Leukemias that harbor translocations involving the mixed lineage leukemia gene (MLL) possess unique biologic characteristics and often have an unfavorable prognosis. Gene expression analyses demonstrate a distinct profile for MLL-rearranged leukemias with consistent high-level expression of select Homeobox genes, including HOXA9. Here, we investigated the effects of HOXA9 suppression in MLL-rearranged and MLL-germline leukemias using RNA interference. Gene expression profiling after HOXA9 suppression demonstrated co–down-regulation of a program highly expressed in human MLL-AML and murine MLL-leukemia stem cells, including HOXA10, MEIS1, PBX3, and MEF2C. We demonstrate that HOXA9 depletion in 17 human AML/ALL cell lines (7 MLL-rearranged, 10 MLL-germline) induces proliferation arrest and apoptosis specifically in MLL-rearranged cells (P = .007). Similarly, assessment of primary AMLs demonstrated that HOXA9 suppression induces apoptosis to a greater extent in MLL-rearranged samples (P = .01). Moreover, mice transplanted with HOXA9-depleted t(4;11) SEMK2 cells revealed a significantly lower leukemia burden, thus identifying a role for HOXA9 in leukemia survival in vivo. Our data indicate an important role for HOXA9 in human MLL-rearranged leukemias and suggest that targeting HOXA9 or downstream programs may be a novel therapeutic option.
Introduction
Translocations involving the mixed lineage leukemia locus (MLL, All-1, HRX) on chromosome 11q23 are found in a variety of hematologic malignancies, including acute myeloid leukemias (AMLs), B-precursor and T-lineage acute lymphoblastic leukemias, and myelodysplastic syndrome. MLL rearrangements are present in most infant leukemias1,,–4 and in secondary leukemias after treatment with topoisomerase inhibitors.5,,–8 Infants diagnosed with lymphoblastic leukemia harboring a MLL translocation respond poorly to current chemotherapy regimens and have a particularly unfavorable prognosis with an overall survival of less than 50%.9,,,–13
The pathophysiologic mechanisms by which MLL translocations cause leukemia and the genes that serve as critical downstream targets during induction and maintenance of the leukemic phenotype are incompletely characterized. Gene expression profiling in human acute myeloid and lymphoblastic leukemias demonstrated a characteristic gene expression pattern for cases with MLL rearrangements14,–16 that may be driven by unique histone methylation programs.17,,–20 A common unifying feature in myeloid and lymphoid leukemias with MLL rearrangements is high-level expression of Homeobox (HOX) genes with a particular emphasis on the 5′-HOXA genes (HOXA5-11).14,–16,21,22 Elevated expression of certain 5′-HoxA cluster genes is also found in murine leukemia models after introduction of various leukemia-associated Mll-fusion proteins.23,,,–27 In a recent murine retroviral transduction/transplantation study, we determined the gene expression profile of leukemia stem cells that were initiated by expression of Mll-Af9 in committed granulocyte macrophage progenitors (GMPs).28 5′-HoxA cluster genes HoxA5, HoxA10, and in particular HoxA9 were prominent members of a gene expression signature found in leukemia stem cells and were immediately induced after Mll-Af9 expression.28 These findings support a hierarchical model of leukemia initiation by the MLL-AF9 fusion where certain HOXA cluster genes belong to a crucial subset of proximate target genes, which are immediately activated by MLL-AF9 expression.28,29 Further evidence that MLL fusion oncoproteins may directly regulate certain HOX genes in hematopoietic cells emerges from recent studies in which wild-type MLL or MLL fusion proteins were found to be part of a multiprotein complex that regulates chromatin structure near the 5′-HOXA cluster.30,,–33
HOXA9 is directly involved in human leukemia caused by the translocation of HOXA9 with the gene encoding NUP98, a nucleoporin protein.34,35 Expression of this fusion protein immortalizes murine bone marrow cells and results in a myeloproliferative disorder and AML in bone marrow transplantation experiments.36,37 Retrovirally enforced overexpression of HOX genes in murine and human marrow cells enhances expansion of hematopoietic cells in short-term in vitro cultures, and murine transplantation studies showed that forced expression of HoxA9 in hematopoietic precursors results in a significant expansion of hematopoietic stem cells and leukemia.38,,,,–43 Moreover, coexpressing HoxA9 suppresses Meis1-mediated apoptosis in a variety of murine and human leukemia cell types.44
During normal hematopoietic development, HOX genes are expressed in the stem cell and immature progenitor compartments of bone marrow but are transcriptionally down-regulated on induction of terminal differentiation.45,–47 Homozygous HoxA9-deficient mice are viable but exhibit a variety of mild hematopoietic defects, including reduced number of committed myeloid and B-cell progenitors and a blunted neutrophilic response to granulocyte colony-stimulating factor.48,49 HoxA9-deficient hematopoietic stem cells (HSCs) demonstrate an impaired proliferative response to cytokines in vitro and defective competitive repopulating activity in vivo.50 Thus, HoxA9 does appear to play a role in normal HSC biology, but is not essential for HSC development.
Although most studies point to some role for HOX genes in myeloid leukemogenesis, it remains unclear whether continued expression of any HOXA cluster gene is necessary for survival of AMLs. Murine studies addressing this issue have provided mixed results. Mll-Enl–transduced murine myeloid progenitors deficient for HoxA9 demonstrate significantly compromised self-renewal potential in vitro and impaired development of leukemias when transplanted in vivo.27 However, HoxA9−/−-deficient progenitors transduced with Mll-Gas7 are capable of initiating leukemia,51 and Mll-Af9+, HoxA9−/− mice develop leukemia with a similar frequency and latency as mice carrying Mll-Af9 with germline HoxA9.24 These studies raise the issue as to whether there is compensation for the lack of HoxA9 during development, and prompt assessment of an acute loss of HOXA9 expression in a fully developed leukemia. In this study, we aimed to further elucidate the role of HOXA9 expression in human MLL-rearranged and nonrearranged leukemias. Through use of a small hairpin RNA (shRNA)–mediated RNA interference approach and subsequent gene expression profiling analysis, we demonstrate a marked impact of HOXA9 suppression on survival of human MLL-rearranged leukemia cells associated with tight co–down-regulation of genes that were part of a MLL signature in primary human samples.
Methods
Generation of murine leukemia stem cells
Murine leukemic granulocyte macrophage progenitors (L-GMPs) were generated as described previously28 (Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). For HOXA9 shRNA knockdown experiments, IL-7R−Lin−Sca-1−c-Kit+CD34+-FcgRII/III+ GMP-like leukemic cells (L-GMPs) sorted from leukemic mice were incubated with lentiviral shRNA vectors and plated in methylcellulose media M3234 or liquid culture (Myelocult M5300; StemCell Technologies, Vancouver, BC) supplemented with 10 ng/mL IL-3 (PeproTech, Rocky Hill, NJ), 1× penicillin/streptomycin (Invitrogen, Carlsbad, CA). All mouse studies were approved by the Dana Farber Cancer Institute Institutional Animal Care and Use Committee.
Leukemia primary patient samples
Pediatric AML samples were obtained from the Erasmus MC, Sophia Children's Hospital (Rotterdam, The Netherlands). Approval was obtained from the Erasmus MC Institutional Review Board for these studies. Informed consent was obtained in accordance with the Declaration of Helsinki. Analyzed samples consisted of cryopreserved cells of AML patients with confirmed rearrangement of the MLL locus determined by either cytogenetic analysis, fluorescence in situ hybridization, or Southern blot and AML patients with MLL germline karyotype. Patient samples 1, 2, 3, 5, and 6 harbored a t(9;11). Sample 4 possessed a t(11;19). Mean percentage of blasts was approximately 90%. Although a variable number of dead cells was observed after thawing in the same range as previously demonstrated,52 culture conditions were optimized to support 50% to 60% viable cells at 72 to 96 hours for subsequent phenotypic assays (see Document S1).
Transfection with shRNA constructs
shRNA cloned into hairpin pLKO.1-lentiviral vector were kindly synthesized by the Harvard/MIT Broad Institute RNA consortium using a complex selection algorithm to minimize off-target effects (http://www.broad.mit.edu/rnai/trc). Lentiviral supernatants were obtained in 293T cells by cotransfection of the shRNA plasmids, packaging plasmids containing gag, tat, and rev genes, and a plasmid expressing VSV-G. For lentiviral infection of leukemia cell lines or primary AML patient cells, 105 cells were seeded in 96-well tissue culture plates (100 μL per well) in the appropriate culture media. Polybrene (hexadimethrine bromide; Sigma-Aldrich, St Louis, MO) was added at a final concentration of 7 μg/mL. After adding 150 μL lentiviral particles (titer of each lentiviral shRNA construct adjusted to 1.6 × 106 transducing units/mL to achieve a multiplicity of infection of 2.4 transducing units/cell), spinoculation was performed at 2250g for 90 minutes at 37°C. Eight hours after infection, the media was replaced with 100 μL of the appropriate fresh culture media. If puromycin selection was desired, puromycin dihydrochloride (Sigma-Aldrich) at a final concentration of 2.5 μg/mL was added. Cells were incubated at 37°C, 5% CO2 until use for downstream applications (see Document S1).
Analysis of cell viability and apoptosis
To assay cell viability, we used the colormetric diphenyltetrazolium bromide (MTT) Cell Proliferation Kit I (Roche Diagnostics, Indianapolis, IN) according to the instructions of the manufacturer and flow cytometric analysis after fluorescein isothiocyanate–annexin V (BioVision, Mountain View, CA)/propidium iodide (Invitrogen) double staining. Cell-cycle analysis was performed by flow cytometric analysis of propidium iodide-stained cells at defined time points after shRNA transduction.
Murine transplantation experiments and in vivo imaging
Luminescent t(4;11) SEMK2-M1 cells were generated, and bioluminescent in vivo imaging was performed as previously described53 (Document S1). For further postmortem analysis of mouse leukemia burden, single-cell suspensions were generated from spleen. Red blood cells were lysed on ice using red blood cell lysis solution (Gentra Systems, Minneapolis, MN) for 5 minutes. After centrifugation, cells were resuspended in phosphate-buffered saline supplemented with 10% fetal bovine serum. A total of 5 × 108 of nucleated cells were incubated 40 minutes on ice with 10 μL of each of the following fluorochrome-conjugated antibodies: mouse anti–human CD19-TC (Invitrogen), anti–mouse CD45.2-fluorescein isothiocyanate (BD Biosciences, San Jose, CA). After a double wash in phosphate-buffered saline, the cell suspension was sorted using FACSAria multicolor cell sorter (BD Biosciences).
Data analysis and statistical methods
After hybridization to Affymetrix Human Genome U133A2.0 microarrays (Santa Clara, CA), data were analyzed using the GenePattern 3.0 software package,54 including the Gene set enrichment analysis (GSEA) module55 (Broad Institute; Document S1). All other statistical calculations were performed using SPSS 14.0 software package (SPSS, Chicago, IL). The original microarray data have been deposited into the Gene Expression Omnibus database, under accession number GSE13714.
Results
Establishment and validation of HOXA9 suppression in human leukemia cells
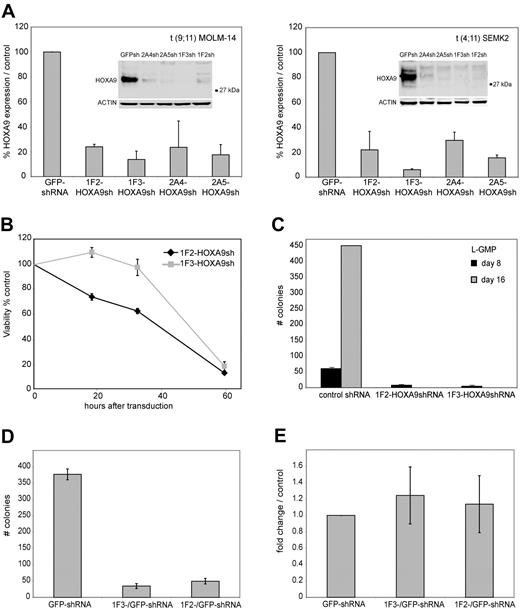
To establish efficient knockdown targeting the human HOXA9 mRNA, 5 different shRNA constructs (1F2-HOXA9shRNA, 1F3-HOXA9shRNA, 2A4-HOXA9 shRNA, 2A5-HOXA9shRNA, and 2A6-HOXA9shRNA) were stably introduced into target cells of interest using a lentiviral vector system. The shRNA sequences were selected using an algorithm that enhances the likelihood of efficient RNA interference (RNAi) and minimizes targeting of other mRNAs.56 Genome-wide blast search demonstrated at least 6 sequence mismatches for any other gene in the human genome database. First, we assessed transduction efficiency using the green fluorescent protein (GFP)–control shRNA construct in 17 different MLL-rearranged and MLL wild-type leukemia cell lines by selection in puromycin and demonstrated a high percentage of puromycin resistance (> 80% transduction efficiency), which was similar in each cell line tested (Figure S1), whereas nontransduced control cells of each type were effectively eliminated (not shown). Thus, all subsequent experiments were performed without puromycin selection. We then introduced each of the HOXA9 targeting shRNAs into t(9;11) MOLM-14 cells and t(4;11) SEMK2-M1 (SEMK2) cells and compared the suppression efficiency to cells that were transduced with either a nontargeting control shRNA directed toward GFP, an empty lentiviral vector, or untreated. Using quantitative real-time polymerase chain reaction (PCR), we found that 3 of 5 HOXA9shRNA constructs (1F2-HOXA9shRNA, 1F3-HOXA9shRNA, and 2A5-HOXA9shRNA) suppressed HOXA9 RNA levels by at least 80% compared with control. The 2A4-HOXA9shRNA construct showed a more variable effect (60%-80% suppression; Figure 1A), and the 2A6-HOXA9shRNA had only minimal effect (not shown) and was therefore excluded from further experiments. The efficacy of HOXA9 suppression by the 4 effective constructs was confirmed by Western blot analysis (Figure 1A). Because introduction of some RNAi constructs has been previously found to unspecifically induce interferon response,57 we next analyzed expression changes in the interferon response genes OAS1, IFITM1, and MX1 by quantitative real-time PCR as suggested in recent guidelines.58 Transduction of any of the lentiviral constructs showed no induction of the interferon response genes analyzed with expression levels similar to untreated control cells (Figure S2). Thus, we have validated 4 independent shRNA constructs that suppress HOXA9 expression in MLLAF9 and MLLAF4 human leukemia cell lines.
ShRNA-mediated knockdown of HOXA9 expression in human t(9;11) MOLM-14 and t(4;11) SEMK2 cells and murine leukemia stem cells (L-GMP). (A) Quantitative real-time PCR analysis of HOXA9 expression 72 hours after transduction of MOLM-14 and SEMK2 cells with 1 of 4 different HOXA9 shRNA constructs (1F2-HOXA9shRNA, 1F3-HOXA9shRNA, 2A4-HOXA9shRNA, and 2A5-HOXA9shRNA) without puromycin selection. A highly efficient HOXA9 suppression is demonstrated compared with a nontargeting control shRNA directed toward GFP. Western blot analysis 72 hours after transduction confirmed HOXA9 protein knockdown. (B) HoxA9 depletion in highly purified murine leukemia stem cells (L-GMP) resulted in a progressive induction of apoptosis in liquid culture compared with cells transduced with a control shRNA. (C) Analysis of L-GMP colony formation and replating capacity after HoxA9 suppression. Colonies were counted and replated at day 8. A significant decrease in colony number and replating capacity is demonstrated for L-GMPs transduced with HoxA9-directed shRNA without puromycin selection. (D) Analysis of L-GMP colony formation after HOXA9 suppression in a mixed-populations assay. L-GMPs were transduced with 1F2-HOXA9shRNA, 1F3-HOXA9shRNA, or GFP-control shRNA constructs, and populations of HOXA9shRNA or GFP-control shRNA-transduced cells were mixed in a ratio of 9:1 (HOXA9shRNA-transduced cells/GFP-control shRNA-transduced cells) and plated in semisolid medium. The total number of colonies at day 8 is shown for the mixed populations (1F2-HOXA9shRNA/GFP-control-shRNA; 1F3-HOXA9shRNA/GFP-control-shRNA) and the controls containing only GFP-control shRNA-transduced cells. (E) Quantitative real-time PCR further demonstrated similar HOXA9 expression in colonies that arose from the mixed HOXA9shRNA/GFP-control shRNA populations and controls containing only GFP-control shRNA-transduced cells (day 8).
ShRNA-mediated knockdown of HOXA9 expression in human t(9;11) MOLM-14 and t(4;11) SEMK2 cells and murine leukemia stem cells (L-GMP). (A) Quantitative real-time PCR analysis of HOXA9 expression 72 hours after transduction of MOLM-14 and SEMK2 cells with 1 of 4 different HOXA9 shRNA constructs (1F2-HOXA9shRNA, 1F3-HOXA9shRNA, 2A4-HOXA9shRNA, and 2A5-HOXA9shRNA) without puromycin selection. A highly efficient HOXA9 suppression is demonstrated compared with a nontargeting control shRNA directed toward GFP. Western blot analysis 72 hours after transduction confirmed HOXA9 protein knockdown. (B) HoxA9 depletion in highly purified murine leukemia stem cells (L-GMP) resulted in a progressive induction of apoptosis in liquid culture compared with cells transduced with a control shRNA. (C) Analysis of L-GMP colony formation and replating capacity after HoxA9 suppression. Colonies were counted and replated at day 8. A significant decrease in colony number and replating capacity is demonstrated for L-GMPs transduced with HoxA9-directed shRNA without puromycin selection. (D) Analysis of L-GMP colony formation after HOXA9 suppression in a mixed-populations assay. L-GMPs were transduced with 1F2-HOXA9shRNA, 1F3-HOXA9shRNA, or GFP-control shRNA constructs, and populations of HOXA9shRNA or GFP-control shRNA-transduced cells were mixed in a ratio of 9:1 (HOXA9shRNA-transduced cells/GFP-control shRNA-transduced cells) and plated in semisolid medium. The total number of colonies at day 8 is shown for the mixed populations (1F2-HOXA9shRNA/GFP-control-shRNA; 1F3-HOXA9shRNA/GFP-control-shRNA) and the controls containing only GFP-control shRNA-transduced cells. (E) Quantitative real-time PCR further demonstrated similar HOXA9 expression in colonies that arose from the mixed HOXA9shRNA/GFP-control shRNA populations and controls containing only GFP-control shRNA-transduced cells (day 8).
Effects of HoxA9 knockdown in murine leukemia stem cells
In our recent murine Mll-Af9 transduction/transplantation study, gene expression profiling revealed aberrant expression of HoxA9 as part of a stem cell signature in a IL-7R−Lin−Sca-1−c-Kit+CD34+FcgRII/III+ GMP-like (L-GMP) leukemia stem cell population.28 We therefore first explored the effects of HoxA9 depletion in this well-defined and previously characterized murine leukemia stem cell population. To achieve maximum HoxA9 knockdown in murine L-GMP, the 2 most effective HOXA9 shRNA constructs with 100% target sequence homology between the human and murine HOXA9 gene were selected (1F2-HOXA9shRNA, 1F3-HOXA9shRNA). These constructs had already been previously successfully used in murine hematopoietic cells.59 Using quantitative real-time PCR, we found that both shRNA constructs efficiently suppressed HOXA9 RNA levels in L-GMP compared with cells transduced with a nontargeting control shRNA (Figure S3). L-GMP cells were transduced with either the 1F2-HOXA9shRNA or the 1F3-HOXA9shRNA construct, and viability in liquid culture supplemented with cytokines was compared with L-GMP cells that were transduced with a nontargeting control shRNA. Whereas control shRNA-transduced cells survived in liquid culture, L-GMPs transduced with either the 1F2-HOXA9shRNA or 1F3-HOXA9shRNA construct showed a progressive induction of cell death with onset short after transduction, leading to almost complete loss of viable cells at day 3 (Figure 1B). Next, we analyzed colony-forming and replating capacity of L-GMPs transduced with 1F2-HOXA9shRNA, 1F3-HOXA9shRNA, or control shRNA constructs in semisolid medium. At day 8, colony formation was significantly reduced in the 2 L-GMP populations transduced with either the 1F2-HOXA9shRNA or 1F3-HOXA9shRNA construct demonstrating a colony count of less than 10% of the control shRNA treated group (Figure 1C). Moreover, replating and reevaluation at day 16 revealed a more than 10-fold expansion in the control L-GMP population, whereas colony-forming activity was completely abrogated in the HoxA9 shRNA-transduced populations (Figure 1C). Furthermore, analysis of colony-forming capacity of L-GMP in a mixed-populations assay demonstrated similar findings: L-GMPs were transduced with the 1F2-HOXA9shRNA, 1F3-HOXA9shRNA, or GFP-control shRNA construct. Cells were then mixed in a ratio of 9:1 (HOXA9shRNA-transduced cells: GFP-control shRNA-transduced cells) and plated in semisolid medium with cytokines. At day 8, a significantly lower number of colonies was observed in the 1F2-HOXA9shRNA/GFP-control shRNA and 1F3-HOXA9shRNA/GFP-control shRNA mixed populations compared with controls containing only cells transduced with GFP-control-shRNA (Figure 1D). Quantitative real-time PCR further demonstrated HOXA9 expression levels in the mixed populations similar to those of the controls (Figure 1E), indicating that only cells with nonsuppressed HOXA9 expression were capable of colony formation. These findings point toward a central role for HoxA9 in the maintenance of murine Mll-rearranged leukemia stem cells. We therefore sought to further characterize the effects of HOXA9 depletion in human leukemia cell lines and primary leukemia patient samples.
Effects of HOXA9 suppression in human MLLAF4 SEMK2 and MLLAF9 MOLM-14 leukemia cells
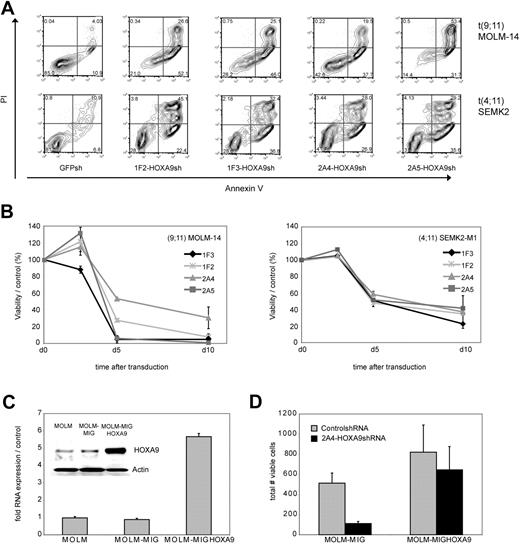
To examine the effect of HOXA9 suppression on the cellular phenotype in human leukemia cells, we first analyzed apoptosis induction as measured by annexin V/PI positivity after transduction of t(9,11) MOLM-14 AML M5 and t(4;11) SEMK2 ALL cells with each of the 4 HOXA9 targeted shRNAs or GFP-control shRNA. HOXA9 suppression led to a significant reduction of cell viability with all 4 HOXA9shRNA constructs compared with control (Figure 2A). In an independent experiment, time course analysis of apoptosis induction further demonstrated progressive reduction of cell viability starting 72 to 84 hours after transduction (Figure 2B). Induction of cell death was preceded by inhibition of proliferative capacity as assessed by MTT assay approximately 24 hours before induction of cell death became evident (48 hours after transduction; Figure S4).
Analysis of phenotypic consequences after shRNA-mediated HOXA9 suppression in t(9;11) MOLM-14 AML and t(4;11) SEMK2 ALL cells. (A) Analysis of cell viability as measured by annexin V/PI positivity at day 7 after transduction of t(9,11) MOLM-14 cells and t(4;11) SEMK2 cells with 1 of 4 HOXA9 targeted shRNAs or GFP control shRNA. The percentage of viable (annexin V/PI negative) cells is shown in the HOXA9shRNA and GFP control shRNA-transduced group. (B) Time course analysis of cell viability as measured by annexin V/PI positivity after HOXA9 suppression in an independent experiment. Cells were transduced with either 1 of 4 HOXA9 targeted shRNAs or GFP-control shRNA. The data are graphed as percentage of viable cells that were transduced with HOXA9shRNA/percentage of viable cells that were transduced with GFP-control shRNA at a given time point. These results were confirmed 3 times. (C) Quantitative real-time PCR and Western blot analysis of ectopic HOXA9 overexpression in t(9;11) MOLM-14 cells compared with untransduced cells and cells transduced with the empty vector construct. (D) MOLM-14 cells ectopically overexpressing HOXA9 were transduced with the 2A4-HOXA9shRNA construct specifically targeting the 3′-untranslated region of endogenous HOXA9 or the GFP-control shRNA construct. Ectopic overexpression of HOXA9 almost completely rescued the phenotype (annexin V/PI staining).
Analysis of phenotypic consequences after shRNA-mediated HOXA9 suppression in t(9;11) MOLM-14 AML and t(4;11) SEMK2 ALL cells. (A) Analysis of cell viability as measured by annexin V/PI positivity at day 7 after transduction of t(9,11) MOLM-14 cells and t(4;11) SEMK2 cells with 1 of 4 HOXA9 targeted shRNAs or GFP control shRNA. The percentage of viable (annexin V/PI negative) cells is shown in the HOXA9shRNA and GFP control shRNA-transduced group. (B) Time course analysis of cell viability as measured by annexin V/PI positivity after HOXA9 suppression in an independent experiment. Cells were transduced with either 1 of 4 HOXA9 targeted shRNAs or GFP-control shRNA. The data are graphed as percentage of viable cells that were transduced with HOXA9shRNA/percentage of viable cells that were transduced with GFP-control shRNA at a given time point. These results were confirmed 3 times. (C) Quantitative real-time PCR and Western blot analysis of ectopic HOXA9 overexpression in t(9;11) MOLM-14 cells compared with untransduced cells and cells transduced with the empty vector construct. (D) MOLM-14 cells ectopically overexpressing HOXA9 were transduced with the 2A4-HOXA9shRNA construct specifically targeting the 3′-untranslated region of endogenous HOXA9 or the GFP-control shRNA construct. Ectopic overexpression of HOXA9 almost completely rescued the phenotype (annexin V/PI staining).
Next, to further assess specificity of the observed correlation between HOXA9 suppression and induction of apoptosis, GFP-HOXA9 was ectopically overexpressed in MOLM-14 cells. GFP-positive cells were sorted and HOXA9 overexpression was confirmed by quantitative real-time PCR and Western blot analysis (Figure 2C). Transduction with the 2A4-HOXA9shRNA construct to specifically target the 3′-untranslated region of endogenous HOXA9 demonstrated almost complete rescue of the phenotype with a high level of protection by ectropic overexpression of nontargetable HOXA9 compared with controls (Figure 2D). Rescue of the phenotype by ectopically expressing a nontargetable HOXA9 construct was also confirmed in an independent experiment in t(9;11) THP-1 cells applying siRNA-mediated HOXA9 suppression (Figure S5).
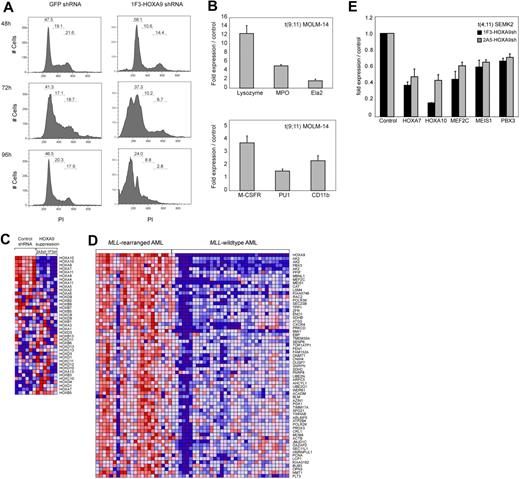
To further assess the effects on proliferation and viability after HOXA9 depletion, we analyzed cell cycle status. A time course analysis of MOLM-14 cells transduced with the 1F3-HOXA9shRNA construct revealed a progressive increase of cells in the G1 phase and a decrease in G2 and S phases, 24 to 36 hours before the prominent induction of cell death (Figure 3A). These findings are suggestive of an accumulation of the HOXA9shRNA-transduced cell population in G1 phase before progressively undergoing apoptosis. We next analyzed whether HOXA9 depletion might induce differentiation before apoptosis. Analysis of mRNA expression of myeloid differentiation markers M-CSF1R, PU.1, and ITGAM (CD11b) and granule molecules associated with terminal myeloid differentiation (lysozyme [LYZ]), myeloperoxidase [MPO]), elastaseA2 [ELA2]) by quantitative real-time PCR demonstrated a significant induction of these markers 72 hours after HOXA9shRNA transduction compared with GFP shRNA-transduced control cells (Figure 3B). Fluorescence-activated cell sorter analysis of the myeloid differentiation marker CD14 in MOLM-14 cells revealed a significant increase in CD14 expression in cells transduced with HOXA9 shRNA constructs compared with controls (Figure S6). Induction of CD14 expression was detectable as early as 48 hours after shRNA transduction and reached a peak at 84 hours (GFP shRNA, 1.62%; 1F3-HOXA9shRNA, 14.61%; 2A5-HOXA9shRNA, 17.58%) shortly before progressive induction of cell death set in (Figure S6). These data indicate that HOXA9 depletion initiates some degree of cellular differentiation before induction of apoptosis.
Effects of HOXA9 suppression in human t(9;11) MOLM-14 and t(4;11) SEMK2 cells. (A) Fluorescence-activated cell sorter analysis of cell-cycle status after transduction of MOLM-14 cells with 1F3-HOXA9shRNA after PI staining. A progressive increase of cells in G1 phase and a decrease of cells in G2 and S phases were noted compared with control GFP-shRNA–transduced cells with onset 24 to 36 hours before induction of apoptosis. (B) Analysis of mRNA expression of myeloid differentiation markers M-CSF1R, PU.1, and ITGAM (CD11b) and the granule molecules lysozyme (LYZ), myeloperoxidase (MPO), and elastaseA2 (ELA2) associated with terminal myeloid differentiation by quantitative real-time PCR 72 hours after HOXA9shRNA transduction. Compared with control GFP-shRNA transduced cells, a significant induction of mRNA expression was observed. (C) Gene expression analysis 44 hours after HOXA9 suppression in t(9;11) MOLM-14 cells with 2 different HOXA9shRNAs (1F3-HOXA9shRNA; 2A5-HOXA9shRNA) in triplicates compared with GFP-shRNA–transduced controls. Analysis of concomitant expression changes in other Homeobox A-D cluster genes demonstrated co–down-regulation of 5′-HOXA cluster gens in the HOXA9 suppression signature. (D) GSEA of the top 500 genes down-regulated after HOXA9 suppression in a gene expression dataset of 56 human AML patients.60 The HOXA9 suppression gene set is significantly enriched in a signature previously identified as more highly expressed in MLL-AML compared with other genetically defined AML subtypes (ES = 0.48; P < .001). (E) Confirmation of co–down-regulation of 5′-HOXA cluster genes as well as the HOXA9 cofactors MEIS1 and PBX3 and the transcription factor MEF2C in t(4;11) SEMK2 cells (quantitative real-time PCR analysis).
Effects of HOXA9 suppression in human t(9;11) MOLM-14 and t(4;11) SEMK2 cells. (A) Fluorescence-activated cell sorter analysis of cell-cycle status after transduction of MOLM-14 cells with 1F3-HOXA9shRNA after PI staining. A progressive increase of cells in G1 phase and a decrease of cells in G2 and S phases were noted compared with control GFP-shRNA–transduced cells with onset 24 to 36 hours before induction of apoptosis. (B) Analysis of mRNA expression of myeloid differentiation markers M-CSF1R, PU.1, and ITGAM (CD11b) and the granule molecules lysozyme (LYZ), myeloperoxidase (MPO), and elastaseA2 (ELA2) associated with terminal myeloid differentiation by quantitative real-time PCR 72 hours after HOXA9shRNA transduction. Compared with control GFP-shRNA transduced cells, a significant induction of mRNA expression was observed. (C) Gene expression analysis 44 hours after HOXA9 suppression in t(9;11) MOLM-14 cells with 2 different HOXA9shRNAs (1F3-HOXA9shRNA; 2A5-HOXA9shRNA) in triplicates compared with GFP-shRNA–transduced controls. Analysis of concomitant expression changes in other Homeobox A-D cluster genes demonstrated co–down-regulation of 5′-HOXA cluster gens in the HOXA9 suppression signature. (D) GSEA of the top 500 genes down-regulated after HOXA9 suppression in a gene expression dataset of 56 human AML patients.60 The HOXA9 suppression gene set is significantly enriched in a signature previously identified as more highly expressed in MLL-AML compared with other genetically defined AML subtypes (ES = 0.48; P < .001). (E) Confirmation of co–down-regulation of 5′-HOXA cluster genes as well as the HOXA9 cofactors MEIS1 and PBX3 and the transcription factor MEF2C in t(4;11) SEMK2 cells (quantitative real-time PCR analysis).
Gene expression analysis after HOXA9 suppression
Next, we used gene expression profiling to characterize changes that occur after HOXA9 suppression in t(9;11) MOLM-14 cells. RNA was purified 44 hours after transduction in triplicates with 2 of the 2 most effective HOXA9shRNA constructs (3 × 1F3-HOXA9shRNA; 3 × 2A5-HOXA9shRNA) or GFP-control shRNA (6 ×) and hybridized to Affymetrix Human Genome U133A2.0 microarrays. Initial comparative marker analysis of expression changes applying either one of the HOXA9shRNAs against GFP-control shRNA identified large groups of genes with decreased expression in cells treated with both HOXA9-directed shRNA constructs. GSEA55 further demonstrated almost complete overlap of the top 500 gene probes with decreased expression after HOXA9 suppression applying the 2 different HOXA9-shRNAs (enrichment score [ES] = 0.82, P < .001; Figure S7A). Having established that both HOXA9-shRNAs led to almost identical expression changes after HOXA9 suppression, both datasets were combined for further analysis. Among the genes with the most significantly decreased expression after HOXA9 suppression were genes previously identified as important in leukemogenesis and self-renewal of leukemia stem cells, such as the HOXA9 cofactor MEIS1, MEF2C, a member of the myocyte enhancer factor 2 gene family of transcription factors and BMI1 (Figure S7B). We then aimed to determine whether HOXA9 suppression leads to concomitant changes in other Homeobox A-D cluster genes. Assessment of expression changes in a panel of Homeobox A-D cluster genes demonstrated concomitant co–down-regulation of a subset of 5′-HOXA cluster genes, including HOXA7 and HOXA10 (Figure 3C). Next, to assess the relevance of the HOXA9 suppression signature in primary human AML, we determined whether there was significant overlap between our data and a gene expression signature in primary human MLL-rearranged AML.60 GSEA demonstrated significant overlap between the top 500 genes that decreased in expression after HOXA9 suppression (Figure S7B) and genes more highly expressed in human MLL-AML compared with other genetically defined AML subtypes (ES = 0.48; P < .001; Figure 3D). These overlapping gene sets further validated genes previously implicated in the pathogenesis of MLL-rearranged leukemias, including the HOXA9 cofactors MEIS1 and PBX3, MEF2C, and BMI1 (Figure 3D). Co–down-regulation of 5′-HOXA-cluster genes and the HOXA9 cofactors MEIS1 and PBX3 as well as the myocyte enhancer factor 2c (MEF2C) after HOXA9 suppression was also demonstrated in t(4;11) SEMK2 cells (Figure 3E). These data demonstrate an important role for HOXA9 as an upstream regulator of an MLL-AF9–associated gene expression program.
Effect of HOXA9 suppression in human leukemia cell lines and primary human leukemias
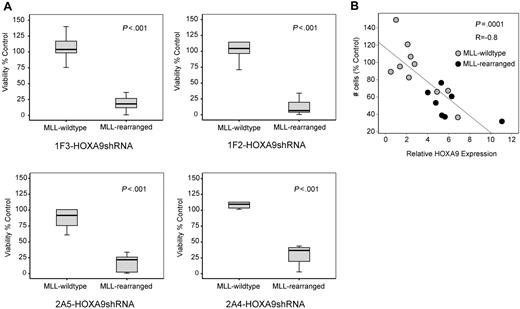
Next, we analyzed whether the observed induction of apoptosis after HOXA9 suppression is specific for t(9;11) MOLM-14 and t(4;11) SEMK2 cells or whether a similar phenotype can also be identified in an extended panel of MLL-rearranged and MLL-wild-type cell lines. In a panel of 11 AML/ALL cell lines (6 MLL-rearranged; 5 MLL-wild-type), a significantly higher induction of cell death was demonstrated in the MLL-rearranged group after transduction with each of the 4 HOXA9shRNA constructs, revealing a differential effect on viability that is significantly more prominent in MLL-rearranged leukemia cells (Figure 4A). Next, we reasoned that induction of apoptosis via specific suppression of HOXA9 might manifest as a correlation between HOXA9 expression and cellular sensitivity. In an independent experiment, determination of baseline HOXA9 expression before HOXA9 suppression (Figure S9A) and analysis of viability after HOXA9shRNA transduction by MTT assay in a panel of AML and ALL cell lines (Figure S9B) identified a significant correlation between cell number and baseline HOXA9 mRNA expression, with the greatest effect on viability in cells expressing the highest HOXA9 levels (R = −0.8, P = .001; Figure 4B). These data demonstrate that continued HOXA9 expression is important for leukemia cells expressing HOXA9 at high levels and further support the specificity of the shRNA-mediated effects.
Induction of apoptosis by HOXA9 suppression is significantly correlated with the presence of the MLL-fusion oncogene. (A) Analysis of cell viability at day 7 after HOXA9 shRNA transduction in a larger panel of 11 AML/ALL cell lines (6 MLL-rearranged; 5 MLL-wild-type) showing significantly higher apoptosis induction in MLL-rearranged cells with all 4 HOXA9shRNA constructs. (B) Independent analysis in a panel of 17 AML/ALL cell lines (7 MLL-rearranged; 10 MLL-wild-type) further demonstrated a significant correlation between cell number and baseline HOXA9 expression after shRNA-mediated suppression, with the greatest effect in cell lines expressing the highest HOXA9 levels (R = −0.8: P = .001; ●, MLL-rearranged cell lines;  , MLL-wild-type cell lines).
, MLL-wild-type cell lines).
Induction of apoptosis by HOXA9 suppression is significantly correlated with the presence of the MLL-fusion oncogene. (A) Analysis of cell viability at day 7 after HOXA9 shRNA transduction in a larger panel of 11 AML/ALL cell lines (6 MLL-rearranged; 5 MLL-wild-type) showing significantly higher apoptosis induction in MLL-rearranged cells with all 4 HOXA9shRNA constructs. (B) Independent analysis in a panel of 17 AML/ALL cell lines (7 MLL-rearranged; 10 MLL-wild-type) further demonstrated a significant correlation between cell number and baseline HOXA9 expression after shRNA-mediated suppression, with the greatest effect in cell lines expressing the highest HOXA9 levels (R = −0.8: P = .001; ●, MLL-rearranged cell lines;  , MLL-wild-type cell lines).
, MLL-wild-type cell lines).
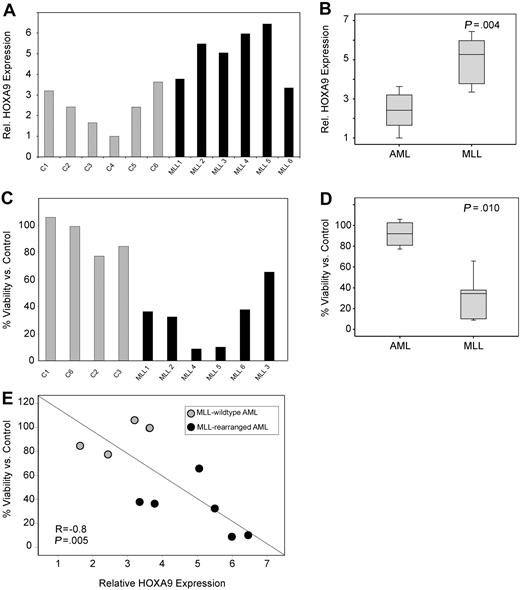
Next, the necessity of HOXA9 expression for survival of primary AML patient samples was assessed. First, we confirmed a high transduction efficiency (∼ 80%) in primary AML cells by puromycin selection after transduction with control shRNA constructs (data not shown) and determined that HOXA9shRNA transduction efficiently suppressed HOXA9 expression compared with control (Figure S10). We then analyzed the effect of HOXA9 knockdown in MLL-rearranged and nonrearranged primary human AML cells (6 MLL-rearranged 6 MLL germline). Similar to our findings in cell lines and to previous publications,14,–16 we find significantly higher HOXA9 mRNA expression in MLL-rearranged AMLs (P = .004; Figure 5A,B). Analysis of apoptosis induction by annexin V staining demonstrated a significant effect on MLL-rearranged leukemias with a mean viability/control of 91% in the MLL-germline group versus 31% in the MLL-rearranged group (P = .010; Figure 5C,D). Moreover, induction of apoptosis was significantly correlated with HOXA9 expression level before lentiviral transduction (R = −0.8, P = .005; Figure 5E). These data were also confirmed in independent repeat experiments and demonstrate that primary human MLL-rearranged AML samples remain dependent on HOXA9 expression for their survival.
HOXA9 suppression induces apoptosis in primary MLL-rearranged human AML cells. (A) Analysis of baseline HOXA9 expression before shRNA transduction by quantitative real-time PCR. (B) The mean HOXA9 expression level was found to be significantly higher in primary AML patient samples bearing an MLL translocation (P = .004). (C) Analysis of apoptosis induction by annexin V staining 72 hours after transduction with the 1F3-HOXA9shRNA or control shRNA. (D) Induction of apoptosis is significantly higher in the MLL-rearranged group (P = .010). (E) A significant correlation between baseline HOXA9 expression and impact of HOXA9 knockdown on viability is demonstrated. Samples expressing high-level HOXA9 were found to be especially susceptible to HOXA9 depletion (R = −0.8; P = .005).
HOXA9 suppression induces apoptosis in primary MLL-rearranged human AML cells. (A) Analysis of baseline HOXA9 expression before shRNA transduction by quantitative real-time PCR. (B) The mean HOXA9 expression level was found to be significantly higher in primary AML patient samples bearing an MLL translocation (P = .004). (C) Analysis of apoptosis induction by annexin V staining 72 hours after transduction with the 1F3-HOXA9shRNA or control shRNA. (D) Induction of apoptosis is significantly higher in the MLL-rearranged group (P = .010). (E) A significant correlation between baseline HOXA9 expression and impact of HOXA9 knockdown on viability is demonstrated. Samples expressing high-level HOXA9 were found to be especially susceptible to HOXA9 depletion (R = −0.8; P = .005).
In vivo assessment of HOXA9 suppression
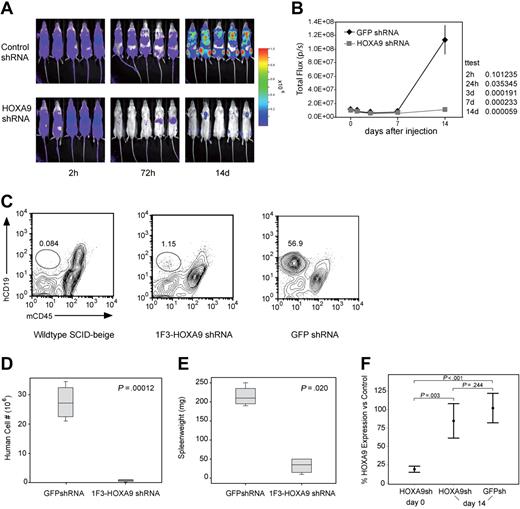
Next, we assessed the in vivo effect of HOXA9 suppression by transplanting luciferase-positive t(4;11) SEMK2 cells into SCID-beige mice followed by subsequent in vivo bioluminescent imaging. In 2 independent experiments, 5 million SEMK2 cells/mouse were transduced with either the 1F3-HOXA9shRNA construct or GFP control shRNA and injected intravenously 24 hours after transduction. Injection of equal numbers of transplanted cells was confirmed by determination of total body luminescence 2 hours after transplantation, and there was no significant difference (Figure 6A). Longitudinal in vivo imaging revealed a significant difference in total body luminescence as early as 24 hours after transplantation (P = .036). This difference increased progressively over time with a maximum at day 14 (P = .001) shortly before the mice in the GFP shRNA control group began to die of leukemia (Figure 6A,B). A subset of mice were killed at day 14 and assessed for the number of human cells present in spleens from each group of mice (Figure 6C). There was an approximately 40-fold difference in the number of human cells found in the spleens of the 2 groups of mice (P = .001; Figure 6D). Furthermore, there was a significant difference in the spleen weights between mice that received control-transduced cells and those that received HOXA9shRNA-transduced cells (Figure 6E). Thus, continued expression of HOXA9 is required for survival/proliferation of SEMK2 cells in vivo.
Assessment of the effects of HOXA9 suppression in vivo using bioluminescent imaging. (A) Before transplantation, SEMK2 cells were transduced with either 1F3-HOXA9 shRNA or GFP control shRNA and injected intravenously 24 hours after transduction. Mice were imaged 2, 24, and 72 hours after injection and then weekly. (B) There was no significant difference in total body luminescence 2 hours after injection confirming that an equal number of treated cells was injected (P = .101). Repetitive longitudinal in vivo bioluminescent imaging at later time points revealed a significant difference of total body luminescence as early as 24 hours after transplantation (P = .036). Fourteen days after transplantation, the differences in total body luminescence reached a significant maximum (P = .001), shortly before the GFP shRNA–treated control group died of overt leukemia. (C) The degree of leukemic organ infiltration in both groups was assessed. The percentage of hCD19+/mCD45− SEMK2 cells was evaluated in spleens from wild-type SCID-beige mice that received 1F3-HOXA9–transduced or GFP-control–transduced cells. (D) The total number of human cells/spleen is graphed for both groups of mice. There is a 40-fold difference in total number of SEMK2 between groups (P = .001). (E) Spleen weights for are shown for mice that received either control or 1F3-HOXA9-transduced SEMK2 cells. (F) Analysis of HOXA9 expression in SEMK2 cells at the day of transplantation (day 0) confirmed efficient HOXA9 suppression (80%) in the 1F3-HOXA9 shRNA-treated group compared with control shRNA-transduced cells. Human cells were isolated from mice that received either control or 1F3-HOXA9 shRNA-transduced SEMK2 cells 14 days after injection, and the HOXA9 expression level was compared with SEMK2 cells growing in culture. The HOXA9 expression level at this time point was similar in the sorted SEMK2 cells from the 1F3-HOXA9shRNA group as in those sorted from the GFP shRNA control group (P = .244).
Assessment of the effects of HOXA9 suppression in vivo using bioluminescent imaging. (A) Before transplantation, SEMK2 cells were transduced with either 1F3-HOXA9 shRNA or GFP control shRNA and injected intravenously 24 hours after transduction. Mice were imaged 2, 24, and 72 hours after injection and then weekly. (B) There was no significant difference in total body luminescence 2 hours after injection confirming that an equal number of treated cells was injected (P = .101). Repetitive longitudinal in vivo bioluminescent imaging at later time points revealed a significant difference of total body luminescence as early as 24 hours after transplantation (P = .036). Fourteen days after transplantation, the differences in total body luminescence reached a significant maximum (P = .001), shortly before the GFP shRNA–treated control group died of overt leukemia. (C) The degree of leukemic organ infiltration in both groups was assessed. The percentage of hCD19+/mCD45− SEMK2 cells was evaluated in spleens from wild-type SCID-beige mice that received 1F3-HOXA9–transduced or GFP-control–transduced cells. (D) The total number of human cells/spleen is graphed for both groups of mice. There is a 40-fold difference in total number of SEMK2 between groups (P = .001). (E) Spleen weights for are shown for mice that received either control or 1F3-HOXA9-transduced SEMK2 cells. (F) Analysis of HOXA9 expression in SEMK2 cells at the day of transplantation (day 0) confirmed efficient HOXA9 suppression (80%) in the 1F3-HOXA9 shRNA-treated group compared with control shRNA-transduced cells. Human cells were isolated from mice that received either control or 1F3-HOXA9 shRNA-transduced SEMK2 cells 14 days after injection, and the HOXA9 expression level was compared with SEMK2 cells growing in culture. The HOXA9 expression level at this time point was similar in the sorted SEMK2 cells from the 1F3-HOXA9shRNA group as in those sorted from the GFP shRNA control group (P = .244).
Even though there was a dramatic difference in the number of human cells present in the 2 groups of mice, some human cells could be found in the mice that received HOXA9shRNA-transduced SEMK2 cells (Figure 6C). We reasoned that the presence of these cells could indicate either the presence of a subpopulation of cells that do not require HOXA9 or the presence of cells that have escaped HOXA9 suppression. Thus, we sorted SEMK2 cells from both groups and assessed HOXA9 expression by quantitative real-time PCR. We compared expression levels to untreated SEMK2-M1 cells growing in culture and to SEMK2 cells 48 hours after transduction with the HOXA9shRNA construct. As expected, assessment before transplantation showed a significant decrease of HOXA9 expression in the transduced cells (Figure 6F). Interestingly, the cells sorted at day 14 from mice transplanted with HOXA9shRNA-transduced SEMK2 cells showed no significant difference compared with cells isolated from mice transplanted with GFP shRNA-transduced cells (P = .244; Figure 6F). This demonstrates that the few human cells found in the mice that received HOXA9shRNA-transduced SEMK2 cells have escaped HOXA9 suppression. Therefore, cells expressing high-level HOXA9 have a growth and survival advantage in vivo, and there does not appear to be a significant population of cells that are less dependent on HOXA9 expression for their survival/proliferation. These results were also confirmed applying a second HOXA9shRNA construct (2A5-HOXA9shRNA) in an independent in vivo experiment (Figure S11A-E). As in the in vivo experiment comparing 1F3-HOXA9shRNA or GFP-control-shRNA transduced SEMK2 cells, human cells present in the spleens of SCID-beige mice transplanted with 2A5-HOXA9shRNA or GFP-control-shRNA-transduced SEMK2 cells were isolated at day 14. Assessment of HOXA9 expression showed no significant difference in both groups (P = .195; Figure S11D). Next, to determine whether the few isolated cells from the 2A5-HOXA9shRNA-transduced group were susceptible to subsequent HOXA9 suppression, cells were transduced with the 1F3-HOXA9shRNA construct or the GFP-control shRNA construct and viability in liquid culture was assessed. Seven days after transduction, a highly significant decreased number of viable cells was observed in cells transduced with the 1F3-HOXA9shRNA construct compared with cells transduced with the GFP-control shRNA (P = .001; Figure S11E). This demonstrates that the cells remained susceptible to HOXA9 suppression and further supports the notion that the small population of human cells found in the mice that received 2A5-HOXA9shRNA-transduced SEMK2 cells was not rendered HOXA9 independent but has rather escaped HOXA9 suppression in vivo.
Discussion
Comparative analysis of gene expression profiles in human acute myeloid and lymphoblastic leukemias implicates up-regulation of a subset of HOX genes as a central mechanism of leukemic transformation by MLL oncoproteins.14,–16,21,22,61,62 Gene expression studies in murine models also demonstrate high-level expression of Hox genes in Mll-fusion initiated murine leukemias and leukemia stem cells.27,28 The central role for HOX genes is further supported by recent studies that have identified HOX genes as direct binding targets for wild-type MLL and MLL-fusion proteins.30,–32 However, experiments in murine leukemia models directly assessing the requirement for specific Hox genes in Mll-rearranged leukemias have produced mixed results, perhaps because the Hox gene of interest is most often deleted in the germline and thus not present during leukemia initiation. Such models assess the necessity for a gene product to initiate leukemia, but they may not assess the continued requirement of a particular gene in a fully developed leukemia.
To further investigate the possibility that HOXA9 is necessary for survival of human leukemias, we used a lentiviral-based shRNA approach to inhibit HOXA9 in human leukemia cell lines and primary AML patient samples. We demonstrate that HOXA9 depletion leads to an almost immediate reduction of proliferative capacity, differentiation, and subsequent progressive induction of apoptosis, which was rescued by ectopic overexpression of HOXA9. The effect on apoptosis induction was significantly more pronounced in cells carrying an MLL-fusion than in cells with MLL germline karyotype; however, even MLL germline cells possessing high-level HOXA9 expression demonstrated enhanced sensitivity to HOXA9 suppression. Furthermore, we demonstrate that HOXA9 suppression is also required for appropriate proliferation/survival in vivo. Global expression analysis after HOXA9 suppression revealed a tight correlation between HOXA9 suppression and subsequent down-regulation of a gene set previously identified to be overexpressed in murine MLL-leukemia stem cells and human MLL-AML including 5′-HOXA cluster genes, the HOXA9 cofactors MEIS1 and PBX3, and the transcription factor MEF2C. Although part of the observed expression changes might be the result of an already initiated apoptosis program rather than a direct effect of HOXA9 suppression, these findings further support HOXA9 as a central downstream target of select MLL-fusion proteins, which contributes to the induction and maintenance of an inappropriate gene expression program also found in primary human AML and in leukemogenic progenitor populations.28,29 The fact that some but not all genes aberrantly expressed in murine MLL leukemia stem cells and primary human MLL leukemias are also part of the HOXA9 suppression signature implies that MLL-fusion oncogene-mediated leukemic transformation is also mediated by other dysregulated pathways independent from HOXA9.
Our data showing that transplantation of HOXA9 depleted, human t(4;11) SEMK2 cells into SCID-beige mice revealed a significant decrease in leukemia burden in vivo supports the concept that HOXA9 is crucial for survival of MLL-rearranged leukemias. It has been proposed previously that there may be functional redundancy within members of the clustered HOX genes such that suppression of any one HOXA cluster gene may be compensated for by the high-level expression of other HOXA genes. In contrast, the data presented here reveal a significant effect of HOXA9 suppression, which could not be compensated by other mechanisms to maintain cell proliferation/survival. We note in our gene expression analysis that expression of other 5′-HOXA cluster genes shows a significant decrease after HOXA9 suppression. Extensive differences in sequence between HOXA9 and other Homeobox genes in the shRNA-targeted regions and similar expression changes with 2 different HOXA9-shRNAs suggest that the shRNAs are not directly suppressing other HOX genes. Therefore, the observed decrease of gene expression of other HOX genes appears to be a result of decreased HOXA9 expression, and sustained HOXA9 gene expression is necessary for growth/survival of human MLL-rearranged leukemias.
Demonstration that continued HOXA9 expression is required for leukemia survival has potential therapeutic implications. Although it is well known that HOX gene expression is a frequent characteristic of both human acute leukemias and some solid tumors, most notably ovarian carcinoma, it has remained unclear whether such expression is required to maintain tumor survival. Therefore, the therapeutic potential of HOX-associated gene expression programs remained unclear. The demonstration that suppression of HOX gene expression leads to growth arrest in human MLL-rearranged leukemia cell lines and primary patient samples and subsequent down-regulation of other leukemia associated genes prompts further study of methods to inhibit HOX gene function and their downstream-associated programs. Having identified a gene signature after HOXA9 suppression that is associated with the abrogation of survival of MLL-rearranged human leukemias, it might be possible to use this “HOXA9-associated leukemia abrogation signature” for gene expression-based screening approaches of small molecule libraries as previously successfully demonstrated.63,–65 Another approach would combine in vivo RNA interference conjugated to monoclonal antibodies to specifically target leukemia cells and suppress HOXA9-dependent pathways necessary for leukemia survival. It has been demonstrated in a recent proof-of-principle experiment that this approach might be feasible.66 Such novel therapeutic approaches might prove particularly useful in high risk MLL-rearranged leukemias. A similar approach can now be used to assess the importance of HOX gene expression in other tumor types.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Elise Porter, Sara Swalnick, and Megan Smith for administrative assistance, other members of the Armstrong laboratory for helpful discussion, and Dr William Hahn and other members of the RNAi consortium for providing the shRNA constructs.
This work was supported by the Leukemia & Lymphoma Society, the Richard and Susan Smith Foundation, and the National Cancer Institute (NCI P01 CA66996).
National Institutes of Health
Authorship
Contribution: J.F. performed and designed research and wrote the paper; A.V.K. performed and designed research; M.C.S., R.W., and T.N.D. performed research; M.v.d.H.-E. and C.M.Z. contributed vital reagents; and A.L.K. and S.A.A. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Scott A. Armstrong, Children's Hospital, Boston, Karp Family Research Laboratories, 1 Blackfan Circle, Boston, MA 02215; e-mail: Scott.Armstrong@childrens.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal