclonal rabbit antibody (see “Methods”) that detects a specific band at 33 kDa. Actin was used as a loading control. (C) Expression of RINF protein in nuclear and cytosolic fractions of NB4 cells treated or not treated with 1 μM ATRA. RINF was detected with the polyclonal antibody. In this experiment, PARP and actin were used as controls to evaluate quality, purity, and loading of the nuclear and cytosolic extracts. (D) FLAG-tagged RINF (green; Alexa Fluor 488–conjugated anti-FLAG monoclonal antibody) localizes in the nucleus (stained in blue with DAPI) of MCF7 cells analyzed by confocal and differential interference contrast (DIC) microscopy. Scale bar, 10 μm. (E) RINF mRNA expression in various myeloid cell lines measured by quantitative RT-PCR with or without ATRA at 4 hours of treatment (see “Methods”). Expression of RINF protein in various myeloid cell lines (F) treated or not treated with 1 μM ATRA during 4 hours and in various human tissues (G). The same amounts of protein (determined with bicinchoninic acid [BCA] assay test) were loaded for each of the myeloid cell line (20 μg) and human tissue extracts (65 μg; see “Methods”). Actin was used as a loading control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/14/10.1182_blood-2008-07-170035/4/m_zh80170933900003.jpeg?Expires=1769194248&Signature=Zve70B7y-uMBfRzagmBAUi1WqaTKQAXS7dYsOOPReP0hPSz1To0kUyqCpK6868cNiMvxZBXM9e7wREQT8iFl0HD1VtJ8pKZkhCTvNVwwr3qEC0qzRhJ7gndIxDOoDUy1UQpQytA6tQR7D~qWyaks4Mn8TmoQrSaE~fN0qYU6sCgc8fAxH7zgxZP-IIDweWWX99L-iVj-EsO0gBb4l4Y~L4~k68vfRRrXCRX1rjuzmpTqLrpzFNX779IqO-IwO~J7-U757Akyv4X2Fg-a6vlN7YKlxA0x1s4REGyjv0mYRLhE43iFzNySgNqw2wxF3IBMqoXKWfjUjgCwewLsLbGXfQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. shRNA-mediated silencing of RINF imparts resistance to ATRA-induced terminal differentiation of NB4 cells. (A) Lentiviral shRNA vectors and their mRNA target sequences used to knock down RINF expression. After infection and selection of NB4 cells (see “Methods”) with the lentiviral vector constructs, their efficiencies to target basal RINF expression were monitored by quantitative RT-PCR measuring basal RINF mRNA expression levels (indicated in percent of mock control, the pLKO.1/empty vector). The most efficient knockdowns were obtained with shRNA/RINF-3 and shRNA/RINF-4 constructs (61% and 85%, respectively, in the absence of ATRA) indicated in red. (B) Cell growth (population doublings) of NB4 cells, stably expressing the shRNA constructs, in the presence of 1 μM ATRA. The presented kinetic experiment (here during 12 days of ATRA treatment) is representative of 3 separate experiments (performed from different batches of infections). † indicates postmaturation cell death of control cells. (C) RINF mRNA expression was assessed by quantitative RT-PCR (percent of untreated mock control at 12 hours of culture), terminal differentiation was assessed by cell morphology at day 4 (scale bar, 25 μm), and NBT reduction assay was assessed at day 2 (scale bar, 25 μm) of NB4 cells infected with empty, shRNA/scramble, shRNA/RINF-3, and shRNA/RINF-4 vectors. Cells were treated or not treated with 1 μM ATRA for 4 days (first round of ATRA, d0-4, left panel). After 2 more weeks of culture in the absence of ATRA, shRNA/RINF-3 and shRNA/RINF-4 cells that escaped the first round of ATRA (indicated by an arrow [→]) were retreated (second round of ATRA, days 20 to 24, right panel) for 4 days with 1 μM ATRA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/14/10.1182_blood-2008-07-170035/4/m_zh80170933900004.jpeg?Expires=1769194248&Signature=xT4rhlinVgOrXoP1-GsBzaR-FTmkyojhhQ6DAvFl65VFTPVD8rPHWZ~u90VAi8FxK6-eOOIg30-v6T639xOjsFZMhXI5hB2K41vDIgKkfRLjGpP5q6JynwiQBaRwvQh-FJjmdnSW4Sv8L3UDp29WzMpVKEc~SJ4UnvTAD7zuWPvZ8tlb0yiAvLi4T3QEhj8cnkcrpyjz4cmIxzBl6buUDICwwti0N-3g1ZZdrlpIvmJ-yul1OUjCYwr8oz9dba-8lPu5ivPmvjuzsrxqIJoVd-KfOz0ilr5ZCoxzX4Z6Wxz-jiZJ5FbhBqHyVaYGbNJplf4kNxJ5x58oC7XFa8d0uw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Functional involvement of RINF during normal myelopoiesis. CD34+ myeloid progenitors isolated from 3 healthy donors (A-C) were infected with each of the 4 lentiviral shRNA constructs, either targeting RINF expression (shRNA/RINF-3 and shRNA/RINF-4) or control vectors (empty and shRNA/scramble). The 12 cell populations were then treated in the same conditions with cytokines (IL3 and G-CSF at 20 ng/mL, SCF at 50 ng/mL) to drive them into granulocytes. For each cell population, myeloid differentiation was evaluated, every 2 to 3 days of culture and during several weeks, by cell morphology analysis after cytospin and MGG staining (scale bar, 25 μm). According to the donor, cell differentiation of cultured progenitors developed with different kinetics, and the figure shows morphology of the cell populations at the most informative days recorded, respectively, after 14, 30, and 18 days of culture for donors A, B, and C. Note that cell cultures infected with shRNA–RINF-3 or shRNA–RINF-4 constructs display more immature cells at the promyelocytic/myelocytic stage (indicated by arrows [↑]) than the controls (cell cultures infected with empty or scramble vectors). For donor A, the kinetic of granulocytic differentiation was fast (until day 10), and only a few adherent monocytes/macrophages persisted in control cultures at day 14.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/14/10.1182_blood-2008-07-170035/4/m_zh80170933900006.jpeg?Expires=1769194248&Signature=g5Bjl2wtvzqrq7qeOfV0cIZKhGa~DA83bf2UjhU8knzanPFnlD-K-~BL6uiR10ee9lguhXPi3vpMGHughcTrv3BD8Ymeit0HYZA0Np9fMRbkZ3pDtaNXIWBVf04Ge9A-8sOSaHFFHoDy2Dx3G1LUa5ibEM9rxu5gFSuJawrW2P77EqLOfVCQuc4q-I7dzGMWZBGWync5Yorbod2sLYHgYMO54MGm1nZXtmFiXirEwJunjc3ISJS61XL~xV~m~SLntYfb5IQhg8i1DM529ZWlL8vzzX6VkxVOZuzC~sUX1vLbbVSU7XYk62n01UCBRBVt2xjkm1Bmx~Jjl-CENq6sHw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal