Abstract
Adult T-cell leukemia (ATL) is a highly aggressive T-cell malignancy caused by human T-cell leukemia virus type 1 (HTLV-1). The activation of NF-κB by Tax has been reported to play a crucial role in HTLV-1–induced transformation. The HTLV-1 bZIP factor (HBZ), which is encoded by an mRNA of the opposite polarity of the viral genomic RNA, is involved in both T cell proliferation and suppression of Tax-mediated viral gene transcription, suggesting that HBZ cooperates closely with Tax. In the present study, we observed that HBZ specifically suppressed NF-κB–driven transcription mediated by p65 (the classical pathway) without inhibiting the alternative NF-κB signaling pathway. In an immunoprecipitation assay, HBZ bound to p65 and diminished the DNA binding capacity of p65. In addition, HBZ induced p65 degradation through increasing the expression of the PDLIM2 gene, which encodes a ubiquitin E3 ligase for p65. Finally, HBZ actually repressed the transcription of some classical NF-κB target genes, such as IL-8, IL2RA, IRF4, VCAM-1, and VEGF. Selective suppression of the classical NF-κB pathway by HBZ renders the alternative NF-κB pathway predominant after activation of NF-κB by Tax or other stimuli, which might be critical for oncogenesis.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) is an oncogenic retrovirus that causes adult T-cell leukemia (ATL).1-4 A unique sequence was found in the viral genome between env and the 3′ long terminal repeat (LTR). Denoted the pX region, this sequence encodes the regulatory and accessory genes: tax, rex, p12, p13, and p30.5 Among the proteins encoded by these genes, Tax has been thought to play a central role in the leukemogenesis of ATL because of its pleiotropic actions.6,7 Another gene, the HTLV-1 bZIP factor (HBZ) gene, is encoded by an mRNA of the opposite polarity of the viral genomic RNA.8-10 Two major isoforms of the HBZ gene transcript have been reported: spliced HBZ (sHBZ) and unspliced HBZ (usHBZ).11-13 Only sHBZ is abundantly expressed.14 HBZ was found to inhibit Tax-mediated transactivation of viral transcription from the 5′LTR by heterodimerizing with c-Jun and CREB2.15-17 In addition, sHBZ RNA promotes proliferation of ATL cells.11
Tax-mediated nuclear factor-κB (NF-κB) activation plays a pivotal role in the transforming activity of HTLV-1.18 Activation of NF-κB by Tax involves both the classical and alternative pathways, which use the NF-κB precursor proteins p105 (NF-κB-1) and p100 (NF-κB-2), respectively. These proteins are processed to the mature p50 NF-κB-1 and p52 NF-κB-2 proteins that heterodimerize with other members of the NF-κB family, RelA (p65), c-Rel, or RelB. Tax activates the classical pathway by interacting with IKKγ,19,20 which stimulates the phosphorylation of IKKα and IKKβ; phosphorylated IKKα and IKKβ then induce the degradation of IκB by the proteasome,21 leading to the release of p50/p65 from IκB. Tax also triggers activation of the alternative pathway downstream of NF-κB–inducing kinase (NIK) by activating IKKα via IKKγ and recruiting IKKα to p100, resulting in enhanced processing to p52.22 In both pathways, the mature dimeric NF-κB proteins translocate to the nucleus and activate genes involved in anti-apoptosis, cell proliferation and angiogenesis. Although NF-κB-1 and NF-κB-2 have similar structures, recent studies strongly suggest that the classical and alternative pathways are involved in the expression of different genes: the classical pathway is mostly involved in innate immunity and inflammatory responses, while the alternative pathway is involved in adaptive immunity and the organogenesis of peripheral lymphoid tissues.23,24
NF-κB is activated in many viral infections, and is thought to be important in the protective response of the host to the viral pathogens. Thus, many viruses have evolved distinct strategies to attenuate NF-κB activation. Known mechanisms of NF-κB suppression in virus-infected cells include (1) inhibition of the IKK activity and IκB phosphorylation,25 (2) inhibition of the NF-κB nuclear translocation,26 (3) induction of the caspase-mediated cleavage of the RelA subunit,27 and (4) inhibition of NF-κB RelA transcriptional activation through protein-protein interaction.28
Although NF-κB activation by Tax has been reported, it remains unknown whether other viral proteins act on the NF-κB pathway. In this study, we report that HBZ inhibits NF-κB activity by inhibiting p65 DNA binding capacity and promoting expression of PDLIM2 E3 ubiquitin ligase, which results in p65 degradation. This HBZ mediated suppression of the classical NF-κB pathway results in decreased expression of some genes associated with innate immunity and inflammatory responses.
Methods
Cell culture
Jurkat cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and antibiotics. Jurkat and Kit 225 cells stably expressing sHBZ were maintained as described previously.11 293FT and Hela cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS and 500 μg/mL G418.
Plasmids
The κB-Luc construct contains 5 tandem repeats of an NF-κB binding site from the IL-2R α-chain gene linked to the firefly luciferase gene. The AP-1-Luc construct contains 2 copies of the IL-8 AP-1 binding site upstream of the IL-8 enhancer-less core promoter linked to the luciferase gene.29 pSRF-Luc was purchased from Stratagene (Heidelberg, Germany). phRL-TK was purchased from Promega (Madison, WI). Expression vectors for usHBZ and sHBZ deletion mutants were generated by PCR using pME18Sneo-HBZ as a template.11 These fragments were subcloned into pME18Sneo and pcDNA3.1/myc-His(-) (Invitrogen, Carlsbad, CA). The coding region of IKKγ and p50 were amplified by RT-PCR from total RNA derived from Jurkat cells and cloned into the vector pCMV-HA. To construct vectors expressing wild type and deletion mutants of p65, we amplified the coding sequence from Jurkat cell cDNA and subcloned it into the pCMV-Tag 2 vector. The pEF-p52 expression vector, pCGN-HA-ubiquitin, pCG-Tax, pCG-Tax M47 (Tax mutant unable to activate the CREB/ATF pathway), and pCG Tax M22 (Tax mutant defective for NF-κB activation) were described elsewhere.30-32
Luciferase assay
Jurkat cells were plated on 6-well plates at 3.5 × 105 cells per well. After 24 hours, cells were transfected with the indicated luciferase reporter plasmid and expression plasmid, and/or empty expression vector (to normalize the DNA dose) mixed with Transfectin (Bio-Rad, Hercules, CA). After 48 hours, cells were collected and luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). Luciferase values were normalized to renilla luciferase and expressed as the mean of a triplicate set of experiments plus or minus SD.
Measurement of phosphorylation of IκBα
The FunctionELISA IκBα assay was performed according to the manufacturer's instructions (Active Motif, Carlsbad, CA) using 100 μg freshly prepared cytoplasmic protein extract from the samples.
Immunoprecipitation and immunoblotting
To examine protein-protein interaction in 293FT cells, subconfluent cells were transfected with the indicated combinations of expression vectors. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer 48 hours later (50 mM Tris-HCl, pH8.0, 1% Triton X-100, 100 mM NaCl, 1 mM MgCl2, 0.5% Nonidet P-40, and protease inhibitors). Lysates were precleared by incubation with 20 μl of 50% slurry of protein G-agarose (GE Healthcare Life Sciences, Piscataway, NJ) for 30 minutes at 4°C. Pre-cleared cell lysates were incubated with anti-HA (clone 3C2) (MBL, Nagoya, Japan), anti–c-Myc (clone 9E10, Sigma-Aldrich, St Louis, MO) or anti-FLAG M2 (Sigma-Aldrich) antibodies for 1 hour at 4°C, and immune complexes were collected by incubation for 1 hour at 4°C with protein G-agarose. After extensive washing, immunoprecipitated proteins were resolved by 5% to 20% SDS-PAGE and analyzed by western blotting with HRP-conjugated anti-FLAG (Sigma-Aldrich), anti–His-Tag (PM002, MBL), anti-Tax or anti-HA (Sigma-Aldrich) antibodies. Membranes were developed with enhanced chemiluminescence (GE Healthcare Life Sciences). Other antibodies used were as follows: anti–mouse ImmunoglobulinG (IgG) and anti–rabbit IgG were from GE Healthcare Life Sciences; anti-Sp1 and anti–lamin B were from Santa Cruz Biotechnology (Santa Cruz, CA).
Immunofluorescence analysis
Hela cells were transfected with expression vectors using Lipofectamine LTX (Invitrogen). Thirty-six hours after transfection, cells were stimulated with phorbal myristate acetate (PMA) plus ionomycin for 8 hours. sHBZ protein was detected using anti–c-MYC Cy3 (clone 9E10′ Sigma-Aldrich). p65 was detected using anti–Flag-biotin (Sigma-Aldrich) and secondary Streptavidin-Alexa 488 antibody (Invitrogen). Fluorescence was observed with a 63×/1.4-0.60 HCX PL APO objective on a DMIRE2-TCS SP2 AOBS confocal microscope system (Leica, Wetzlar, Germany) as described.17 Images were acquired and analyzed using LCS 2.61 (Leica) and processed using Photoshop CS2 (Adobe Systems, San Jose, CA).
NF-κB DNA-binding activity assay
Nuclear extracts were prepared using the NucBuster Protein Extraction Kit (Novagen, Madison, WI) according to the manufacturer's instructions. The DNA-binding activity of NF-κB was assayed colorimetrically, using the NoShift Transcription Factor Assay Kit and NoShift NF-κB (p65) reagents (Novagen) according to the manufacturer's instructions. To assess sequence-specific binding activity, we incubated 15 μg of nuclear extract with various combinations of biotinylated NF-κB wild-type dsDNA, specific NF-κB competitor dsDNA lacking biotin end labels, and nonspecific, nonbiontinylated dsDNA with a mutant NF-κB consensus binding motif. All assays were performed in triplicate.
Measurement of apoptotic cell death
For detection of apoptosis, the annexin V-binding capacities of the treated cells were examined by flow cytometry using an annexin V–PE Apoptosis Detection Kit (Biovision, Mountain View, CA), according to the manufacturer's instructions.
Ubiquitination assay
To analyze the ubiquitination of p65, we transfected 293FT cells with expression plasmids encoding FLAG-tagged p65, HA-tagged ubiquitin and mycHis-tagged wild type or mutant sHBZ. Extracts were incubated with anti-FLAG antibodies plus protein G-agarose, washed 5 times, and analyzed by immunoblot with anti-HA antibody.
Synthesis of cDNA and semiquantitative RT-PCR
Total RNA was isolated using Trizol Reagent (GIBCO, Grand Island, NY) according to the manufacturer's instructions. We reverse transcribed 1 μg of total RNA into single-stranded cDNA with SuperScript II reverse transcriptase (Invitrogen). Using the forward (F) and reverse (R) primers specific to the target genes, the cDNA was amplified by increasing number of PCR cycles. The primers used for this study are shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The intensity of PCR-amplified band was quantified using ATTO densitography 4.0 (Atto Instruments, Tokyo, Japan). The mRNA level of each product was normalized to the level of GAPDH and calculated as a ratio to the level of the control. Representative results of 3 independent experiments are shown.
Small interfering RNA (siRNA) transfection
siRNA targeted to human PDLIM2 was synthesized according to a previous report.33 293FT cells were transfected with expression vectors and siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Western blotting with anti-PDLIM2 antibody (Abnova, Taipei, People's Republic of China) detected PDLIM2 protein 48 hours after transfection.
sHBZ transgenic mice
Transgenic mice expressing the spliced HBZ gene under control of CD4 specific promoter/enhancer/silencer have been reported previously.11 CD4+ cells were isolated from mice thymus using anti–mouse CD4 particles-DM (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. The experiments and protocols used were approved by the Animal Use and Care Committee of the Institute for Virus Research at Kyoto University.
Statistical analyses
Statistical analyses were performed using the unpaired Student t test.
Results
HBZ inhibits the classical NF-κB signaling pathway
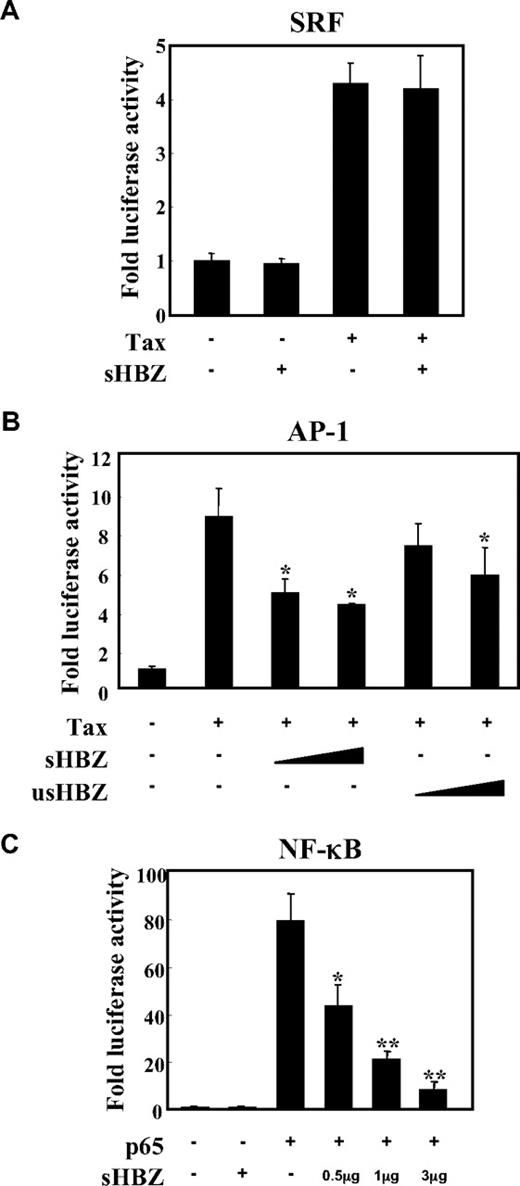
To analyze the possible involvement of HBZ in several signaling pathways in ATL, we first evaluated whether HBZ could influence the AP-1, SRF and NF-κB pathways, which are activated by the Tax protein,34,35 using a transient transfection assay. There are 2 isoforms of the HBZ gene: spliced and unspliced HBZ. In most of the experiments, we used sHBZ, because the protein derived from sHBZ is more abundant and potent than that of usHBZ.14 As shown in Figure 1A, the expression of sHBZ did not affect Tax-mediated effects using SRF reporter. Consistent with the previous report, sHBZ has ability to suppress Tax-induced AP-1 activation, and its inhibitory effect was more significant than that of usHBZ (Figure 1B). In addition, sHBZ suppressed p65 mediated NF-κB activation in a dose-dependent manner (Figure 1C).
sHBZ inhibited NF-κB and AP-1 activation but did not influence the SRF pathway. Jurkat cells were cotransfected with phRL-TK and reporter plasmid pSRF-Luc (A), AP-1-Luc (B), or κB-Luc (C), respectively, with or without 1 or 3 μg pME18Sneo-sHBZ and 1 μg pCG-Tax (A,B) or 1 μg pCMV-Tag 2-p65 (C). The total amount of DNA for transfection was equalized by adding empty vectors. After 48 hours, a dual luciferase reporter assay was preformed as described in “Methods”. All the data shown are relative values of firefly luciferase normalized to Renilla luciferase and expressed as mean of a triplicate set of experiments (± SD). *P < .05; **P < .01.
sHBZ inhibited NF-κB and AP-1 activation but did not influence the SRF pathway. Jurkat cells were cotransfected with phRL-TK and reporter plasmid pSRF-Luc (A), AP-1-Luc (B), or κB-Luc (C), respectively, with or without 1 or 3 μg pME18Sneo-sHBZ and 1 μg pCG-Tax (A,B) or 1 μg pCMV-Tag 2-p65 (C). The total amount of DNA for transfection was equalized by adding empty vectors. After 48 hours, a dual luciferase reporter assay was preformed as described in “Methods”. All the data shown are relative values of firefly luciferase normalized to Renilla luciferase and expressed as mean of a triplicate set of experiments (± SD). *P < .05; **P < .01.
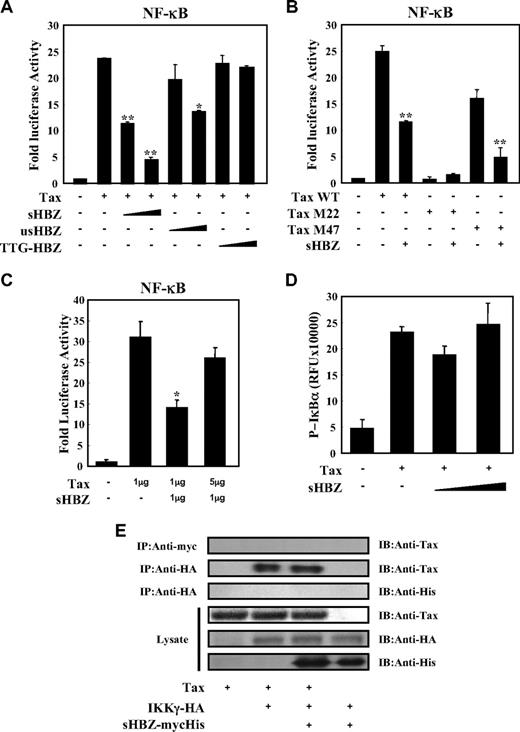
Tax has been reported to activate NF-κB by several mechanisms.36 sHBZ might have a negative regulatory effect on Tax-mediated NF-κB activation. When co-expressed, sHBZ dramatically repressed κB-Luciferase expression activated by wild type Tax and Tax M47 mutant (Figure 2A,B). However, because Tax M22 mutant did not activate NF-κB, HBZ expression had no effect. The NF-κB suppressive effect mediated by usHBZ was much weaker than that mediated by sHBZ. Although we previously reported that sHBZ RNA was responsible for growth-promoting activity,11 sHBZ RNA did not influence NF-κB (Figure 2A, TTG-HBZ), indicating that HBZ protein has the suppressive function on NF-κB. On the other hand, overexpression of Tax overcame the sHBZ-mediated suppression of NF-κB transcriptional activation (Figure 2C).
sHBZ suppressed Tax-mediated NF-κB activation but did not interfere with the Tax/IKKγ complex. (A,B) sHBZ repressed Tax-induced NF-κB activation. Jurkat cells were cotransfected with κB-Luc and phRL-TK, with or without 1 μg pCG-Tax, pCG-Tax M47, pCG Tax M22, and with 1 or 3 μg pME18Sneo-sHBZ, usHBZ, or TTG-HBZ. After 48 hours, luciferase activity was measured. (C) Tax overexpression overcame sHBZ-mediated suppression of NF-κB activation. Jurkat cells were cotransfected with κB-Luc and phRL-TK, with or without pCG-Tax and pME18Sneo-sHBZ. The total amount of DNA was equalized by adding empty vectors. (D) sHBZ could not modulate the Tax-driven phosphorylation of IκBα. Jurkat cells were transfected with pCG-Tax and pME18Sneo-sHBZ. Cell lysates were subjected to FunctionELISA IκBα assay. (E) sHBZ did not influence the interaction between Tax and IKKγ. 293FT cells were transfected with the indicated cDNA expression constructs. Cell lysates were subjected to immunoprecipitation (IP) with anti–c-Myc and anti-HA followed by immunoblotting (IB) using anti-Tax or anti-His, respectively. The expression levels of Tax, IKKγ and sHBZ were analyzed. *P < .05; **P < .01.
sHBZ suppressed Tax-mediated NF-κB activation but did not interfere with the Tax/IKKγ complex. (A,B) sHBZ repressed Tax-induced NF-κB activation. Jurkat cells were cotransfected with κB-Luc and phRL-TK, with or without 1 μg pCG-Tax, pCG-Tax M47, pCG Tax M22, and with 1 or 3 μg pME18Sneo-sHBZ, usHBZ, or TTG-HBZ. After 48 hours, luciferase activity was measured. (C) Tax overexpression overcame sHBZ-mediated suppression of NF-κB activation. Jurkat cells were cotransfected with κB-Luc and phRL-TK, with or without pCG-Tax and pME18Sneo-sHBZ. The total amount of DNA was equalized by adding empty vectors. (D) sHBZ could not modulate the Tax-driven phosphorylation of IκBα. Jurkat cells were transfected with pCG-Tax and pME18Sneo-sHBZ. Cell lysates were subjected to FunctionELISA IκBα assay. (E) sHBZ did not influence the interaction between Tax and IKKγ. 293FT cells were transfected with the indicated cDNA expression constructs. Cell lysates were subjected to immunoprecipitation (IP) with anti–c-Myc and anti-HA followed by immunoblotting (IB) using anti-Tax or anti-His, respectively. The expression levels of Tax, IKKγ and sHBZ were analyzed. *P < .05; **P < .01.
It has been reported that activation of the classical NF-κB pathway by Tax is mediated by its ability to physically bind to IKKγ, which results in enhanced phosphorylation and subsequent degradation of IκB.21 We analyzed whether sHBZ could modulate the Tax-driven phosphorylation of IκBα using a FunctionELISA kit (Figure 2D). No effects on the Tax-up-regulated phosphorylation level of IκBα were observed when cells were cotransfected with sHBZ. In addition, we investigated whether HBZ influenced the interaction between Tax and IKKγ. 293FT cells were transfected with various combinations of Tax, HA-tagged IKKγ and mycHis-tagged sHBZ expression vectors. As shown in Figure 2E, sHBZ did not interfere with the formation of Tax/IKKγ complexes, and there was no obvious binding of sHBZ to IKKγ or Tax.
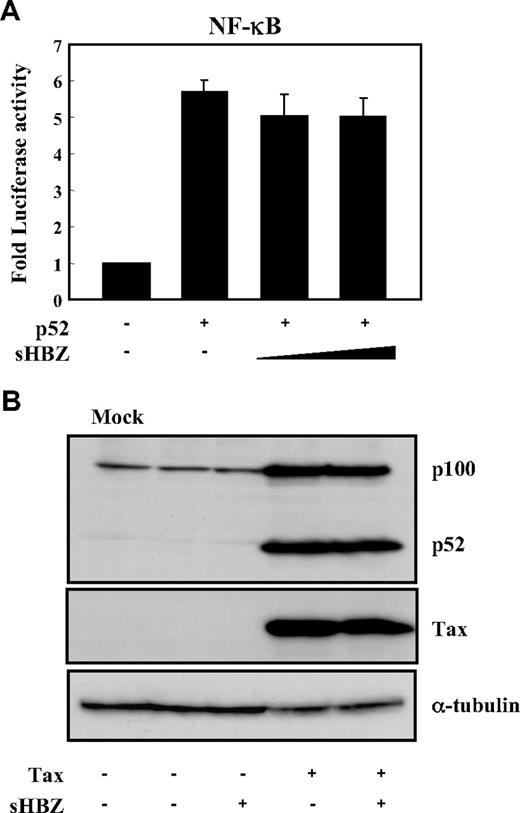
Two main pathways control the nuclear translocation of NF-κB: the classical and alternative NF-κB pathways.36 Recently, constitutive activation of the alternative NF-κB pathway has been reported in several lymphoid malignancies, where it may play a critical role.37 We next analyzed the effect of sHBZ on the alternative (p52 mediated) pathway of NF-κB activation. Figure 3A demonstrates that sHBZ was not capable of suppressing p52-mediated NF-κB activation. In addition, sHBZ had no remarkable effect on Tax-mediated up-regulation of p52 and its precursor protein p100 (Figure 3B). Thus, HBZ-mediated suppression of NF-κB activation occurs via inhibition of the classical pathway.
sHBZ did not affect alternative NF-κB pathway. (A) The effect of sHBZ on p52-mediated alternative NF-κB activation. Jurkat cells were cotransfected with κB-Luc, phRL-TK, pEF-p52, and pME18Sneo-sHBZ. Luciferase levels were measured after 48 hours. (B) sHBZ did not influence p52 and p100 expression. Jurkat cells were cotransfected with vectors that express Tax and sHBZ. After 48 hours, cell lysates were then subjected to immunoblot with anti-p52, Tax, and α-tubulin.
sHBZ did not affect alternative NF-κB pathway. (A) The effect of sHBZ on p52-mediated alternative NF-κB activation. Jurkat cells were cotransfected with κB-Luc, phRL-TK, pEF-p52, and pME18Sneo-sHBZ. Luciferase levels were measured after 48 hours. (B) sHBZ did not influence p52 and p100 expression. Jurkat cells were cotransfected with vectors that express Tax and sHBZ. After 48 hours, cell lysates were then subjected to immunoblot with anti-p52, Tax, and α-tubulin.
Domains of sHBZ responsible for suppression of NF-κB p65
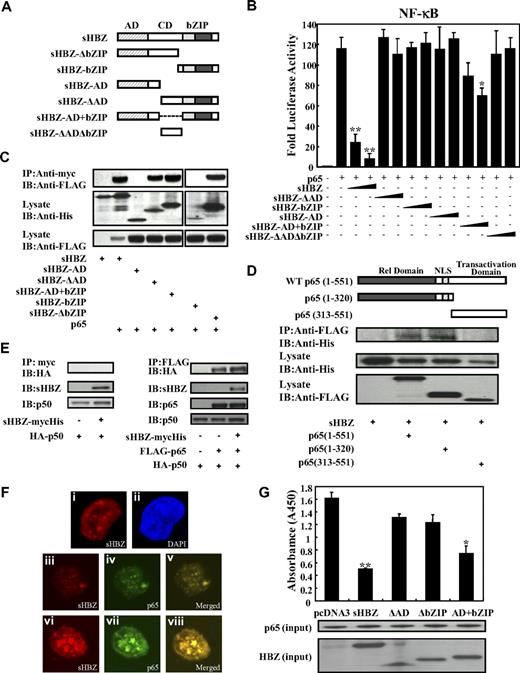
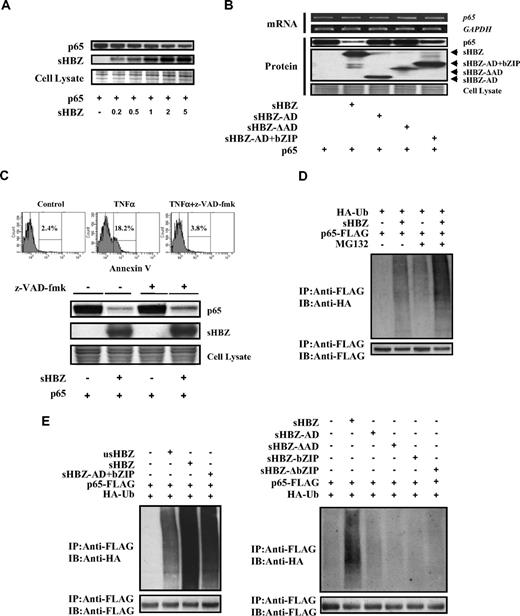
Next, we sought to identify the region of sHBZ responsible for the repression of NF-κB activation. To this end, we tested the sHBZ deletion mutants shown in Figure 4A. As shown in Figure 4B, wild-type sHBZ dramatically down-regulated p65-mediated NF-κB activation. Compared with other mutants, only the sHBZ-AD+bZIP mutant exhibited weak suppressive activity.
Domains of sHBZ responsible for suppression of NF-κB p65. (A) Schematic diagram of HBZ and its mutants used in this study. Characteristic domains of HBZ are indicated as follows: activation domain (AD), central domain (CD) and basic leucine zipper domain (bZIP). (B) Analysis of HBZ deletion mutants for the effect on p65-mediated NF-κB activation. Jurkat cells were cotransfected with κB-Luc and phRL-TK, with or without 1 μg of pCMV-Tag 2-p65, and with 1 or 5 μg pME18Sneo-sHBZ or sHBZ mutant. After 48 hours, luciferase levels were measured. *P < .05; **P < .01. Error bars represent SD. (C) Determination of the region of HBZ responsible for the interaction with p65. 293FT cells were transfected with the indicated mycHis-sHBZ mutants along with the FLAG-p65 vector. Cell lysates were subjected to immunoprecipitation (IP) using anti–c-Myc followed by immunoblotting (IB) using anti-FLAG. The expression levels of p65 and sHBZ mutants were detected. (D) Mapping the region of the p65 protein necessary for the interaction with sHBZ. The schema of p65 deletion mutants has been shown. The locations of the Rel homology domain, the nuclear localization signal (NLS), and the transactivation domain are indicated. 293FT cells were transfected with mycHis-sHBZ along with full-length or mutant FLAG-p65. At 48 hours after transfection, total cell lysates were subjected to IP using anti-FLAG followed by IB using anti-His. (E) HBZ did not influence p65/p50 interaction. 293FT cells were transfected with the indicated expression vectors. Cell lysates were subjected to IP using anti–c-Myc or anti-FLAG followed by IB using anti-HA. The expression levels of p65, p50, and sHBZ were detected. (F) sHBZ colocalizated with p65 in the cell nucleus. HeLa cells were transfected with mycHis-sHBZ together with (panels iii-viii) or without (panels i,ii) FLAG-p65. sHBZ was detected using anti-MYC Cy3 antibody (panels i,iii,vi). p65 was detected using anti–Flag-biotin and secondary Streptavidin-Alexa 488 antibody (panels iv,vii). The overlay of sHBZ and p65 is shown (panels v,viii). DAPI (4,6 diamidino-2-phenylindole) was used to counterstain the nucleus (panel ii). (G) sHBZ decreased p65 DNA binding capability. 293FT cells were transfected with FLAG-p65 together with either mycHis-sHBZ or one of its mutants. Cell lysates were subjected to the enzyme-linked immunosorbent assay (ELISA)–based NoShift assay to measure the DNA binding capability of p65. The absorbance at 450 nm indicated the binding ability of p65 (top panel). The bottom panel shows the amount of p65 and sHBZ in the 20% of input for analysis. *P < .05; **P < .01. Error bars represent SD.
Domains of sHBZ responsible for suppression of NF-κB p65. (A) Schematic diagram of HBZ and its mutants used in this study. Characteristic domains of HBZ are indicated as follows: activation domain (AD), central domain (CD) and basic leucine zipper domain (bZIP). (B) Analysis of HBZ deletion mutants for the effect on p65-mediated NF-κB activation. Jurkat cells were cotransfected with κB-Luc and phRL-TK, with or without 1 μg of pCMV-Tag 2-p65, and with 1 or 5 μg pME18Sneo-sHBZ or sHBZ mutant. After 48 hours, luciferase levels were measured. *P < .05; **P < .01. Error bars represent SD. (C) Determination of the region of HBZ responsible for the interaction with p65. 293FT cells were transfected with the indicated mycHis-sHBZ mutants along with the FLAG-p65 vector. Cell lysates were subjected to immunoprecipitation (IP) using anti–c-Myc followed by immunoblotting (IB) using anti-FLAG. The expression levels of p65 and sHBZ mutants were detected. (D) Mapping the region of the p65 protein necessary for the interaction with sHBZ. The schema of p65 deletion mutants has been shown. The locations of the Rel homology domain, the nuclear localization signal (NLS), and the transactivation domain are indicated. 293FT cells were transfected with mycHis-sHBZ along with full-length or mutant FLAG-p65. At 48 hours after transfection, total cell lysates were subjected to IP using anti-FLAG followed by IB using anti-His. (E) HBZ did not influence p65/p50 interaction. 293FT cells were transfected with the indicated expression vectors. Cell lysates were subjected to IP using anti–c-Myc or anti-FLAG followed by IB using anti-HA. The expression levels of p65, p50, and sHBZ were detected. (F) sHBZ colocalizated with p65 in the cell nucleus. HeLa cells were transfected with mycHis-sHBZ together with (panels iii-viii) or without (panels i,ii) FLAG-p65. sHBZ was detected using anti-MYC Cy3 antibody (panels i,iii,vi). p65 was detected using anti–Flag-biotin and secondary Streptavidin-Alexa 488 antibody (panels iv,vii). The overlay of sHBZ and p65 is shown (panels v,viii). DAPI (4,6 diamidino-2-phenylindole) was used to counterstain the nucleus (panel ii). (G) sHBZ decreased p65 DNA binding capability. 293FT cells were transfected with FLAG-p65 together with either mycHis-sHBZ or one of its mutants. Cell lysates were subjected to the enzyme-linked immunosorbent assay (ELISA)–based NoShift assay to measure the DNA binding capability of p65. The absorbance at 450 nm indicated the binding ability of p65 (top panel). The bottom panel shows the amount of p65 and sHBZ in the 20% of input for analysis. *P < .05; **P < .01. Error bars represent SD.
Accumulating evidence shows that the interaction between bZIP proteins and Rel family proteins affects their subcellular localization and modulates transcription activation.38,39 Therefore, we investigated whether sHBZ can physically interact with NF-κB p65. 293FT cells were transfected with vectors expressing sHBZ and p65. Figures 4C through E illustrate the physical binding between sHBZ and p65 or p50. HBZ has 3 domains, an activation domain (AD), a central domain (CD) and a basic leucine zipper domain (bZIP). To determine which portion of sHBZ is necessary for the binding with NF-κB p65 protein, we performed a coimmunoprecipitation assay. Three mutants (sHBZ-ΔAD, sHBZ-ΔbZIP, and sHBZ-AD+bZIP) could bind to p65 (Figure 4C), indicating that at least 2 of 3 main domains in sHBZ were necessary for the binding between sHBZ and p65. However, only sHBZ-AD+bZIP could suppress NF-κB activation as shown in Figure 4B, indicating the significance of AD and bZIP domains for binding with p65.
To determine which part of p65 protein is necessary for binding with sHBZ, we tested the ability of a series of FLAG-tagged truncated p65 proteins to interact with the full length of sHBZ (Figure 4D). The p65 (313-551) mutant, which did not contain the Rel homology domain, was incapable of interacting with the sHBZ protein. The truncated p65, 1 to 320, which lacked the transactivation domain but remained the Rel homology domain, still interacted efficiently with sHBZ. We conclude that sHBZ interacts with the Rel homology domain of p65. Next, we studied the binding of sHBZ to p50 by immunoprecipitation. As shown in Figure 4E, HBZ did not bind to p50, and did not interfere with the binding between p65 and p50.
The HBZ-p65 interaction was further investigated by confocal microscopy. HBZ exhibited a granular speckles pattern as previously reported15 (Figure 4Fi). After stimulating with PMA/ionomycin, the cotransfected cells showed nuclear spots representing colocalization of sHBZ and p65 (Figure 4F iii-v, vi-viii).
Because sHBZ interacts with p65, we studied whether sHBZ inhibits the ability of p65 to bind the κB binding site. As expected, sHBZ dramatically decreased p65 DNA binding capability (Figure 4G). Furthermore, analysis using deletion mutants of sHBZ revealed that sHBZ-AD+bZIP could also decrease the binding of p65 protein to DNA. The suppressive effects of deletion mutants lacking AD or bZIP were not statistically significant. This result suggests that both AD and bZIP domains are important for the inhibition of p65 binding to its target sequence, and coincides with the results of the luciferase assay.
Taken together, these observations demonstrate that sHBZ represses p65-induced transcription through direct physical association between sHBZ and p65 via the AD and bZIP domains, and this interaction inhibits the binding of p65 to target sites in DNA.
HBZ promotes p65 degradation through a ubiquitination-dependent pathway
A previous report showed that HBZ could bind to c-Jun, and decrease c-Jun protein via proteasome dependent mechanism.17 Therefore, we analyzed whether sHBZ could also affect the turnover of NF-κB p65 protein. Expression of sHBZ repressed the level of p65 protein in a dose-dependent manner (Figure 5A). To determine which domain of sHBZ is responsible for the down-regulation of p65 protein levels, we analyzed the level of p65 when co-expressed with each of the sHBZ mutants (Figure 5B). The level of p65 was slightly decreased when the sHBZ-AD+bZIP mutant was expressed, again suggesting that the AD and bZIP domains of sHBZ are required for suppression of p65. By semiquantitative RT-PCR, we confirmed that the level of p65 mRNA did not differ when co-expressed with sHBZ and its mutants (Figure 5B). Taken together, these results demonstrated that sHBZ reduced the level of p65 protein by acting at a posttranscriptional level.
sHBZ promotes p65 degradation through a ubiquitination-dependent pathway. (A) sHBZ repressed the level of p65 in a dose-dependent manner. 293FT cells were transfected with 1 μg pEF-p65 and various amounts of mycHis-sHBZ (0.2, 0.5, 1, 2, and 5 μg). After 36 hours, the cell lysates were subjected to immunoblotting. (B) Activation and leucine-zipper domains of sHBZ were necessary for suppression of p65. 293FT cells were transfected with 50 ng pEF-p65 and 250 ng either mycHis-sHBZ or its mutants. At 36 hours after transfection, the level of p65 mRNA was analyzed by semiquantitative RT-PCR. The levels of GAPDH mRNA are shown as internal control (top panel). Whole cell lysates were subjected to immunoblotting (bottom panel). (C) sHBZ-mediated suppression of p65 protein is caspase independent. Top panel: 293FT cells were transfected with FLAG-p65 together with mycHis-sHBZ. The caspase inhibitor z-VAD-fmk was added 2 hours before transfection. At 48 hours after transfection, cell lysates were subjected to immunoblotting. Bottom panel: Jurkat cells were cultured in the presence of indicated drugs for 24 hours. Cell death was analyzed by annexin V staining. (D,E) sHBZ accelerated the ubiquitination of p65. 293FT cells were transfected with FLAG-p65, HA-ubiquitin, and either mycHis-sHBZ or its mutants. After 24 hours, cells were treated with or without MG132 for 12 hours. Cell lysates were subjected to IP using anti-FLAG followed IB using anti-HA.
sHBZ promotes p65 degradation through a ubiquitination-dependent pathway. (A) sHBZ repressed the level of p65 in a dose-dependent manner. 293FT cells were transfected with 1 μg pEF-p65 and various amounts of mycHis-sHBZ (0.2, 0.5, 1, 2, and 5 μg). After 36 hours, the cell lysates were subjected to immunoblotting. (B) Activation and leucine-zipper domains of sHBZ were necessary for suppression of p65. 293FT cells were transfected with 50 ng pEF-p65 and 250 ng either mycHis-sHBZ or its mutants. At 36 hours after transfection, the level of p65 mRNA was analyzed by semiquantitative RT-PCR. The levels of GAPDH mRNA are shown as internal control (top panel). Whole cell lysates were subjected to immunoblotting (bottom panel). (C) sHBZ-mediated suppression of p65 protein is caspase independent. Top panel: 293FT cells were transfected with FLAG-p65 together with mycHis-sHBZ. The caspase inhibitor z-VAD-fmk was added 2 hours before transfection. At 48 hours after transfection, cell lysates were subjected to immunoblotting. Bottom panel: Jurkat cells were cultured in the presence of indicated drugs for 24 hours. Cell death was analyzed by annexin V staining. (D,E) sHBZ accelerated the ubiquitination of p65. 293FT cells were transfected with FLAG-p65, HA-ubiquitin, and either mycHis-sHBZ or its mutants. After 24 hours, cells were treated with or without MG132 for 12 hours. Cell lysates were subjected to IP using anti-FLAG followed IB using anti-HA.
To further elucidate the mechanism by which sHBZ reduces the amount of p65, we measured p65 levels after blocking caspase activity because caspase-mediated cleavage of the NF-κB p65/RelA subunit has been reported as a mechanism of decreasing p65 levels.27 The activity of z-VAD-fmk caspase inhibitor was confirmed by the finding that z-VAD-fmk could block apoptosis induced by TNFα (Figure 5C). Treatment with z-VAD-fmk caspase inhibitor did not change the down-regulation of p65 by sHBZ (Figure 5C top panel), indicating that caspase-mediated cleavage of p65 was not involved in this inhibitory effect.
Protein ubiquitination is a crucial modification, which induces degradation of proteins in the proteasome and controls the activity of many signaling molecules, including transcription factors such as p53, c-Jun, and p65.40-42 We therefore studied whether expression of sHBZ could induce the polyubiquitination of p65 and trigger the degradation of p65 protein. Immunoprecipitation studies showed that expression of sHBZ induced heavy ubiquitination of p65 (Figure 5D). In the presence of a proteasome inhibitor, sHBZ remarkably accelerated the ubiquitination of p65 (Figure 5D). This effect was far stronger than that of usHBZ (Figure 5E). Analysis of sHBZ deletion mutations showed that only sHBZ-AD+bZIP induced polyubiquitination of p65, while other mutants had no effect on p65 ubiquitination (Figure 5E).
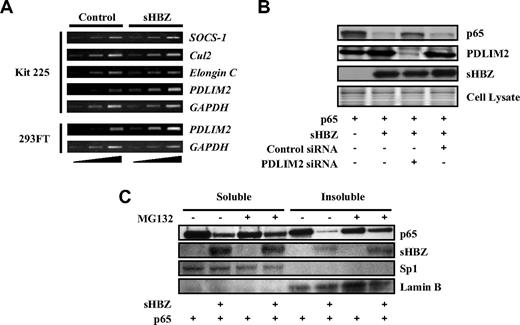
Several ubiquitin E3 ligases have been shown to specifically induce the ubiquitination of p65, including SOCS-1, Cul2, Elongin B/C, and PDLIM2.41,43-45 To identify the ubiquitin E3 ligase responsible for polyubiquitination of p65 in Kit 225 cells, we analyzed transcriptional profiles of these ubiquitin E3 ligase genes in sHBZ-transfected and control cells. Expression of these candidate genes was analyzed by semiquantitative RT-PCR with different cycles of amplification, which indicated that transcription of the PDLIM2 gene was up-regulated in sHBZ-transfected Kit 225 and 293FT cells (Figure 6A).
HBZ up-regulates expression of PDLIM2 gene. (A) sHBZ up-regulates PMLIM2. Total RNA was extracted from sHBZ-expressing or control Kit 225 and 293FT cells. The levels of SOCS-1, Cul2, Elongin C, PDLIM2, and GAPDH mRNA were measured by semiquantitative RT-PCR. The ramp on the left represented an increasing PCR cycle number. (B) Reducing PDLIM2 expression by siRNA recovered sHBZ-mediated suppression of p65. 293FT cells were transfected with expression vectors together with PDLIM2 siRNA or control siRNA. Protein expression was analyzed by western blotting. (C) sHBZ induced the degradation of insoluble p65. 293FT cells, untreated or treated with MG132, were transfected with mycHis-sHBZ along with FLAG-p65. After 48 hours, soluble and insoluble nuclear fractions were subjected to immunblotting. The expression levels of p65, sHBZ, Sp1, and Lamin B were detected.
HBZ up-regulates expression of PDLIM2 gene. (A) sHBZ up-regulates PMLIM2. Total RNA was extracted from sHBZ-expressing or control Kit 225 and 293FT cells. The levels of SOCS-1, Cul2, Elongin C, PDLIM2, and GAPDH mRNA were measured by semiquantitative RT-PCR. The ramp on the left represented an increasing PCR cycle number. (B) Reducing PDLIM2 expression by siRNA recovered sHBZ-mediated suppression of p65. 293FT cells were transfected with expression vectors together with PDLIM2 siRNA or control siRNA. Protein expression was analyzed by western blotting. (C) sHBZ induced the degradation of insoluble p65. 293FT cells, untreated or treated with MG132, were transfected with mycHis-sHBZ along with FLAG-p65. After 48 hours, soluble and insoluble nuclear fractions were subjected to immunblotting. The expression levels of p65, sHBZ, Sp1, and Lamin B were detected.
To confirm whether increased PDLIM2 expression is associated with degradation of p65, we suppressed PDLIM2 expression by siRNA. When PDLIM2 expression was inhibited, p65, decreased by sHBZ, partially recovered, indicating that PDLIM2 acts in the degradation of p65 induced by sHBZ (Figure 6B). A previous report showed that PDLIM2 targets p65 to discrete intranuclear compartments (insoluble nuclear fraction) where polyubiqutinated p65 is degraded by the proteasome.41 We measured p65 levels in soluble and insoluble nuclear fractions of 293FT cells transfected with p65 and sHBZ expression vectors. The down-regulation of p65 protein in the insoluble nuclear fraction was more significant than that in the soluble fraction. Moreover, p65 levels in the insoluble fraction, but not in the soluble fraction, were partially restored by MG132 treatment (Figure 6C). These observations suggested that the insoluble p65 mainly underwent proteasomal degradation and were consistent with the idea that sHBZ acts on p65 levels via PDLIM2. It remains to be elucidated whether HBZ accelerates the ubiquitination of p65 by PDLIM2 in addition to increasing the expression of the PDLIM2 gene.
sHBZ partially represses selected classical NF-κB target genes
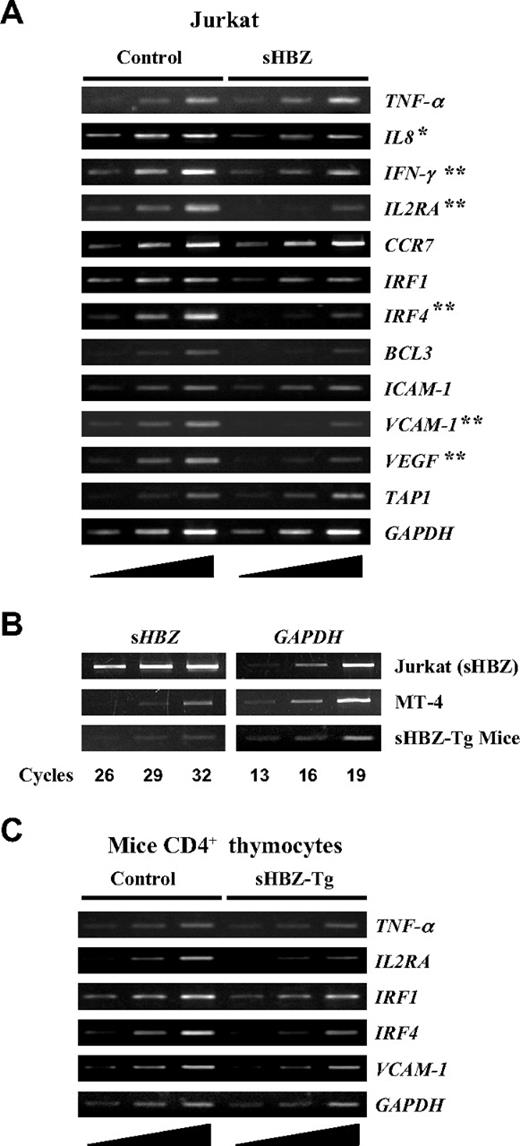
It is well established that the activation and translocation of NF-κB is associated with increased transcription of genes encoding chemokines, cytokines, adhesion molecules, and inhibitors of apoptosis. Recent studies showed that the classical and alternative pathways of NF-κB influence the expression of different sets of genes.24 Because HBZ suppresses the classical NF-κB pathway, we checked the effect of sHBZ on the expression of p65-specific target genes using Jurkat cells stably expressing sHBZ. Expression of genes was analyzed by semiquantitative RT-PCR (Figure 7A). Expression of sHBZ was associated with suppression of some selected target genes, which are normally up-regulated after PMA/ionomycin treatment, such as IL-8, IFN-γ, IL2RA, IRF4, VCAM-1, and VEGF. Because PMA/ionomycin treatment activates NF-κB mainly by the classical pathway, this observation indicates that HBZ expression modulates the transcription of genes activated by the classical pathway. However, overexpression of sHBZ gene might influence these results. Indeed, the level of sHBZ mRNA in sHBZ expressing Jurkat cells was much higher than that in an ATL cell line, MT-4 (Figure 7B). However, the level of sHBZ expression in MT-4 cells is comparable to that of CD4+ T cells from HBZ-transgenic mice. In this transgenic mouse, sHBZ gene is expressed by a mouse CD4-specific promoter/enhancer/silencer. To investigate HBZ-mediated suppression of genes activated by the classical NF-κB pathway in vivo, we studied the expression of p65-specific target genes in thymus CD4+ cells from sHBZ transgenic mice. After stimulating the cells with PMA/ionomycin, expressions of IL2RA, IRF4, and VCAM-1 genes were suppressed in sHBZ transgenic mice as observed in sHBZ transfected Jurkat cells (Figure 7C).
Suppressed expression of selected classical NF-κB target genes in vitro and in vivo by sHBZ. (A) Transcriptional changes of selected classical NF-κB target genes in sHBZ-expressing Jurkat cells. After stimulating the cells with PMA plus ionomycin, the levels of TNF-α, IL-8, IFN-γ, IL2RA, CCR7, IRF1, IRF4, BCL3, ICAM-1, VCAM-1, VEGF, TAP1, and GAPDH mRNA were analyzed by increasing cycles of semiquantitative RT-PCR, represented by the ramp on the left. (B) Comparison of the HBZ gene transcripts in an ATL cell, MT-4, in HBZ-transfected Jurkat cells, and in CD4+ thymocytes from HBZ transgenic mice. All samples were amplified over the same number of PCR cycles as shown. (C) Transcriptional changes of selected classical NF-κB target genes in CD4+ thymocytes from sHBZ transgenic mice. After stimulating the cells with PMA plus ionomycin, the levels of TNF-α, IL2RA, IRF1, IRF4, VCAM-1, and GAPDH mRNA were analyzed by increasing cycles of semiquantitative RT-PCR, represented by the ramp on the left. *P < .05; **P < .01.
Suppressed expression of selected classical NF-κB target genes in vitro and in vivo by sHBZ. (A) Transcriptional changes of selected classical NF-κB target genes in sHBZ-expressing Jurkat cells. After stimulating the cells with PMA plus ionomycin, the levels of TNF-α, IL-8, IFN-γ, IL2RA, CCR7, IRF1, IRF4, BCL3, ICAM-1, VCAM-1, VEGF, TAP1, and GAPDH mRNA were analyzed by increasing cycles of semiquantitative RT-PCR, represented by the ramp on the left. (B) Comparison of the HBZ gene transcripts in an ATL cell, MT-4, in HBZ-transfected Jurkat cells, and in CD4+ thymocytes from HBZ transgenic mice. All samples were amplified over the same number of PCR cycles as shown. (C) Transcriptional changes of selected classical NF-κB target genes in CD4+ thymocytes from sHBZ transgenic mice. After stimulating the cells with PMA plus ionomycin, the levels of TNF-α, IL2RA, IRF1, IRF4, VCAM-1, and GAPDH mRNA were analyzed by increasing cycles of semiquantitative RT-PCR, represented by the ramp on the left. *P < .05; **P < .01.
Discussion
Activation of the NF-κB signaling pathway, which has been reported in various cancer cells, plays an important role in the development and progression of tumor cells.46,47 In oncogenesis by HTLV-1, the tax gene has been extensively studied. Tax can activate various transcription pathways and functionally inhibit p53 and MAD1.7 Furthermore, various tumors have been observed in tax gene transgenic animals, depending on the promoter used.48,49 These findings show the oncogenic potential of Tax in vivo. Although Tax can transform Rat-1 cells in vitro, a Tax mutant lacking the ability to activate NF-κB lost its transforming activity, indicating that NF-κB activation is indispensable for Tax mediated transformation.18 Tax can activate NF-κB by both the classical and alternative pathways via its interactions with IKKγ20 and p100.22 Although they are often activated concurrently, the classical and alternative NF-κB pathways have distinct regulatory functions. Accumulating evidence suggests that the alternative NF-κB pathway is more important in several cancers.37 It has been reported that the classical and alternative NF-κB pathways differentially control genes with anti-apoptotic functions in lymphoma cell lines.50 In transformation by Tax, it has been reported that the alternative pathway is critical.51 This study demonstrates the selective suppression of the classical NF-κB pathway by HBZ, a phenomenon that selectively modulates NF-κB activation by Tax. In many ATL cells, Tax is not expressed, while the HBZ gene is expressed in all ATL cases. Even in ATL cells without Tax expression, NF-κB is constitutively activated.52 Recently, elevated expression of NIK has been reported in ATL cells.53 Because NIK activates both the classical and alternative NF-κB pathways, HBZ might modulate the classical NF-κB pathway even in the absence of Tax, leading to predominant activation of alternative pathway, and perhaps to oncogenesis.
The classical NF-κB pathway is still potently activated by Tax in the HTLV-1 transformed T-cell lines, such as MT-4 and MT-2, regardless of HBZ expression. Because these cell lines express large amount of Tax, it is likely that HBZ does not have strong suppressive effect on classical pathway of NF-κB due to excess Tax expression. However, the effect of HBZ on the classical NF-κB pathway may be more pronounced when the level of Tax expression is down-regulated or silenced as in chronically infected T cells in infected people and ATL cells.
Many viruses have developed strategies to manipulate NF-κB signaling through the use of multifunctional viral proteins. For example, the HIV-1 encoded Tat protein enhances NF-κB mediated LTR activation while HIV-1 Nef induces the expression of NF-κB inhibitor, IκBα, to suppress this pathway.54 In Epstein-Barr virus, the LMP-2 viral protein activates the NF-κB pathway by the recruitment of cellular adaptor proteins, TNF receptor–associated factor families and TNF receptor–associated death domain, to the C-terminal domain. Like HBZ, the EBV bZIP protein inhibits the classical NF-κB pathway through interacting with p65.28 Similar suppression of NF-κB has been reported for other viruses, including African swine fever virus, hepatitis C virus, and human herpesvirus-8. These findings show that NF-κB suppressive activities are common among different viruses, suggesting that these activities are important for viral infection. In this regard, it is noteworthy that transcription of the IFNγ and IRF4 genes, which is induced by the classical pathway, is suppressed by HBZ as shown in this study. A virus might facilitate escape from the host immune system by suppressing the classical NF-κB pathway in such a manner.
Viruses have evolved to sneak through the innate and adaptive antiviral response both at the cellular and whole organism levels, for survival and successful spread of infection. One mechanism used by some viruses to avoid immune surveillance is to control the level of cellular transcription factors by sorting them for degradation through ubiquitination. In all instances of which we are currently aware, this modulation process takes place at the level of E3 ligase, that is, at the step where the substrate specificity is critically defined. Some viral proteins act as E3 ligases, and others redirect host ubiquitin E3 ligases to target new substrate proteins. For example, the E6 oncoprotein of human papilloma virus binds the tumor suppressor p53 through its interaction with another cellular protein, E6-associated protein, leading to the degradation of p53 via the ubiquitin-mediated pathway.55 VIF encoded by HIV-1 connects APOBEC3G and APOBEC3F as a substrate to the multisubunit E3 ligase for polyubiquitination and degradation.56 Our study is the first to report that a viral protein enhances expression of the cellular ubiquitin E3 ligase, PDLIM2, resulting in the degradation of p65. In T cells, PDLIM2 can interact with STAT and p65 transcription factors and promote their polyubiquitination and subsequent degradation, thereby negatively regulating STAT and NF-κB–dependent signaling.41,57 We have not yet clarified whether HBZ can negatively regulate the JAK/STAT pathway. Because PDLIM2 is expressed not only in T cells but also in innate immune cells,57 we speculate that the positive effect of HBZ on PDLIM2 expression might also influence T-cell proliferation and immune responses.
There are 2 transcripts of the HBZ gene. The transcript of the sHBZ gene is more abundant than that of usHBZ as reported.14 Inhibitory effect of sHBZ on Tax mediated transcription from 5′LTR was much stronger than that of usHBZ, and sHBZ has a much longer half-life than usHBZ. Therefore, the protein level of sHBZ is much higher than that of usHBZ,32 which leads to stronger inhibitory activity of the classical NF-κB pathway as shown in this study. Taken together, sHBZ, rather than usHBZ, is more important in HTLV-1 infected cells.
As shown in this study, HBZ downmodulates the classical NF-κB pathway by 2 mechanisms, (1) inhibition of DNA binding by p65 and (2) enhanced degradation of p65, leading to decreased expression of some RelA specific target genes. Such a function of HBZ in cooperation with Tax-mediated activation might be beneficial for proliferation of infected cells and oncogenesis. Further studies are necessary to clarify the significance of HBZ in proliferation of infected cells and oncogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan, a grant from the Uehara Memorial Foundation, a grant from the Naito Foundation, and a grant from the Sumitomo Foundation to M.M.
Authorship
Contribution: T.Z., J.Y., Y.S., M.N., M.F., and M.M. designed the research; T.Z., J.Y., Y.S., and M.T. performed the research; T.Z., M.N., M.F., and M.M. analyzed the data; and T.Z., J.Y., M.N., M.F., and M.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masao Matsuoka, MD, PhD, Laboratory of Virus Control, Institute for Virus Research, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; e-mail: mmatsuok@virus.kyoto-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal